SUMMARY
The p53 transcription factor participates in diverse cellular responses to stress including cell cycle arrest, apoptosis, senescence and autophagy. The molecular mechanisms defining the ultimate outcome to p53 activation remain poorly characterized. We performed a genome-wide genetic screen in human cells to identify pathway-specific coregulators of the p53 target genes CDKN1A (p21), an inhibitor of cell cycle progression, versus BBC3 (PUMA), a key mediator of apoptosis. Our screen identified numerous factors whose depletion creates an imbalance in the p21:PUMA ratio upon p53 activation. The transcription factor TCF3/E2A drives p21 expression while repressing PUMA across cancer cell types of multiple origins. Accordingly, TCF3/E2A depletion impairs the cell cycle arrest response and promotes apoptosis upon p53 activation by chemotherapeutic agents. In contrast, TRIAP1 is a specific repressor of p21 whose depletion slows down cell cycle progression. Our results reveal strategies to drive cells toward specific p53-dependent responses.
INTRODUCTION
The p53 transcription factor functions as a signaling hub during the cellular response to stress. p53 is activated by various signaling cascades elicited by myriad stress stimuli including oncogene hyperactivation, DNA damage and nutrient deprivation (Vousden and Prives, 2009). These signaling pathways relieve the inhibitory effects of the p53 repressors MDM2 and MDM4, which otherwise target p53 for proteasomal degradation and mask its transactivation domain. Upon activation, p53 induces transcription of genes involved in varied cellular responses such as cell cycle arrest, apoptosis, senescence and autophagy (Riley et al., 2008). Although many p53 target genes participating in each pathway have been identified, the mechanisms defining which cellular response is adopted remain poorly characterized. A thorough understanding of these mechanisms of cell fate choice will be required for the effective deployment of p53-based therapies in the clinic. Currently, small molecule inhibitors of MDM2 and MDM4 are being tested in clinical trials for cancer treatment (Brown et al., 2009). However, the cellular response elicited by these compounds varies greatly across cancer cell types (Paris et al., 2008; Sullivan et al., 2012b; Tovar et al., 2006). Thus, the identification of factors that regulate cell fate choice upon p53 activation would reveal strategies to enhance the therapeutic application of these drugs.
Many efforts in the p53 field have been devoted to the characterization of p53 post-translational modifications and p53 cofactors as well as p53-autonomous mechanisms driving gene-specific regulation within the p53 transcriptional program (Vousden and Prives, 2009). With the advent of functional genomics, it is now possible to perform genetic screens in human cells for the unbiased identification of novel pathway-specific coregulators of p53 target genes. We report here the results of a genome-wide short hairpin RNA (shRNA) genetic screen to identify factors that regulate the expression ratio between CDKN1A (p21), one of the key mediators of p53-dependent cell cycle arrest, and BBC3 (PUMA), a BH3-only protein that mediates much of the apoptotic effects of p53. Using a flow cytometry assay to isolate cells with altered expression of the endogenous p21 and PUMA proteins and a DNA deep-sequencing protocol to identify the shRNAs expressed in these cells, we found numerous factors that affect the p21:PUMA ratio. Most prominent among these was TCF3 (transcription factor 3, also known as E2A), a basic helix-loop-helix (bHLH) DNA binding protein that is required for p21 induction, yet functions as a repressor of PUMA. Depletion of TCF3/E2A leads to lower p21 accumulation and higher PUMA expression across cancer cell types of diverse tissue origin, thus promoting the apoptotic response upon p53 activation. Additionally, we identified TRIAP1 (TP53 regulated inhibitor of apoptosis, also known as p53 cell survival factor or p53CSV) as a gene-specific repressor of p21. TRIAP1 knockdown leads to augmented p21 expression before and during p53 activation and slows down cell proliferation. Overall, our research identified multiple factors that function as gene-specific coregulators within the p53 network and reveals several strategies to manipulate the cellular response upon p53 activation.
RESULTS
A genetic screen in human cells to identify factors regulating the p21:PUMA expression ratio upon p53 activation
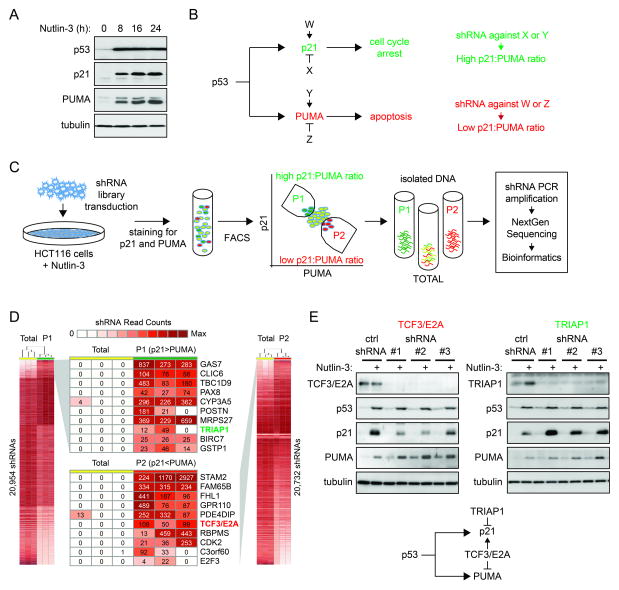
p53 activation commonly leads to concurrent induction of target genes in distinct functional pathways, yet the precise cellular response adopted varies widely across cell types. In HCT116 colorectal cancer cells, p53 activation with Nutlin-3, a small molecule inhibitor of the p53-MDM2 interaction, leads to strong induction of both p21 and PUMA (Figure 1A). In this scenario, HCT116 cells undergo cell cycle arrest, but become highly sensitized to additional apoptotic stimuli (Henry et al., 2012; Sullivan et al., 2012b). We hypothesized the existence of pathway-specific coregulators with the ability to control the p21:PUMA expression ratio in this setting and thus affect the cell fate choice upon p53 activation (factors W, X, Y and Z in Figure 1B). These coregulators would function as gene-specific coactivators or repressors of p21 or PUMA. According to this hypothesis, whereas an shRNA against factors X or Y would lead to a greater p21:PUMA expression ratio, an shRNA against factors W or Z would decrease this ratio (Figure 1B). In order to identify these factors, we designed a genome-wide shRNA screen as depicted in Figure 1C. First, we transduced a lentiviral shRNA library targeting >16,000 human genes (SBI library) into HCT116 cells. After antibiotic selection of stable integrants and clearance of cells carrying shRNAs against essential genes during two weeks of culture, we activated p53 with Nutlin-3 and isolated cells expressing different ratios of endogenous p21 and PUMA by fluorescence activated cell sorting (FACS). After intensive testing of multiple fixation/staining conditions and antibodies, we obtained a protocol that robustly detects a p53-dependent increase in the levels of the endogenous p21 and PUMA proteins by flow cytometry (Figure S1A-B, see Extended Experimental Procedures), which enabled us to sort cells with varying p21:PUMA ratios during the screen. Whereas the population dubbed P1 stained strongly for p21 and weakly for PUMA, the P2 population stained strongly for PUMA and weakly for p21. We then isolated DNA from P1, P2 and the input population (Total) and amplified shRNAs by PCR with primers against the flanking sequences. In a nested PCR reaction we tagged the shRNA cassettes for Illumina deep sequencing and sequenced three biological replicates of each population. Using a novel bioinformatics pipeline dubbed BiNGS (Bioinformatics for Next Generation Sequencing) (Kim and Tan, 2012), we identified shRNAs whose abundance was significantly different between P1 vs. Total and P2 vs. Total (Figure 1D). Of note, we found many shRNAs that fell below the level of detection in the Total population yet could be counted hundreds of times in either P1 or P2. Importantly, BiNGS takes into account how many shRNAs against a given mRNA display the same behavior, a concept dubbed ‘reagent redundancy’ in RNAi screens, which reduces the chances of false positives due to ‘off target’ effects of individual shRNAs (Echeverri et al., 2006). The information on shRNA multiplicity is then condensed into a p(wZ) value for each gene (Figure S1C) (see Extended Experimental Procedures for details). Using a cut-off of p(wZ) < 0.01 we identified 491 candidate genes whose knockdown would create an increase in the p21:PUMA ratio and 523 candidate genes exerting the opposite effect (Figure S1C and Table S1). Next, we selected ~40 candidate genes in each category for validation and established stable knockdowns in HCT116 cells for each of them (see Extended Experimental Procedures for selection criteria and Table S2). Importantly, during these validation efforts we utilized shRNAs from a different library (TRC, The RNAi Consortium). Since the seed sequences in the TRC shRNAs are different from those in the SBI shRNAs used in the initial screen, this exercise significantly increased our confidence in positively validated results. In order to test the effect of the 81 individual knockdowns on the p21:PUMA ratio, we employed a dual-fluorescence Western blot assay that enabled us to monitor p21 and PUMA protein expression within the same blot (see example in Figure S1D). Because this assay bypasses the fixation steps involved in the flow cytometry assay used for the screen, which could potentially create artifacts, it increased the stringency of our validation efforts. When using a cut-off of 20% change in the p21:PUMA protein ratio relative to a non-targeting shRNA control, this assay led to the identification of 12 genes whose knockdown tips the balance toward more PUMA (e.g. TCF3/E2A, C3orf60, GPR110) and 7 genes with the opposite effect (e.g. TRIAP1, GSPT1, POSTN) (Figure 1E, Figure S1E and Table S2).
Figure 1. A genome-wide shRNA screen identifies pathway-specific regulators within the p53 network.
A. Western blot assays showing that activation of p53 with Nutlin-3 leads to concurrent induction of p21 and PUMA in HCT116 cells. B. Hypothetical factors W, X, Y and Z function as gene-specific coregulators of p21 and PUMA. C. Experimental design for shRNA screen leading to identification of gene-specific coregulators of p21 and PUMA. D. Heatmaps showing differential counts of shRNAs in the sorted populations dubbed P1 (high p21:PUMA ratio) and P2 (low p21:PUMA ratio) versus the Total population. Numbers represent the shRNA count in the three sets of triplicates. E. Different shRNAs against TCF3/E2A and TRIAP1 change the p21:PUMA expression ratio as predicted by the genetic screen. TCF3/E2A depletion reduces p21 expression and increases PUMA expression without affecting p53 accumulation. TRIAP1 knockdown leads to increased p21 expression with no effects on PUMA or p53 accumulation. See also Figure S1.
From these validation efforts we selected two factors for further study, TCF3/E2A and TRIAP1, whose depletion produced the most pronounced changes in p21:PUMA expression. Knockdown of TCF3/E2A with three different shRNAs leads to decreased p21 induction and increased PUMA expression upon Nutlin-3 treatment, without affecting p53 accumulation (Figure 1E). In contrast, depletion of TRIAP1 with three different shRNAs leads to stronger p21 induction with negligible effects on PUMA expression or p53 accumulation. Thus, TCF3/E2A works concurrently as a co-activator of p21 and repressor of PUMA, while TRIAP1 behaves as a gene-specific repressor of p21 (Figure 1E, diagram).
Taken together, these results demonstrate that our genetic screen successfully identified novel regulators of the p21:PUMA expression ratio, which could potentially modulate the cellular response to p53 activation.
TCF3/E2A and TRIAP1 are conserved specific coregulators of the p21:PUMA expression ratio
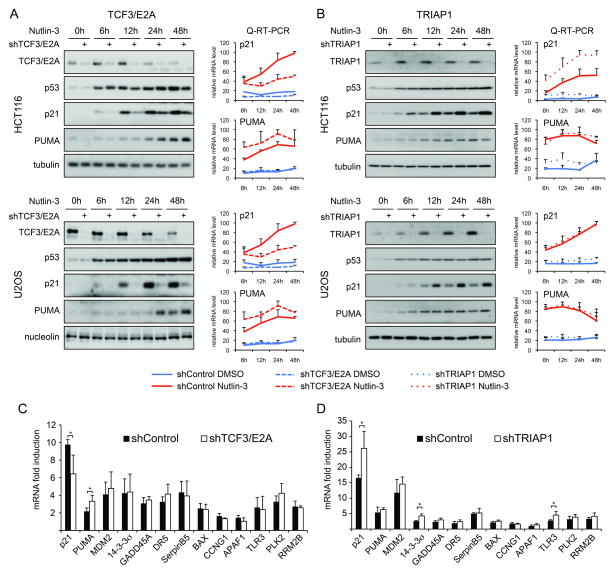
The cellular response to p53 activation varies greatly across cell types. Several gene-specific regulators of p53 target genes have been identified, but their action is often restricted to specific cell types (Sullivan et al., 2012a; Vousden and Prives, 2009). In order to test for the conservation of TCF3/E2A and TRIAP1 effects, we defined their impact across three cancer cell types of diverse tissue origins during a time course of Nutlin-3 treatment between 6 and 48 hours. Importantly, TCF3/E2A knockdown leads to decreased p21 induction and increased PUMA expression in HCT116 cells (colorectal carcinoma), U2OS cells (osteosarcoma) and A549 cells (lung carcinoma) at multiple time points (Figure 2A and Figure S2A). Of note, TCF3/E2A depletion had no significant effects on the extent and kinetics of p53 accumulation, suggesting that this factor acts downstream or in parallel to p53. Interestingly, TCF3/E2A expression decreases upon Nutlin-3 treatment in all three cell types tested, but not in HCT116 p53−/− cells (Figure 2A and S2A, C). Of note, TCF3/E2A levels are also downregulated over prolonged time in cell culture regardless of p53 status, which correlates with decreased proliferation rates (Figure S2C-D), and this downregulation is also observed at the mRNA level (Figure S2E-F). Overall, these results indicate that TCF3/E2A expression is higher in proliferating cells and downregulated by p53-dependent and -independent cell cycle arrest. Next, we investigated the effect of TCF3/E2A depletion on expression of the p21 and PUMA mRNAs using quantitative RT-PCR. In fact, TCF3/E2A knockdown decreases p21 mRNA accumulation while enhancing PUMA mRNA induction at multiple time points in every cell line tested (Figure 2A and Figure S2A). Of note, TCF3/E2A knockdown also affects the basal expression levels of p21 mRNA (see Figure S2G for re-plotting of the basal expression data at a lower scale). Interestingly, TCF3/E2A does not affect the induced expression of several additional p53 target genes in various functional pathways, thus demonstrating its highly specific effects toward p21 and PUMA in this setting (Figure 2C). Altogether, our results demonstrate that TCF3/E2A is a conserved and specific coregulator of the p21:PUMA expression ratio upon p53 activation acting primarily at the mRNA expression level.
Figure 2. TCF3/E2A and TRIAP1 control the p21:PUMA expression ratio in diverse cancer cell types.
A. Western blot and Q-RT-PCR assays show that TCF3/E2A functions as a coactivator of p21 expression and a repressor of PUMA expression in HCT116 colorectal carcinoma cells and U2OS osteosarcoma cells. See Figure S2 for data on A549 lung carcinoma cells B. TRIAP1 functions as a gene-specific repressor of p21. C,D. Q-RT-PCR analysis showing the effects of TCF3/E2A and TRIAP1 knockdown on diverse p53 target genes. All Q-RT-PCR experiments are expressed as averages of three biological replicates −/+ s.d. See also Figure S2.
TRIAP1 was previously characterized as a p53 target gene with pro-survival functions (Park and Nakamura, 2005). Indeed, we observed increased TRIAP1 expression upon p53 activation in HCT116, U2OS and A549 cells (Figure 1E, 2B and Figure S2B). TRIAP1 was identified in our screen as a factor whose depletion leads to a higher p21:PUMA ratio. In fact, TRIAP1 knockdown increased p21 protein levels in all three cell lines tested throughout the time course of Nutlin-3 treatment (Figure 2B and S2B). In contrast, PUMA expression was not affected by TRIAP1 knockdown. As observed for TCF3, TRIAP1 does not affect p53 accumulation. In HCT116 cells, Q-RT-PCR analysis demonstrates that TRIAP1 depletion produces an increase in p21 mRNA expression, both basal and induced, with no effects on PUMA mRNA (Figure 2D and S2H). However, this effect on p21 mRNA was not observed in U2OS and is weak in A549 cells (Figure 2B and Figure S2B), suggesting that TRIAP1 may influence p21 protein expression by multiple mechanisms. Q-RT-PCR analysis of multiple p53 target genes in HCT116 cells showed that TRIAP1 depletion leads to a modest but statistically significant increase in the expression of 14-3-3σ and TLR3 (Figure 2D). Overall, these data indicate that TRIAP1 functions as a p53-inducible repressor of p21 expression across diverse cancer cell types.
TCF3/E2A does not impair p53 binding or RNAPII transactivation at the p21 locus, but affects p21 mRNA stability
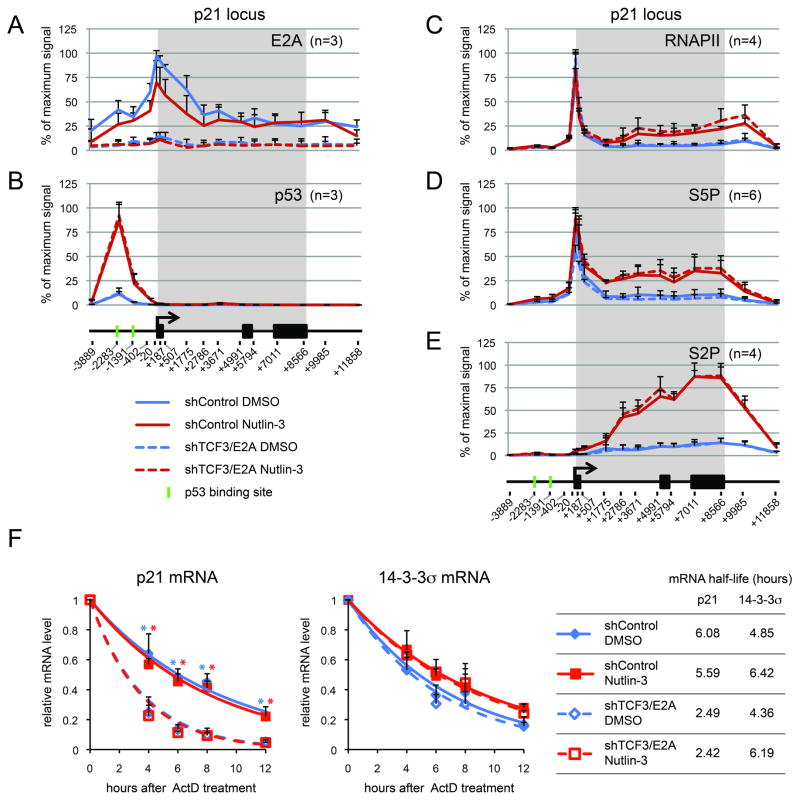
TCF3/E2A has been previously reported to be a positive regulator of p21 expression (Prabhu et al., 1997), but its mechanism of action has not been elucidated. Chromatin Immunoprecipitation (ChIP) analysis of the p21 locus demonstrates that TCF3/E2A binds to the p21 locus with peak signals around the transcription start site (Figure 3A). Expectedly, TCF3/E2A binding is much reduced in shTCF3 cells. A modest decrease in TCF3/E2A binding is observed upon Nutlin-3 treatment, which reflects the lower cellular levels of this factor in this condition (Figure 2A). Importantly, p53 binding to the upstream enhancers in the p21 locus is not affected by TCF3/E2A knockdown, indicating that TCF3/E2A does not modulate p53 DNA binding activity or chromatin accessibility (Figure 3B). Next, we tested whether TCF3/E2A affects RNA polymerase II (RNAPII) transactivation at the p21 locus. Toward this end we performed ChIP assays of total RNAPII, initiating RNAPII (i.e. phosphorylated on Serine 5 of the C-terminal domain repeats, S5P) and elongating RNAPII (Serine 2 phosphorylated, S2P). In agreement with previous reports (Beckerman et al., 2009; Gomes et al., 2006), we observed that p53 activation leads to increased levels of elongating RNAPII at the p21 locus, as denoted by higher levels of total RNAPII preferentially in the intragenic region, as well as marked increases in S5P- and S2P-RNAPII at the 5′ and 3′ ends of the gene, respectively (Figure 3C–E). Interestingly, TCF3/E2A depletion does not affect the basal or induced levels of total, S5P- and S2P-RNAPII, indicating that this factor is not required for p53-dependent transactivation of RNAPII at the p21 locus. Intrigued by this result, we investigated whether the impact of TCF3/E2A depletion on p21 mRNA expression could be explained by effects on mRNA half-life. Indeed, mRNA half-life assays demonstrate that the p21 mRNA is more rapidly degraded in shTCF3/E2A cells, both before and after Nutlin-3 treatment (Figure 3F). This effect is specific, as the half-life of the 14-3-3σ (SFN) mRNA is not affected by TCF3/E2A depletion. Overall, these results indicate that TCF3/E2A does not affect p53 binding, RNAPII initiation or elongation, but acts instead at downstream steps. This could be due to direct action at the p21 locus to facilitate co-transcriptional RNA processing or indirect action by affecting the expression of post-transcriptional regulators of p21 mRNA stability. Future studies will discriminate among these possibilities.
Figure 3. TCF3/E2A affects p21 mRNA stability.
A–E. TCF3/E2A binds to the p21 proximal promoters and its depletion does not affect p53 binding or RNAPII transactivation. ChIP analysis of the p21 locus with antibodies against TCF3/E2A (A), p53 (B), total RNAPII (C), Serine 5-phosphorylated RNAPII (S5P, D) and Serine 2-phoshoryalted RNAPII (E), in HCT116 cells expressing control shRNA of shRNA against TCF3/E2A, treated with DMSO or 10 μM Nutlin-3 (24 hours). The p21 gene map is shown at the bottom, with black boxes representing exons and the transcribed region highlighted by a gray box. Numbers at the bottom indicate the position of PCR amplicons used to analyze ChIP-enriched DNA relative to the transcription start site. n indicates the number of ChIPs performed with each antibody. F. TCF3/E2A depletion decreases p21 mRNA half life. mRNA half life assays were performed for the p21 and 14-3-3s (SFN) mRNAs by collecting RNA samples at the indicated times after Actinomycin D treatment in HCT116 cells expressing control or TCF3/E2A shRNAs, treated with DMSO or 10 μM Nutlin-3 (24 hours). Asterisks indicate significant different (p<0.05) between shControl and shTCR3/E2A samples treated with DMSO (blue) or Nutlin-3 (red) within a given time point. Data are expressed as averages of three experiments −/+ s.d.
TCF3/E2A modulates cell fate choice upon p53 activation by DNA damage in a p21- and PUMA-dependent fashion
The ultimate goal of our genetic screen was to identify strategies to manipulate the cellular response to p53 activation via the identification of factors that modulate the p21:PUMA ratio. We found that TRIAP1 represses p21 expression prior to p53 activation (see Figures S2H for RNA data and S3A for protein). Accordingly, we observed decreased levels of BrdU incorporation upon TRIAP1 knockdown in HCT116 and U2OS cells, indicative of slow cell cycle progression, as well as decreased doubling time of the cell cultures (Figure S3B–C). Interestingly, TRIAP1 has been previously characterized as a pro-survival factor (Park and Nakamura, 2005). In agreement with this report, we found that TRIAP1 depletion leads to increased levels of cell death upon DNA damage by doxorubicin (Figure S3D). Thus, although TRIAP1 functions as a repressor of p21, which is often considered an anti-apoptotic factor, the overall effects of TRIAP1 on cellular viability can not be solely explained by its effects on p21 expression.
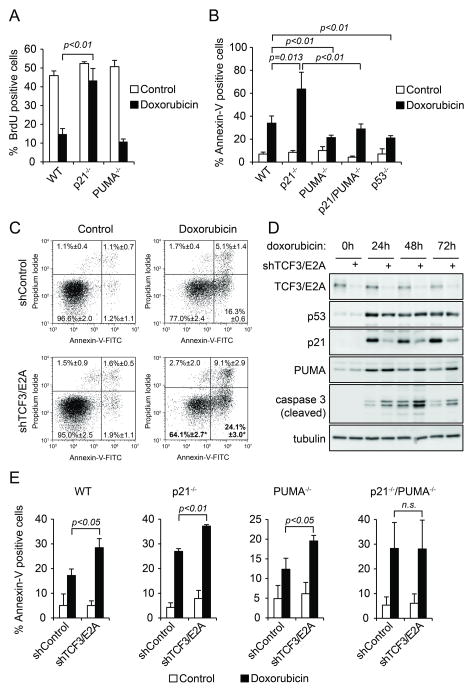
Next, we focused our efforts on TCF3/E2A. Interestingly, independent previous work from our group identified TCF3 as a candidate ‘Synthetic Lethal with Nutlin-3’ gene whose knockdown would sensitize cells to Nutlin-3-induced cell death (Sullivan et al., 2012b). In fact, we confirmed that TCF3/E2A depletion increases the apoptotic index upon Nutlin-3 treatment (Figure S3E), which agrees with the observed decrease in the p21:PUMA ratio. In order to further test the impact of TCF3/E2A on cell fate choice upon p53 activation, we employed doxorubicin, a topoisomerase II inhibitor commonly used in the clinic that induces a DNA damage response leading to a combination of p53/p21-dependent cell cycle arrest and p53/PUMA-dependent apoptosis (Bunz et al., 1998; Yu et al., 2003). Using an isogenic panel of HCT116 cell lines of varied p21 and PUMA status, we confirmed that p21 knockout significantly impairs the cell cycle arrest response upon doxorubicin treatment while sensitizing to apoptosis (Figure 4A-B). In this system, PUMA knockout has no significant effects on cell cycle arrest but clearly decreases doxorubicin-induced apoptosis (Figure 3A-B). Thus, the cellular response to doxorubicin could be modulated by the p21:PUMA ratio. Importantly, TCF3/E2A depletion in HCT116 cells leads to a significant increase in the number of apoptotic cells upon doxorubicin treatment as measured by Annexin V staining without significant effects on the basal levels of apoptosis (Figure 4C, E). This effect is also observed in U2OS cells (Figure S3F). TCF3/E2A knockdown leads to decreased p21 expression, increased PUMA induction and increased levels of active caspase 3 at 24, 48 and 72 hours of doxorubicin treatment without significant effects on p53 accumulation (Figure 4D). Since TCF3/E2A could possibly modulate the expression of hundreds of target genes in the genome beyond p21 and PUMA, we decided to investigate to what extent its effects on cell fate choice are dependent on these two p53 target genes (Figure 4E). Expectedly, p21 knockout increases the apoptotic index upon doxorubicin treatment while PUMA knockout decreases it (compare Y axis values for control shRNA cells). Interestingly, TCF3/E2A knockdown further increases apoptosis in p21−/− and PUMA−/− cells, but has no significant effects in the apoptotic index of the double knockout cells. Thus, we conclude that TCF3/E2A regulates the cell fate choice upon p53 activation by doxorubicin in a p21/PUMA-dependent manner.
Figure 4. TCF3/E2A controls the cellular response to p53 activation upon DNA damage in a p21/PUMA-dependent fashion.
A. BrdU incorporation assays demonstrate that p21 is required for the cell cycle arrest response observed upon doxorubicin treatment (0.5 μM, 12 h) in HCT116 cells. B. Apoptotic index assays show that p21 attenuates p53/PUMA-dependent apoptosis in response to doxorubicin treatment. C. TCF3/E2A knockdown increases the apoptotic index of HCT116 cells upon doxorubicin treatment. D. Western blot assays show that TCF3/E2A depletion leads to a lower p21:PUMA ratio concurrently with increased levels of activated executioner caspase 3 upon doxorubicin treatment. E. TCF3/E2A knockdown enhances the apoptotic index only in cells expressing p21 and/or PUMA. Data in A, B, C and E are expressed as averages of three experiments −/+ s.d. See also Figure S3.
DISCUSSION
Pleiotropy is a hallmark of ubiquitously expressed master transcriptional regulators such as p53, NF-kB and various nuclear hormone receptors, which participate in a plethora of cellular processes by regulating genes involved in distinct functional pathways. Accordingly, the cellular response elicited by activation of these regulators varies greatly with the context, and such pleiotropy often becomes an obstacle to effectively harnessing their activities for therapeutic purposes. In the case of p53, its pleiotropic character limits the therapeutic potential of p53-activating agents. In theory, non genotoxic MDM2/4 inhibitors could be employed to treat the ~11 million cancer patients currently carrying tumors with wild type p53, as these agents could trigger selective elimination of cancer cells via p53-dependent apoptosis (Brown et al., 2009). However, these compounds trigger p53-dependent apoptosis only in select cancer cell types (Paris et al., 2008; Tovar et al., 2006). Thus, there is a clear need to identify and characterize the molecular mechanisms defining cell fate choice in response to p53 activation. Here we report a novel experimental approach to identify pathway-specific coregulators within a transcriptional program. We employed p53 as the master regulator of choice and its target genes p21 and PUMA as the pathway-specific effectors. Our efforts led to the discovery of many factors modulating the p21:PUMA expression ratio, including TCF3/E2A and TRIAP1.
TCF3/E2A is a member of the Class I helix-loop-helix (HLH) transcription factors, or E proteins, not to be confused with TCF7L1 (also commonly referred to as TCF3, a member of the T cell factor/lymphoid enhancer family of transcription factors activated by β-catenin). E proteins are encoded by three genes: TCF3 (E2A), TCF12 (HEB) and TCF4 (E2-2) (de Pooter and Kee, 2010). Additionally, the TCF3 locus encodes two highly similar splicing variants of E2A, known as E12 and E47, which were first cloned based on their ability to bind an ‘E-box’ sequence in the immunoglobulin κ (Igκ) light chain enhancer (Murre et al., 1989). Although E protein transcription factors are ubiquitously expressed, they play unique essential roles in B and T lymphocyte development (de Pooter and Kee, 2010). Mice lacking TCF3/E2A display profound defects in B lymphopoiesis, decreased thymus size and development of aggressive T-cell lymphomas by 3–6 months of age (Engel et al., 2001; Yan et al., 1997; Zhuang et al., 1994). Interestingly, in the absence of TCF/E2A, hematopoietic stem cells (HSCs) and their progeny show increased cell cycle progression which has been linked to decreased expression of p21 and other CDK inhibitors (Semerad et al., 2009), thus leading to eventual exhaustion of the HSC pool.
Although TCF3/E2A is widely expressed across tissues, its roles outside the immune system are poorly understood. Previous studies have shown that TCF3/E2A binds several E-boxes in the p21 proximal promoter region and drives p21 expression in HeLa and HEK293T cells with no active p53 (Prabhu et al., 1997). Here we show that TCF3/E2A is required for full p21 induction upon p53 activation by both non-genotoxic and genotoxic agents in multiple cancer cell types expressing wild type p53. Thus, p53 and TCF3/E2A act coordinately to enforce p21 expression and cell cycle arrest in response to p53 activating agents. This effect is exquisitely gene-specific, as TCF3/E2A knockdown does not affect induction of many other p53 target genes in diverse pathways. Interestingly, TCF3/E2A seems to act at steps downstream of p53 binding to the p21 enhancers and RNAPII transactivation at this locus, affecting instead p21 mRNA stability. Strikingly, TCF3/E2A also acts as a gene-specific repressor of PUMA. The PUMA locus has an unusual chromatin architecture with constitutive intragenic transcription and internal chromatin boundaries defined by the insulator factor CTCF (Gomes and Espinosa, 2010). We found that TCF3/E2A binds at several sites within the PUMA locus (data not shown, Z.A. and J.M.E.). Of note, there is evidence that TCF3/E2A can function as a transcriptional repressor in B cells (Greenbaum et al., 2004). Future studies will focus on elucidating the molecular mechanism by which TCF3/E2A exerts opposite effects on p21 and PUMA expression.
We found that TCF3/E2A depletion leads to increased apoptosis in response to doxorubicin treatment, suggesting that increased expression of TCF3/E2A may protect cancer cells from the killing effects of this commonly used chemotherapeutic agent. Indeed, TCF3/E2A is expressed at high levels in prostate cancer cells where it confers resistance to doxorubicin-induced apoptosis (Patel and Chaudhary, 2012). The TCF3 locus is a common target of chromosome rearrangements in diverse leukemias leading to E2A chimeric proteins. Interestingly, the chimeric E2A-HLF transcription factor was shown to abrogate p53-induced apoptosis in myeloid leukemia cells by an undefined mechanism (Altura et al., 1998). Our observation that TCF3/E2A is a repressor of PUMA may explain these observations and provide a mechanistic basis for the oncogenic properties of TCF3/E2A chimeras.
Very little is known about TRIAP1, a factor identified in our screen as a specific repressor of p21. Interestingly, TRIAP1 was first characterized as a p53-inducible cell survival factor (Park and Nakamura, 2005). Whereas knockdown of TRIAP1 lead to increased levels of cell death in response to doxorubicin treatment, its overexpression protected against cell death. We confirmed that TRIAP1 functions as a survival factor but also found that TRIAP1 reduces basal p21 protein expression, thus allowing for increased cell cycle progression as measured by BrdU incorporation (Figure S3A-C). We have also confirmed that TRIAP1 is a p53-activated gene as seen by Western blot upon Nutlin-3 treatment in multiple cell types (Figure 2B and Figure S2B). Thus, TRIAP1 enables an incoherent feed forward loop within the p53 network, where p53 induces transcription of both p21 and a p21 repressor. Curiously, whereas TRIAP1 repression of p21 could be explained by effects on p21 mRNA accumulation in HCT116 cells, TRIAP1 seems to regulate p21 levels by other mechanisms in U2OS and A549 cells. Regardless of mechanism, it is clear that the overall effects of TRIAP1 as a pro-survival factor in the p53 network can not be explained by its function as a p21 repressor, but may be due to other roles. In fact, TRIAP1 was previously found to interact with APAF1 and prevent activation of caspase 9 (Park and Nakamura, 2005). Thus, the inhibitory effect of TRIAP1 on apoptosome activity may override its repressive effect on p21 expression.
In summary, our functional genomics approach has identified new gene-specific regulators within the p53 network, which opens up new research venues not only to define their mechanism of action, but also to design strategies for manipulating the cellular response to p53 activation.
EXPERIMENTAL PROCEDURES
Cell culture and knockdown cell lines preparation
HCT116 cells were cultivated in McCoy’s medium (Sigma) and U2OS and A549 cells in DMEM (Sigma) supplemented with 10% fetal bovine serum and antibiotic-antimycotic mixture (Life Technologies). Nutlin-3 (Cayman) was solubilized in DMSO and used at 10 μM, doxorubicin (Sigma) was used at 0.5 μM. Individual knockdown cell lines were generated using Sigma Mission shRNA lentiviral plasmids (pLKO.1-puro, see Extended Experimental Procedures) as described previously (Sullivan et al., 2012b).
Apoptotic index, BrdU incorporation, proliferation and cell cycle profile assays
For apoptosis assays, cells were treated with doxorubicin for 72 h, both adherent and floating cells were collected by trypsinization, labeled with Annexin-V-FITC (Invitrogen) and propidium iodide (10 μg/ml) for 15 min in the dark at room temperature and analyzed by flow cytometry (BD Accuri C6, Becton Dickinson). For BrdU incorporation analysis, cells were exposed to doxorubicin for 12 h. One hour prior to harvest, 1 μg/ml of BrdU was added to the medium and trypsinized cells were fixed in 70% ethanol. DNA denaturation was performed for 10 min in 2 M HCl with 0.5 % Triton X-100 at 37°C. After neutralization with 0.1 M NaBO4 cells were labeled with anti-BrdU antibody and analyzed by flow cytometry. See Extended Experimental Procedures for antibody information, proliferation assays and cell cycle profile analysis. Student’s T-test was used for statistical analysis.
Western Blotting
Protein samples were subjected to SDS-PAGE and transferred to PVDF membrane (Millipore). Following incubation in blocking solution and with primary antibody (see Extended Experimental Procedures for antibody information), HRP-conjugated secondary antibodies and enhanced chemiluminescence solution (Millipore) were used to visualize proteins of interest. Alternatively, for the detection of p21:PUMA ratio, membranes were stained with both antibodies simultaneously. After incubation with fluorescent dye-labeled secondary antibodies, membranes were scanned by LiCor system and band intensities were quantified in Odyssey 3.0.30 software (LiCor).
Q-RT-PCR
Total RNA was extracted using TRIreagent (SIGMA) and template cDNA was prepared with the qScript cDNA Synthesis Kit (Quanta Biosciences). 10 ng of cDNA was used per PCR reaction with SYBR Green PCR Master Mix (Applied Biosystems, ABI) and quantified with the Absolute Quantification Method in 7900HT ABI instrument (see Extended Experimental Procedures for primer information).
See Extended Experimental Procedures for shRNA screen strategy, ChIP assays and mRNA half life assays.
Supplementary Material
HIGHLIGHTS.
A novel genomics approach to identify gene-specific regulators is described
An shRNA screen identifies factors regulating the p21:PUMA ratio upon p53 activation
TCF3/E2A drives p21 expression while repressing PUMA
TRIAP1 is a gene-specific repressor of p21
Acknowledgments
We are grateful to all members of the Espinosa lab for invaluable discussion and reagents, in particular to Dr. Kelly Sullivan for preparing the HCT116 cells transduced with the shRNA library. This work was supported by grant NIH 2RO1CA117907-07 to J.M.E. and grants from the Cancer League of Colorado and Golfers Against Cancer/AMD Fund to A.C.T.. J.M.E. is an HHMI Early Career Scientist. We thank Dr. Luis Fonrouge for inspiration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altura RA, Inukai T, Ashmun RA, Zambetti GP, Roussel MF, Look AT. The chimeric E2A-HLF transcription factor abrogates p53-induced apoptosis in myeloid leukemia cells. Blood. 1998;92:1397–1405. [PubMed] [Google Scholar]
- Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, Prives C. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- de Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunological reviews. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nature methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. The Journal of experimental medicine. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Espinosa JM. Disparate chromatin landscapes and kinetics of inactivation impact differential regulation of p53 target genes. Cell Cycle. 2010;9:3428–3437. doi: 10.4161/cc.9.17.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S, Lazorchak AS, Zhuang Y. Differential functions for the transcription factor E2A in positive and negative gene regulation in pre-B lymphocytes. J Biol Chem. 2004;279:45028–45035. doi: 10.1074/jbc.M400061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RE, Andrysik Z, Paris R, Galbraith MD, Espinosa JM. A DR4:tBID axis drives the p53 apoptotic response by promoting oligomerization of poised BAX. EMBO J. 2012;31:1266–1278. doi: 10.1038/emboj.2011.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tan AC. BiNGS!SL-seq: a bioinformatics pipeline for the analysis and interpretation of deep sequencing genome-wide synthetic lethal screen. Methods in molecular biology. 2012;802:389–398. doi: 10.1007/978-1-61779-400-1_26. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Paris R, Henry RE, Stephens SJ, McBryde M, Espinosa JM. Multiple p53-independent gene silencing mechanisms define the cellular response to p53 activation. Cell Cycle. 2008;7:2427–2433. doi: 10.4161/cc.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WR, Nakamura Y. p53CSV, a novel p53-inducible gene involved in the p53-dependent cell-survival pathway. Cancer research. 2005;65:1197–1206. doi: 10.1158/0008-5472.CAN-04-3339. [DOI] [PubMed] [Google Scholar]
- Patel D, Chaudhary J. Increased expression of bHLH transcription factor E2A (TCF3) in prostate cancer promotes proliferation and confers resistance to doxorubicin induced apoptosis. Biochem Biophys Res Commun. 2012;422:146–151. doi: 10.1016/j.bbrc.2012.04.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci U S A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Gallant-Behm CL, Henry RE, Fraikin JL, Espinosa JM. The p53 circuit board. Biochim Biophys Acta. 2012a doi: 10.1016/j.bbcan.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Padilla-Just N, Henry RE, Porter CC, Kim J, Tentler JJ, Eckhardt SG, Tan AC, DeGregori J, Espinosa JM. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nature chemical biology. 2012b;8:646–654. doi: 10.1038/nchembio.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci U S A. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.