Abstract
Background
Human cardiac troponin I is known to be phosphorylated at multiple amino acid residues by several kinases. Advances in mass spectrometry allow sensitive detection of known and novel phosphorylation sites and measurement of the level of phosphorylation simultaneously at each site in myocardial samples.
Methods and Results
On the basis of in silico prediction and liquid chromatography/mass spectrometry data, 14 phosphorylation sites on cardiac troponin I, including 6 novel residues (S4, S5, Y25, T50, T180, S198), were assessed in explanted hearts from end-stage heart failure transplantation patients with ischemic heart disease or idiopathic dilated cardiomyopathy and compared with samples obtained from nonfailing donor hearts (n=10 per group). Thirty mass spectrometry–based multiple reaction monitoring quantitative tryptic peptide assays were developed for each phosphorylatable and corresponding nonphosphorylated site. The results show that in heart failure there is a decrease in the extent of phosphorylation of the known protein kinase A sites (S22, S23) and other newly discovered phosphorylation sites located in the N-terminal extension of cardiac troponin I (S4, S5, Y25), an increase in phosphorylation of the protein kinase C sites (S41, S43, T142), and an increase in phosphorylation of the IT-arm domain residues (S76, T77) and C-terminal domain novel phosphorylation sites of cardiac troponin I (S165, T180, S198). In a canine dyssynchronous heart failure model, enhanced phosphorylation at 3 novel sites was found to decline toward control after resynchronization therapy.
Conclusions
Selective, functionally significant phosphorylation alterations occurred on individual residues of cardiac troponin I in heart failure, likely reflecting an imbalance in kinase/phosphatase activity. Such changes can be reversed by cardiac resynchronization.
Keywords: heart failure, mass spectrometry, phosphorylation, troponin
Cardiac TnI (cTnI) plays a key role in the regulation of contraction and relaxation. As part of the thin filament, the troponin complex (cTnI, troponin T [TnT], and troponin C [TnC]), along with tropomyosin, regulates the actin filament interaction with the thick filament (composed primarily of myosin) in a Ca2+-dependent manner. Human cTnI can be phosphorylated at multiple amino acid residues, S22, S23, T30, S41, S43, S76, T77, T142, S149, and S165 (the numbering excludes the initiating methionine), and studies have shown that phosphorylation at different residues can alter function.1,2 For example, phosphorylation of S22/S23 located in the cardiac-specific N terminus of cTnI (residues 1–32) by protein kinase A (PKA),3 protein kinase C (PKC),4 protein kinase D,5 or protein kinase G6 alters its interaction with TnC, reduces myofilament Ca2+ sensitivity, enhances sliding velocity, and thus contributes to the acceleration of relaxation.7 Similarly, phosphorylation at known PKC sites significantly affects function with effects distinct from S22/S23 phosphorylation.2,7 Other sites demonstrated in vitro or in animal models include T30 (mammalian sterile 20-like kinase 1),8 S41/S43 (PKC),9 S76/T77 (kinases undetermined),10 T142 (PKC-βII),11 S149 (p21-activated kinase),12,13 and S165 (PKA).14 However, the roles of these phosphorylatable residues are not well understood.
In end-stage failing hearts, a number of studies have found that the phosphorylation state of S22 and/or S23 is significantly reduced compared with control myocardium, a change that may lead to an increase in Ca2+ sensitivity.3,15,16 The phosphorylation state of the other sites is less clear. Although several phospho-specific antibodies are available (against S22/23, S23, S43, and T142), most have not been fully validated for specificity and cross-reactivity with neighboring phosphorylatable amino acid residues.
Mass spectrometry (MS)–based methods are able to simultaneously measure the state of multiple phosphorylation sites.10,17 Our group has used the newly developed multiplex quantitative proteomics approach, multiple reaction monitoring (MRM),18 which allows site-specific quantification of each phosphorylatable residue. MRM assays have many desirable characteristics, including (1) quantification of both the phosphorylated and unphosphorylated peptides; (2) monitoring of the precursor (Q1) and several fragment (Q3) ions of the peptide, providing a high degree of specificity; (3) sensitivity that is greater than in other MS-based techniques because of the 2-stage filtering of Q1 and Q3, which increases the signal-to-noise ratio; and (4) the ability to provide absolute quantification of each targeted phosphorylatable residue by incorporating stable isotope-labeled peptides as internal standards. Because both isotopes are chemically and physically identical, the labeled peptides (N15) will coelute with the endogenous peptides (N13), and integration of the area beneath both peaks permits accurate quantification of the phosphorylation level.
Using MRM, for the first time, we quantified levels of phosphorylation of 14 sites on cTnI, including 6 novel sites (S4, S5, Y25, T50, T180, S198) in human left ventricles from failing ischemic (ISHD) and dilated (IDCM) cardiomyopathy hearts and donor hearts from apparently healthy individuals. This is the first determination of the relative phosphorylation of the cTnI PKC site T142, PKA site S165, and several novel sites in human heart in vivo. Overall, there is a switch in phosphorylation from the PKA to the PKC sites in ISHD and IDCM, as well as modulation of several novel sites, a finding that was recapitulated in a canine pacing model of heart failure (HF). The present study provides important insight into the status of cTnI regulation in heart disease.
Methods
Cardiac Myofibril and cTnI Protein Preparation
Human left ventricular free wall transmural tissue samples were obtained from explanted end-stage failing hearts with ISHD or IDCM during heart transplant surgery, as well as from unused healthy donor hearts (n=10 per group). Tissue was rapidly frozen and stored in liquid nitrogen (clinical information is shown in Table I in the online-only Data Supplement). Ethics approval was provided by St. Vincent’s Hospital (No. H03/118), Sydney, Australia, and by the University of Sydney (No. 09–2009-12146).
Canine Model of HF and Reverse Remodeling After Cardiac Resynchronization Therapy
Left ventricular tissue was collected from adult mongrel dogs; groups included normal (control), dyssynchronous pacing-induced HF (HFdys), and cardiac resynchronization therapy (CRT) as previously published (n=4 per group).19 All protocols were approved by the Animal Care and Use Committee of the Johns Hopkins Medical Institutions. At the conclusion of the pacing protocol, hearts were extracted under ice-cold cardioplegia and frozen in liquid nitrogen (see Methods in the online-only Data Supplement for details).
MRM Development and Optimization
Possible phosphorylation sites on human cTnI were selected by use of an in silico phosphorylation prediction algorithm (NetPhos 2.0) and MS data obtained from the LTQ Orbitrap liquid chromatography/MS/MS analysis of the isolated cTnI. Three to 5 MRM transitions per peptide were designed on the basis of either MS/MS identification–acquired data or prediction by MRMPilot software 1.0 (AB SCIEX). Each MRM assay was optimized manually on a 4000 QTRAP MS instrument (see Methods in the online-only Data Supplement).
Synthesized Internal Standards
Synthesized internal standards peptides (unphosphorylated peptides had an N15 stable isotope label, whereas phosphorylated peptides were unlabeled and contained N13) were produced by solid-phase peptide synthesis (New England Peptide). For each phosphorylation site, a series dilution of light/heavy peptides was made to produce 6-point calibration curves at 0.125, 0.25, 0.5, 1, 5, and 10 fmol/μL with and without matrix comprising a pool digest of all tissue samples used (see Methods in the online-only Data Supplement). A control peptide (NITEIADLTQK), which does not have any known posttranslational modifications (confirmed by MS and prediction software), was used to determine the total quantity of cTnI (femtomoles) in the sample.
Q-Trap Nano–Liquid Chromatography/MS/MS Analysis
The mixture of peptides from the in-gel digestion of cTnI proteins was reconstituted with 20 μL high-performance liquid chromatography water containing 0.1% formic acid. MRM analyses were performed on a 4000 QTRAP hybrid triple quadrupole/linear IT mass spectrometer (AB SCIEX) operating with Analyst 1.4.2 software–scheduled experiments in positive ion mode (see Methods in the online-only Data Supplement).
Validation of MRM Quantification by Immunoblotting Analysis
The phosphorylation status of cTnI for subset of sites was verified by Western blotting (see Methods in the online-only Data Supplement).
Statistical Analysis
Peak detection and quantification of peak area were determined with Multiquant software version 2.0 (AB SCIEX) and inspected manually to ensure correct peak identification and quantification. Measurements were performed in triplicate and then averaged to reduce technical variation. The quantity (femtomoles) of each peptide was calculated from the linear standard curve, and then the phosphorylated peptide was normalized to the total quantity of cTnI.
For human samples, data were analyzed with 1-way ANOVA on ranks, followed by the Dunnett multiple-comparisons post hoc test. For canine samples, data were analyzed by use of a 1-way ANOVA, followed by Bonferroni multiple-comparisons post hoc test. All calculations were done with SigmaPlot version 11 (Systat), with a value of P<0.05 denoting significance. Human data are presented as median, first quartile, and third quartile; canine data are presented as mean±SEM.
Results
Human cTnI contains 209 amino acids, including 12 Ser, 8 Thr, and 3 Tyr residues. Using NetPhos 2.0, a phosphorylation prediction algorithm, we predicted 16 potential phosphorylatable residues. Fourteen of these predicted phosphorylatable residues were experimentally verified with LTQ Orbitrap MS analysis. The 8 known phosphorylation sites (S22, S23, S41, S43, S76, T77, T142, S165) and additional 6 novel phosphorylation sites (S4, S5, Y25, T50, T180, S198) were observed in the 30 different human heart tissues. To verify and quantify all phosphorylated amino acids, MRM assays were developed for the corresponding tryptic peptides that contained 1 or more of these residues.
MRM Workflow Development and Optimization and Phosphorylation Mapping
The workflow used in this study is outlined in Figure I in the online-only Data Supplement, which schematically illustrates the process for developing and using MRM assays to quantify site-specific cTnI phosphorylation. The steps necessary to develop the pipeline were the following: development of MRM assays for each unphosphorylated and phosphorylated tryptic peptide containing a potential modified residue(s) and for a control peptide (NITEIADLTQK) not predicted to be modified that could be used to estimate the total quantity of cTnI in the sample, optimization of the sample preparation to isolate cTnI from left ventricular tissue, and optimization of tryptic digestion conditions to consistently generate the endogenous tryptic cTnI peptides for measurement by the MRM assays.
For each MRM assay, the MS parameters for each peptide were optimized and transitions were selected to achieve the greatest sensitivity (individual peptide data shown in Figures II–XV in the online-only Data Supplement). Figure 1 shows examples of in vivo MRM assays for representative cTnI peptides. Tables II and III in the online-only Data Supplement list all peptides and transitions used for the MRM assays to monitor and calibrate each phosphorylation site. To test the matrix effect in vivo, we used composite matrix composed of a tryptic digest of a pooled sample (10 μg each of the donor, ISHD, and IDCM), and the coefficient of variation was <1% for the calibration curves with or without matrix. From this, we determined the lower limits of detection and quantification. Figure 2 shows the data for one of the novel sites, S198 (Figures II–XV in the online-only Data Supplement provide the MS/MS spectrum for all peptides and their MRM optimization).
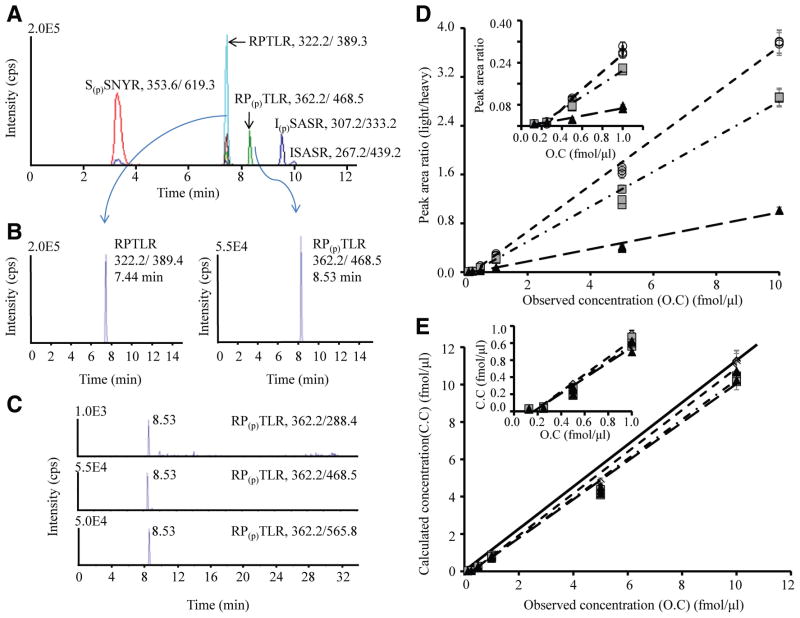
Figure 1.
Development of multiple reaction monitoring assays for the representative cardiac troponin I (cTnI) peptide and its corre-sponding phosphorylated peptide. A typical total ion chromatogram is shown for the multiplex consisting of the peptides S(p)SNYR, RPTLR, RP(p)TLR, ISASR, and I(p)SASR (A). The extracted ion chromatogram for the unphosphorylated peptide (RPTLR with m/z 322.2/389.4) and its corresponding phosphorylated peptide (RP(p)TLR with m/z 362.2/468.5) eluting at 7.44 and 8.53 minutes (B). Three transitions for the unphosphorylated and phosphorylated peptide corresponding to RPTLR at m/z 362.2/288.4, 362.2/468.5, and 362.2/565.8 (C); Table II in the online-only Data Supplement lists the other transitions used. The plot of peak area ratio vs observed concentration (D) or calculated versus observed concentration (E) with linear calibration curve slopes for the 3 transitions of the phosphorylated peptide RP(p)TLR. The ratio is the peak area of phosphorylated peptide RP(p)TLR (unlabeled peptide) divided by the corresponding unphosphorylated (heavy labeled peptide) at 6-point different dilution ratio of concentrations 0.125, 0.25, 0.5, 1, 5, and 10 fmol/μL. The concentration of unlabeled phosphorylation site T142 peptide ranges from subfemtomole to tens of femtomole levels, whereas the concentration of heavy labeled internal standard is kept constant at 1 fmol/μL. Three replicate measurements are represented at each concentration point. Circle indicates transition 1 (m/z 362.2/288.4, y2=0.3789x−0.0931, R2=0.9975); square, transition 2 (m/z 362.2/468.5, y3=0.2868x−0.0745, R2=0.9940); and triangle, transition 3 (m/z 362.2/565.8, y4=0.1005x−0.0267, R2=0.9915). The plots demonstrates good linearity, with slopes falling close to the diagonal black line (theoretical slope=1) and good agreement between the 3 transitions at each concentration point. Inset plots show the lower end of the concentration range. The lower limits of detection and quantification calibration curve generated for this peptide in buffer were 0.1 and 0.3 fmol/μL, respectively.
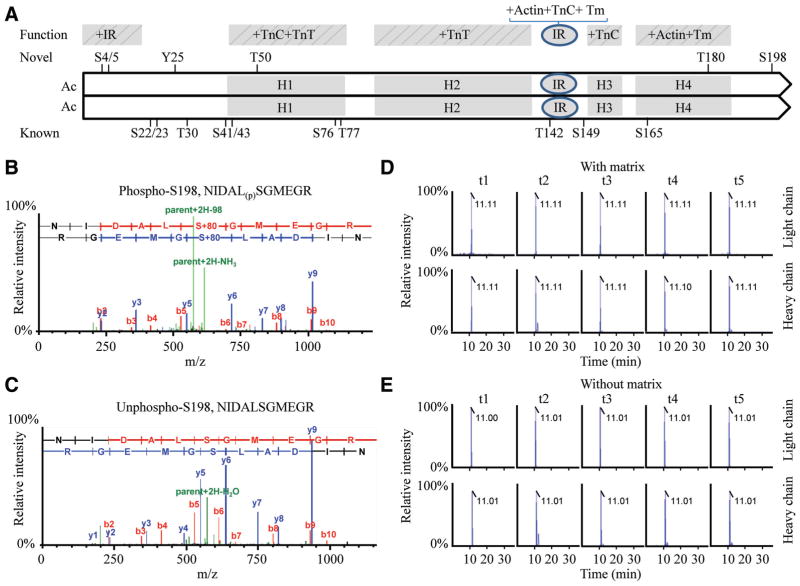
Figure 2.
Alignment of cardiac troponin I (cTnI) sequence and the representative peptide containing a novel phosphorylation site S198 in peptide NIDAL(p)SGMEGR. A schematic of the human cTnI sequence (1–209 amino acid) illustrating function domains and linear positions of the known and novel phosphorylation sites20–26; (A). Residues 1 through 15 interact with the inhibitory region (IR). The domain H1–H4 stands for 4 helixes of cTnI protein. The H1 binds the C terminal of cardiac troponin C (cTnC) and cardiac troponin T (cTnT); H2 binds TnT; H3 binds the N domain of TnC in response to Ca2+ and is referred to as the switch region; and H4 binds actin-tropomyosin. The IR binds both TnC and actin-TM tropomyosin in a Ca2+-dependent manner. The representative mass spectrometry spectrum of the phosphorylated peptide NIDAL(p)SGMEGR (B) containing the site S198 and the corresponding unphosphorylated peptide NIDALSGMEGR (C) of cTnI in left ventricular tissues. Representative multiple reaction monitoring (MRM) spectrum of the phosphorylated peptide NIDAL(p)SGMEGR in matrix (D) or without matrix (E). A 6-point dilution ratio of light/heavy peptide was run for each MRM assay, illustrating the coelution time and relative intensity for the native light peptide phosphorylated S198 peptides, respectively. MRM transitions of t1 to t5 were arranged for the native peptide as in an order (Q1>Q2): t1, 622.8+2>549.7+1 (y5); t2, 622.8+2>573.7+2 (y11–98); t3, 622.8+2>716.8+1 (y6); t4, 622.8+2>918.8+1 (y9–98); and t5, 622.8+2>1016.9+1 (y9); and for the heavy labeled peptide as in an order: t1, 627.8+2>242.4+1(y2); t2, 627.8+2>559.6+1 (y5); t3, 627.8+2>726.8+1 (y6); t4, 627.8+2>839.7+2 (y7); and t5, 627.8+2>928.8+1 (y8), respectively.
Next, we determined the optimal sample processing to ensure consistency and preservation of phosphorylation status. As shown in Figure XVI in the online-only Data Supplement, there is essentially no difference between samples processed with a gel-based (SDS-PAGE, followed by in-gel digestion) or direct protein digestion of the isolated myofibrils. Samples were prepared by SDS-PAGE for the subsequent analysis of the 30 human tissues (see details in Figure XVII and Table IV in the online-only Data Supplement).
The final step required for optimization of the pipeline was to ensure efficient and complete tryptic digestion of cTnI. Importantly, despite efforts at optimization, by either extending the reaction time or adding a second batch of trypsin after 12 hours, several peptides were consistently miscleaved. For example, the peptide containing T142 is surrounded by basic residues, and this resulted in the consistent generation of a miscleaved 5–amino acid peptide. Therefore, to ensure accurate quantification at this PKC site, MRM assays were developed for both the nontryptic and single-tryptic miscleaved phosphorylated and unphosphorylated peptides.
Quantitation of Levels of Phosphorylation
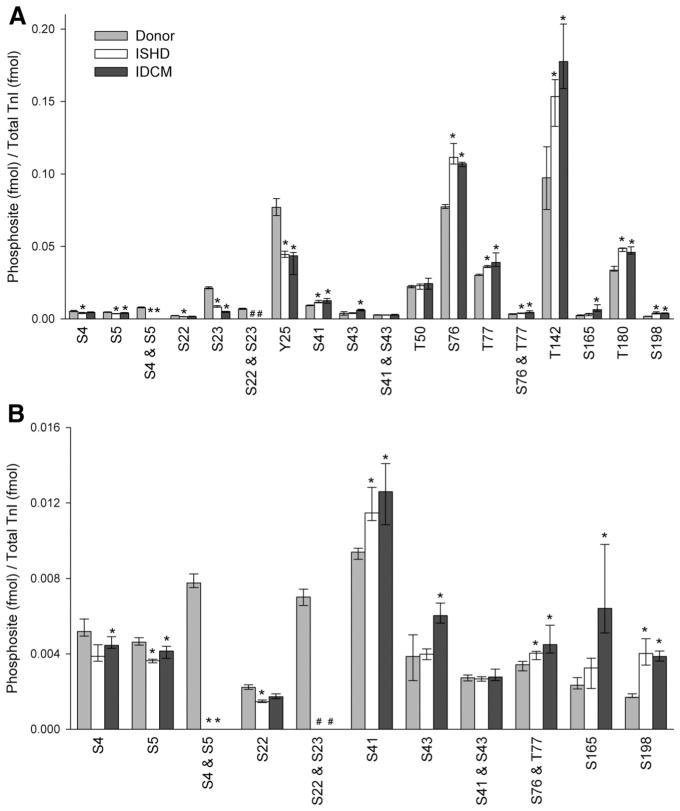
In total, we quantified levels of phosphorylation of 14 cTnI sites in left ventricle obtained from ISHD, IDCM, and donor hearts (n=10 per group; clinical data are presented in Table I in the online-only Data Supplement). Each assay was carried out in triplicate. The MRM quantitative data showed that phosphorylation status is decreased or increased in a site-specific manner (Figure 3 and Table V and Figure XVIII in the online-only Data Supplement). Figure 3 shows the phosphorylation occupancy of each amino acid residue (femtomole phosphorylation per femtomole cTnI). Figure XVIII in the online-only Data Supplement summarizes the fold change for each site. Compared with the donor samples, the amount of phosphorylation of the N-terminus cTnI sites was significantly reduced in ISHD and IDCM. The monophosphorylated S4 or S5 form was 25% and 21% lower in ISHD and 14% (not significant) and 10% lower in IDCM, respectively, compared with cTnI isolated from the control donor myocardium (Figure XVIII in the online-only Data Supplement). In the donor hearts, these adjacent sites exist primarily as diphosphorylated species, but this species was not detected in the failing heart groups (Figure 3 and Table V in the online-only Data Supplement).
Figure 3.
Quantification of phosphorylation sites in myocardium obtained from donor and failing hearts. The stoichiometric quantity (fmol phosphorylation/fmol protein) phosphorylation of cardiac troponin I (cTnI) by multiple reaction monitoring assay for donor (light gray), ischemic heart failure (ISHD; white), and dilated cardiomyopathy (IDCM; dark gray; n=10 per group) for all of the sites (A) and the 11 least abundant sites (B). All raw data were calibrated by the synthesized internal standard peptides, converted for femtomole using standard curves, and normalized to the total quantify of TnI in the sample (fmol). Values are median, first quartile, and third quartile. *P<0.05 for ISHD or IDCM vs donor. #Values were below the lower limit of quantification. Phosphorylated T30 and S149 were not detected in any of the 3 groups and were omitted. See Table V and Figure XVIII in the online-only Data Supplement for complementary data.
The PKA-mediated monophosphorylated and diphosphorylated forms of S22/S23 were also reduced in ISHD and IDCM compared with donor hearts. Monophosphorylated forms were 34% and 61% lower in ISHD and 22% (not significant) and 76% lower in IDCM. Although the diphosphorylated species was quantified in the donor hearts, levels were too low to quantify in the failing heart groups. Notably, the monophosphorylation of S23 was 857% greater than that of S22 in the control hearts, suggesting a potential priority of S23 phosphorylation over S22. There is also greater S23 phosphorylation than S22 phosphorylation in the failing hearts, but the ratio is reduced to 472% in ISHD and 195% in IDCM. Quantification of the phosphorylation was verified by immunoblot for the PKA sites using the anti-S22 and/or anti-S23 phospho-sensitive antibody (Figure XIX in the online-only Data Supplement) versus an antibody against total cTnI, 8I-7. The immunoblot results support the MRM data showing a decrease in phosphorylation of the PKA sites in HF.
The N-terminal novel site Y25 is located near S22/S23 and is the only phosphorylatable Tyr residue in cTnI. Residue Y25 was more phosphorylated (Figure 3) than S22 or S23 and dephosphorylated in ISHD (42%) and IDCM (43%) compared with nonfailing donor samples (Figure 3). Immunoblotting with an anti–phospho-Tyr antibody verified the reduction in phosphorylation of cTnI in HF (Figure XIX in the online-only Data Supplement).
Compared with donor samples, the phosphorylation states of sites just downstream from the N-terminus were increased significantly in ISHD and IDCM. Monophosphorylation of PKC site S41 was increased in ISHD (22%) and IDCM (34%), as was S43 in IDCM (56%; see Table V in the online-only Data Supplement). T50 was phosphorylated, but there was no significant change between groups. The other sites located within the same functional domain of cTnI also had increased phosphorylation (for ISHD: S76, 44%; T77, 20%; diphosphorylated, 18%; for IDCM: S76, 38%; T77, 29%; diphosphorylated, 31%). Interestingly, similar to the other pairs of adjacent phosphorylatable residues in cTnI, there was a preference for one of the residue pair (S41>S43 and S76>T77; Figure 3).
In the important inhibitory domain of cTnI, the sole phosphorylated residue (T142) was increased in ISHD (58%) and IDCM (82%) compared with the donor heart. Remarkably, this site had the greatest amount of phosphorylation of all sites (Figure 3). The p21-activated kinase target site S149 was not phosphorylated because only the nonphosphorylated peptide was observed in the samples. It is possible, however, that the phosphorylated peptide is present but below the detectable limits of this assay (Table V in the online-only Data Supplement). Similarly, the phosphorylated form of T30 was not observed.
Finally, the 3 novel phosphorylation sites in the C-terminal region of cTnI were all increased in HF compared with donor hearts: S165, which was significantly greater only in IDCM (174%), T180 (44% and 36% greater for ISHD and IDCM), and S198 (136% and 127% greater for ISHD and IDCM).
Phosphorylation of Novel Sites in Canine Model of HF and CRT
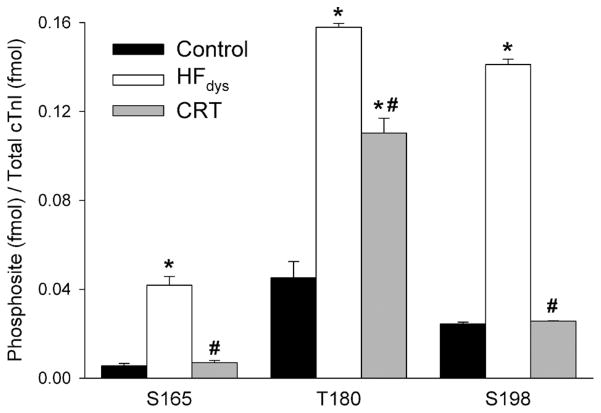
The extent of phosphorylation of the 3 novel residues S165, T180, and S198 was quantified in a well-characterized canine model of HFdys and after treatment with CRT. These 3 sites were chosen because the tryptic peptides encompassing these phosphorylatable residues are conserved between dog and human. As shown in Figure 4, HFdys led to greater phosphorylation of these 3 amino acid residues (S165, 659% increase; T180, 249% increase; S198, 479% increase). This was reversed in dogs treated with CRT, with phosphorylation levels falling relative to HFdys conditions by 83%, 30%, and 82% for S165, T180, and S198, respectively.
Figure 4.
Quantification of phosphorylated sites in a canine model of heart failure (HF). The stoichiometric quantity (fmol phosphorylation/fmol protein) of phosphorylation sites on cardiac troponin I (cTnI) by multiple reaction monitoring assay in control (black), dyssynchronous HF (HFdys; white), and cardiac resynchronization therapy (CRT; gray; n=4 per group). All raw data were calibrated by the synthesized internal standard peptides and then normalized to total TnI. Error bars indicate the SEM. *P<0.001, HFdys or CRT vs control. #P<0.001, CRT vs HFdys by 1-way ANOVA, followed by Bonferroni multiple-comparisons post hoc test.
Discussion
This study provides novel insights into site-specific phosphorylation of cTnI in human myocardial samples. It establishes that human cTnI is more extensively phosphorylated than previously reported, with 14 modifiable amino acid residues located throughout its entire length. We also provide evidence of HF-related phosphorylation changes for the established PKA- and PKC-modifiable amino acid residues and several newly identified phosphorylated sites, specifically S4, S5, Y25, T50, T180, and S198. Finally, data were provided indicating that increased phosphorylation levels of 3 novel sites located in the C terminus of cTnI are observed in an animal model of HF with dyssynchrony but intriguingly reversed by CRT.
Structure and Function of cTnI Domains and Impact on Contractile Function
To provide context for the impact of altered phosphorylation in HF, a brief review of cTnI domains and function is necessary (the Table).20–37 cTnI, the inhibitory protein of troponin, forms a complex with TnT and TnC and is responsible for inhibiting activation of muscle via its interaction with actin-tropomyosin at diastolic levels of calcium. When free calcium levels increase during systole, calcium rapidly binds to site II of cTnC, and the affinity of cTnI for TnC is increased as a result of opening of the hydrophobic patch on the N-terminal lobe of TnC. The cTnI–actin-tropomyosin interaction occurs at 2 key domains of cTnI, the inhibitory region (128–147 in human cTnI) and a more C-terminal region (164–209) with a helical switch peptide (147–163) bridging these 2 regions. Unique to cardiac muscle, the opening of the hydrophobic patch on TnC with calcium binding is facilitated by binding of the switch peptide of cTnI to the N-terminal lobe of TnC.20 This is discussed in detail in numerous excellent reviews.2,21
Table.
Summary of the Known and Novel Phosphorylation Sites in This Study and the Putative Effect of Altered Phosphorylation in Heart Failure
| Residue | Kinase(s) | Domain Within cTnI20 –26 | Physiological Effect of Phosphorylation | Putative Effect of Altered Phosphorylation in HF |
|---|---|---|---|---|
| S4 and S5 | Unknown | Acidic region of N-terminal extension and interacts with inhibitory region when S22 and S23 are phosphorylated | Unknown | May inhibit interaction of acidic region of N terminus with inhibitory region22,25,26; speculated to have an impact similar to that of a decrease in S22 and S23 phosphorylation3 |
| S22 and S23 | PKA3 PKC (β and δ),4 PKD,5 PKG6 | N-terminal extension; phosphorylation results in bending of hinge region and cTnI more compact; promotes acidic region at extreme N-terminus interaction with inhibitory region | Desensitizes to Ca2+; may accelerate cross-bridge kinetics; decreases Ca2+ affinity of cTnC; increases contractile power and relaxation and length-dependent activation | Inhibits lusitropic response to β-adrenergic receptor stimulation; impairs relaxation and force frequency response; may inhibit length-dependent activation (Frank-Starling law)3,4,7,24,27 |
| Y25 | Unknown | N-terminal extension | Unknown | Putatively similar to S22,S23 |
| S41 and S43 | PKC (β and δ) | IT arm | Decreases Ca2+ sensitivity; slows kinetics in motility assays and stabilizes inhibition of activation of thin filament9; pseudophosphorylation decreases contractility and relaxation in vivo7,28 | Impairs relaxation and force frequency response7,9,21,22,28 |
| T50 | Unknown | IT arm | Unknown | Unknown |
| S76* and T77* | Unknown | IT-arm region; interacts with cTnT | Unknown | Unknown |
| T142 | PKC | Within inhibitory region | Conflicting evidence on effect on Ca2+ sensitivity; decreases cooperativity of activation9; pseudophosphorylation in vivo impairs relaxation28 | Impairs relaxation9,28; impairs contractile force |
| S165† | PKA14 | Switch peptide between 2 actin-binding regions; adjacent to key residue for acidosis impact on contractile function30–34 | Unknown | Potential impact on response to acidosis and switching of binding from actin to TnC with activation29,30; reduced proteolysis31 |
| T180 and S198 | Unknown | C-terminal region involved in position of troponin along tropomyosin; also location of HCM mutants | Unknown | Could affect the position of troponin along tropomyosin, effects on activation31,35–37; reduced proteolysis31 |
PKA indicates protein kinase A; PKC, protein kinase C; PKD, protein kinase D; PKG, protein kinase G; cTnI, cardiac troponin I; cTnC, cardiac troponin C; and HCM, hypertrophic cardiomyopathy.
These sites had been noted by researchers10; however it was unclear which of the 2 sites were phosphorylated.
Previously noted only in vitro.
Other key domains of cTnI include an N-terminal extension, unique to the cardiac isoform. Phosphorylation at the S22/S23 sites alters the shape of the cTnI, resulting in a bend at a hinge domain in this region, which permits an intramolecular interaction between the acidic region at the most N-terminal residues and the inhibitory domain.22 Physiological studies speak to an important role of phosphorylation of cTnI at the S22/S23 sites in augmenting relaxation, accelerating force frequency response, and potentially increasing contractile power and length-dependent contraction (Frank-Starling effects).27
The PKC sites lie within the IT-arm of cTnI (S41 and S43), a region that structural studies suggest is relatively rigid and is involved in stabilization of cTnI to the thin filament, including interaction with TnT and the C lobe of TnC. In contrast, T142 lies within the key inhibitory region that switches from actin-tropomyosin to TnC binding as part of the activation process. Accumulating information from structural and biochemical in vitro and in vivo studies suggests that PKC phosphorylation at S41, S43, and T142 opposes effects produced by phosphorylation at the N-terminal extension residues S22 and S23 on relaxation kinetics.7,21 In addition to the PKC sites S41 and 43, sites T50, S76, and T77 lie within the IT-arm domain important for stabilizing cTnI on the actin filament. The most C-terminal region of cTnI contains a second actin-binding site that, although not directly involved in inhibiting the acto-myosin interaction, may facilitate the transition to the activated state.29,30 Most of the C-terminal domain was not mapped in the crystal structure of troponin, suggesting that it is highly mobile. Recent studies indicate that this domain of cTnI influences the azimuthal position of troponin on tropomyosin and that changes in this region (193–209) resulting from mutation or proteolysis during ischemia/reperfusion or stretch injury impairs muscle activation or relaxation, respectively.31,35–37
Sites Within the N-Terminal Extension of cTnI
The data from the MRM assays confirm previous studies that have reported a significant reduction in basal cTnI phosphorylation at S22/S23 in human HF,3,15–17 most likely reflecting the downregulation of β-adrenergic receptors and desensitization of signaling in the PKA pathway with HF. Interestingly, our data show that phosphorylation of S23 predominates over S22 or S22/S23 in donors and patient samples. In agreement with this finding, several in vitro studies using synthetic peptides38 reported different phosphorylation kinetics between S22 and S23 residues by PKA, with phosphorylation of S23 occurring 10-fold faster than that of S22. Zhang and colleagues39 also found that the murine equivalent of S23 was phosphorylated several-fold more rapidly than S22 by PKA. In contrast, a top-down MS analysis of human cTnI10,17 isolated from human hearts revealed that S22 rather than S23 was the only monophosphorylated form and that S23 was found only in diphosphorylated cTnI. Taken together, these studies suggest potential differences in the biological status of individuals and may reflect alterations in turnover rate of the phosphorylation/dephosphorylation for S22 and S23 in vivo.
There are 3 novel phosphorylatable residues in the N terminus of cTnI: Y25, S4, and S5. Y25 is of particular interest because it is the only Tyr residue modified, indicating that a novel kinase signaling pathway may regulate TnI function. Y25 is one of the most highly phosphorylated residues in control hearts. Its close proximity to the well-studied S22/S23 phosphorylatable residues raises the intriguing possibility of synergy between these sites and that the phosphorylation state of 1 site could influence the degree of modification of the other sites. The other 2 novel phosphorylatable (S4/S5) residues are located at the extreme N terminus of cTnI, a region unique to cardiac isoform and not present in either slow or fast skeletal TnI. A series of sequential N-terminal deletion mutants suggest that residues 1 through 15 play a role, albeit minor, in transmitting the phosphorylation signal to other myofilament proteins but are not directly involved in binding to cTnC.40 Potentially, S4 and S5 phosphorylation could enhance the interaction of the N terminus and the internal inhibitory region of cTnI that occurs specifically with phosphorylation30 of S22/S23. Interestingly, the S4/S5 diphosphorylated form is found exclusively in donor myocardium.
PKC Sites of cTnI
Our data demonstrate, for the first time, increased phosphorylation of the PKC sites S41, S43 (in IDCM only), and T142 of cTnI in human HF and a preference for the phosphorylation of S41 over S43. As debated by Solaro and van der Velden,2 the functional role of these sites in human heart and HF is not clear. However, in vitro and mouse mutant data (the Table) suggest that increased phosphorylation at these sites would be expected to slow the kinetics of contraction, to decrease Ca2+ sensitivity, and potentially to depress both relaxation and contractility in vivo.
Although T142 is a substrate of PKC in vitro, phosphorylation of this site has not been documented previously in the human heart. Strikingly, the extent of phosphorylation of T142 and the large change observed in ISHD and IDCM compared with donor hearts suggest that phosphorylation at this site could have the largest functional impact among the PKC sites and those altered in HF. A transgenic mouse model with a pseudophosphorylated mutant of all 3 PKC sites (S43E, S45E, and T144E, equivalent to S41, S43, and T142 in human) resulted in depressed contractility and relaxation and decreased force generation in cardiac muscle even though the mutant is expressed at very low levels.28 These authors used computational analysis to propose that the effect of mimicking enhanced phosphorylation at these PKC sites in this model is related to delayed entry into the cross-bridge cycle and persistence in the activated state. Thus, markedly enhanced phosphorylation at this site is likely to have significant impact in the human heart.
Phosphorylation Sites in the Near–Amino Terminal Domain
Phosphorylated sites in this region (T50, S76, and T77) could affect the interaction with the C lobe of cTnC and C-terminal regions of cTnT.21,30 S41 and S43, when phosphorylated, reduce acto-S1-ATPase activity.9,30 Whereas prior top-down analysis was unable to differentiate S76 and T77 phosphorylation,10,17 the present study was able to define that both S76 and T77 are phosphorylated. The extent of monophosphorylation and diphosphorylation of S76 and T77 is increased in both forms of HF studied here.
Phosphorylation of Sites at the C Terminus of cTnI
Three phosphorylation sites (S165, T180, S198) are located within the C-terminal region of cTnI. The domain includes residues 152 through 199 and is essential for full inhibitory activity and Ca2+ sensitivity of myofibrillar ATPase activity.32 The importance of this region is suggested by the fact that mutations in cTnI S165 and S198 cause hypertrophic cardiomyopathy.33 MRM data indicate that T180 is more highly phosphorylated than the other 2 residues in this region; however, phosphorylation of all 3 sites is increased in disease. S165 in human cTnI is phosphorylated by PKA in vitro,14 but our report is the first to show its phosphorylation in vivo. S165 is in the C-terminal region of cTnI that contains the Ca2+-sensitive TnC and actin-binding sites and influences the affinity of the cTnI inhibitory region and TnC.30 In vitro phosphorylation of S165 reduces the affinity of cTnI for TnC.14 S165 is close to A163, which is a His in the skeletal isoforms. This residue has been implicated in the regulation of the acidosis responsiveness conferred by cTnI.32,34 Mutation of the Ala to His at this position (163 in human or 164 in mice) protects against the deleterious effects of acidosis in a murine model of myocardial infarction.34
Residue S198 is localized within the second actin-binding site30 implicated in the structural reorganization associated with Ca2+ activation that is called the fly-casting mechanism.41 We and others have shown that the truncation of the C terminus occurs in experimental global ischemia, ischemia/reperfusion, volume overload, and ischemic human myocardium.31,35–37 In vivo and in vitro studies showed that truncation resulted in decreased maximal force activation, a divergent increase in maximum Ca2+-activated actin-tropomyosin–myosin S1 ATPase activity, and a structural change in the location of actin-tropomyosin filament.35,37 Phosphorylation of S198 could protect against proteolysis, as previously suggested,31 which would reduce these potential negative functional effects of C-terminal proteolysis. Thus, we expect functional and structural alterations resulting from the phosphorylation on 1 or more of the C-terminal region residues of cTnI.
Conclusions
We developed 30 peptide MRM assays to monitor the multiple phosphorylatable residues of cTnI and provided the first site-specific quantification of the level of phosphorylation of each site in human diseased and control samples. Numerous functional studies have indicated that the HF-associated changes in phosphorylation, including reduced phosphorylation at the S22/S23 sites and enhanced phosphorylation at S41, S43, and T142, are likely to negatively affect function. These effects include impaired contractile power, impaired relaxation, and impaired responses to elevated heart rate. In addition, there may be functional implications of altered phosphorylation status of several residues not previously known to be phosphorylated. Overall, with HF, there is a shift in phosphorylation involving decreases in novel residues in the N-terminal extension (S4, S5, S4/S5, Y25) and increases at others in the near–N-terminal region (S76, T77, S76/T77) and within the C-terminal domains (S165, T180, S198). Prior experiments in vitro and in vivo suggest that any disturbance in the balance of phosphorylation of cardiac proteins may lead to substantial functional consequences in heart disease, but it will be residue dependent. This implies that selectivity of various kinases/phosphatases for each residue will affect function. It is of interest that 3 novel sites located in the C-terminus were dephosphorylated on CRT, reversing the elevation in phosphorylation induced in HFdys. This suggests their potential clinical importance. Finally, this study may spur the development of novel biomarkers. Because cTnI is a valuable diagnostic marker for myocardial infarction, measurement of modified forms of circulating cTnI may provide a sensitive assay for the functional status of the myocardium.
Supplementary Material
CLINICAL PERSPECTIVE.
Phosphorylation of cardiac troponin I, a myofilament protein, regulates myocyte function. Using mass spectrometry, we established that human cardiac troponin I is more extensively phosphorylated than previously reported, with 14 modifiable amino acid residues located throughout its entire length. Six novel phosphorylated amino acid residues were identified in functionally important regions of cardiac troponin I. Furthermore, sensitive mass spectrometry–based assays were developed to simultaneously quantify the extent of phosphorylation of each modifiable residue in control and human failing myocardium. Along with the heart failure–related dephosphorylation of known protein kinase A–targeted sites, we found increased phosphorylation of protein kinase C sites and several of the newly identified sites found in the C terminus of cardiac troponin I. In a canine model of heart failure with dyssynchrony, these C-terminal sites were hyperphosphorylated, and this change shifted back toward normal with cardiac resynchronization. These findings suggest correlation of phosphorylation of cardiac troponin I with functional status and potential utility as a tissue biomarker and therapeutic target.
Acknowledgments
Sources of Funding
This work was supported by the National Heart Lung and Blood Institute: Proteomic Initiative contracts NHLBI-HV-10-05(2) (Dr Van Eyk) and HHSN268201000032C (Drs Van Eyk, Murphy, and Kass), P01HL081427 (Dr Van Eyk), P01HL77189-01 (Drs Kass and Van Eyk), and R01 HL63038 (Dr Murphy); Johns Hopkins Clinical Translational Science Award; and American Heart Association Postdoctoral Fellowships 10POST4000001 (Dr Zhang) and 11POST7210031 (Dr Kirk).
Footnotes
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/lookup/suppl/doi:10.1161/CIRCULATIONAHA.112.096388/-/DC1.
Disclosures
Drs Zhang, Murphy, and Van Eyk have a patent application pending on “Novel Phosphorylation of Cardiac Troponin I as a Monitor for Cardiac Injury.” The other authors report no conflicts.
References
- 1.Murphy AM. Heart failure, myocardial stunning, and troponin: a key regulator of the cardiac myofilament. Congest Heart Fail. 2006;12:32–38. doi: 10.1111/j.1527-5299.2006.04320.x. [DOI] [PubMed] [Google Scholar]
- 2.Solaro RJ, van der Velden J. Why does troponin I have so many phosphorylation sites? Fact and fancy. J Mol Cell Cardiol. 2010;48:810–816. doi: 10.1016/j.yjmcc.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zakhary DR, Moravec CS, Stewart RW, Bond M. Protein kinase A (PKA)-dependent troponin-I phosphorylation and PKA regulatory subunits are decreased in human dilated cardiomyopathy. Circulation. 1999;99:505–510. doi: 10.1161/01.cir.99.4.505. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T, Yang X, Walker LA, Van Breemen RB, Solaro RJ. A non-equilibrium isoelectric focusing method to determine states of phosphorylation of cardiac troponin I: identification of Ser-23 and Ser-24 as significant sites of phosphorylation by protein kinase C. J Mol Cell Cardiol. 2005;38:213–218. doi: 10.1016/j.yjmcc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Haworth RS, Cuello F, Herron TJ, Franzen G, Kentish JC, Gautel M, Avkiran M. Protein kinase D is a novel mediator of cardiac troponin I phosphorylation and regulates myofilament function. Circ Res. 2004;95:1091–1099. doi: 10.1161/01.RES.0000149299.34793.3c. [DOI] [PubMed] [Google Scholar]
- 6.Layland J, Li J-M, Shah AM. Role of cyclic GMP-dependent protein kinase in the contractile response to exogenous nitric oxide in rat cardiac myocytes. J Physiol. 2002;540:457–467. doi: 10.1113/jphysiol.2001.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramirez-Correa GA, Cortassa S, Stanley B, Gao WD, Murphy AM. Calcium sensitivity, force frequency relationship and cardiac troponin I: critical role of PKA and PKC phosphorylation sites. J Mol Cell Cardiol. 2010;48:943–953. doi: 10.1016/j.yjmcc.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You B, Yan G, Zhang Z, Yan L, Li J, Ge Q, Jin JP, Sun J. Phosphorylation of cardiac troponin I by mammalian sterile 20-like kinase 1. Biochem J. 2009;418:93–101. doi: 10.1042/BJ20081340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkart EM, Sumandea MP, Kobayashi T, Nili M, Martin AF, Homsher E, Solaro RJ. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J Biol Chem. 2003;278:11265–11272. doi: 10.1074/jbc.M210712200. [DOI] [PubMed] [Google Scholar]
- 10.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Unraveling molecular complexity of phosphorylated human cardiac troponin I by top down electron capture dissociation/electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2008;7:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Grant JE, Doede CM, Sadayappan S, Robbins J, Walker JW. PKC-βII sensitizes cardiac myofilaments to Ca2+ by phosphorylating troponin I on threonine-144. J Mol Cell Cardiol. 2006;41:823–833. doi: 10.1016/j.yjmcc.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Buscemi N, Foster DB, Neverova I, Van Eyk JE. p21-Activated kinase increases the calcium sensitivity of rat triton-skinned cardiac muscle fiber bundles via a mechanism potentially involving novel phosphorylation of troponin I. Circ Res. 2002;91:509–616. doi: 10.1161/01.res.0000035246.27856.53. [DOI] [PubMed] [Google Scholar]
- 13.Ke Y, Wang L, Pyle WG, de Tombe PP, Solaro RJ. Intracellular localization and functional effects of P21-activated kinase-1 (Pak1) in cardiac myocytes. Circ Res. 2004;94:194–200. doi: 10.1161/01.RES.0000111522.02730.56. [DOI] [PubMed] [Google Scholar]
- 14.Ward DG, Ashton PR, Trayer HR, Trayer IP. Additional PKA phosphorylation sites in human cardiac troponin I. Eur J Biochem. 2001;268:179–185. doi: 10.1046/j.1432-1327.2001.01871.x. [DOI] [PubMed] [Google Scholar]
- 15.Bodor GS, Oakeley AE, Allen PD, Crimmins DL, Ladenson JH, Anderson PA. Troponin I phosphorylation in the normal and failing adult human heart. Circulation. 1997;96:1495–1500. doi: 10.1161/01.cir.96.5.1495. [DOI] [PubMed] [Google Scholar]
- 16.Kooij V, Saes M, Jaquet K, Zaremba R, Foster DB, Murphy AM, Dos Remedios C, van der Velden J, Stienen GJ. Effect of troponin I Ser23/24 phosphorylation on Ca2+-sensitivity in human myocardium depends on the phosphorylation background. J Mol Cell Cardiol. 2010;48:954–963. doi: 10.1016/j.yjmcc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Guy MJ, Norman HS, Chen YC, Xu Q, Dong X, Guner H, Wang S, Kohmoto T, Young KH, Moss RL, Ge Y. Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J Proteome Res. 2011;10:4054–4065. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Q, Schoenhoff F, Savage W, Zhang P, Van Eyk JE. Multiplex assays for biomarker research and clinical application: translational science coming of age. Proteom Clin Appl. 2010;4:271–284. doi: 10.1002/prca.200900217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spragg DD, Leclercq C, Loghmani M, Faris OP, Tunin RS, DiSilvestre D, McVeigh ER, Tomaselli GF, Kass DA. Regional alterations in protein expression in the dyssynchronous failing heart. Circulation. 2003;108:929–932. doi: 10.1161/01.CIR.0000088782.99568.CA. [DOI] [PubMed] [Google Scholar]
- 20.Li MX, Spyracopoulos L, Sykes BD. Binding of cardiac troponin-I147–163 induces a structural opening in human cardiac troponin-C. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 21.Solaro RJ, Kobayashi T. Protein phosphorylation and signal transduction in cardiac thin filaments. J Biol Chem. 2011;286:9935–9940. doi: 10.1074/jbc.R110.197731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howarth JW, Meller J, Solaro RJ, Trewhella J, Rosevear PR. Phosphorylation-dependent conformational transition of the cardiac specific N-extension of troponin I in cardiac troponin. J Mol Biol. 2007;373:706–722. doi: 10.1016/j.jmb.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 23.Takeda S, Yamashita A, Maeda K, Maéda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu Rev Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 25.Heller WT, Finley NL, Dong WJ, Timmins P, Cheung HC, Rosevear PR, Trewhella J. Small-angle neutron scattering with contrast variation reveals spatial relationships between the three subunits in the ternary cardiac troponin complex and the effects of troponin I phosphorylation. Biochemistry. 2003;42:7790–7800. doi: 10.1021/bi0341509. [DOI] [PubMed] [Google Scholar]
- 26.Ward DG, Brewer SM, Calvert MJ, Gallon CE, Gao Y, Trayer IP. Characterization of the interaction between the N-terminal extension of human cardiac troponin I and troponin C. Biochemistry. 2004;43:4020–4027. doi: 10.1021/bi036128l. [DOI] [PubMed] [Google Scholar]
- 27.Van der Velden J, de Jong JW, Owen VJ, Burton PB, Stienen GJ. Effect of protein kinase A on calcium sensitivity of force and its sarcomere length dependence in human cardiomyocytes. Cardiovasc Res. 2000;46:487–495. doi: 10.1016/s0008-6363(00)00050-x. [DOI] [PubMed] [Google Scholar]
- 28.Kirk JA, MacGowan GA, Evans C, Smith SH, Warren CM, Mamidi R, Chandra M, Stewart AF, Solaro RJ, Shroff SG. Left ventricular and myocardial function in mice expressing constitutively pseudophosphorylated cardiac troponin I. Circ Res. 2009;105:1232–1239. doi: 10.1161/CIRCRESAHA.109.205427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T, Solaro RJ. Increased Ca2+ affinity of cardiac thin filaments reconstituted with cardiomyopathy-related mutant cardiac troponin I. J Biol Chem. 2006;281:13471–13477. doi: 10.1074/jbc.M509561200. [DOI] [PubMed] [Google Scholar]
- 30.Tripet B, Van Eyk JE, Hodges RS. Mapping of a second actin-tropomyosin and a second troponin C binding site within the C terminus of troponin I, and their importance in the Ca2+-dependent regulation of muscle contraction. J Mol Biol. 1997;271:728–750. doi: 10.1006/jmbi.1997.1200. [DOI] [PubMed] [Google Scholar]
- 31.McDonough JL, Arrell DK, Van Eyk JE. Troponin I degradation and covalent complex formation accompanies myocardial ischemia/reperfusion injury. Circ Res. 1999;84:9–20. doi: 10.1161/01.res.84.1.9. [DOI] [PubMed] [Google Scholar]
- 32.Ball KL, Johnson MD, Solaro RJ. Isoform specific interactions of troponin I and troponin C determine pH sensitivity of myofibrillar Ca2+ activation. Biochemistry. 1994;33:8464–8471. doi: 10.1021/bi00194a010. [DOI] [PubMed] [Google Scholar]
- 33.Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–451. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- 34.Day SM, Westfall MV, Fomicheva EV, Hoyer K, Yasuda S, La Cross NC, D’Alecy LG, Ingwall JS, Metzger JM. Histidine button engineered into cardiac troponin I protects the ischemic and failing heart. Nat Med. 2006;12:181–189. doi: 10.1038/nm1346. [DOI] [PubMed] [Google Scholar]
- 35.Foster DB, Noguchi T, VanBuren P, Murphy AM, Van Eyk JE. C-terminal truncation of cardiac troponin I causes divergent effects on ATPase and force: implications for the pathophysiology of myocardial stunning. Circ Res. 2003;93:917–924. doi: 10.1161/01.RES.0000099889.35340.6F. [DOI] [PubMed] [Google Scholar]
- 36.Murphy AM, Kogler H, Georgakopoulos D, McDonough JL, Kass DA, Van Eyk JE, Marban E. Transgenic mouse model of stunned myocardium. Science. 2000;287:488–491. doi: 10.1126/science.287.5452.488. [DOI] [PubMed] [Google Scholar]
- 37.Feng J, Schaus BJ, Fallavollita JA, Lee TC, Canty JM., Jr Preload induces troponin I degradation independently of myocardial ischemia. Circulation. 2001;103:2035–2037. doi: 10.1161/01.cir.103.16.2035. [DOI] [PubMed] [Google Scholar]
- 38.Keane NE, Quirk PG, Gao Y, Patchell VB, Perry SV, Levine BA. The ordered phosphorylation of cardiac troponin I by the cAMP-dependent protein kinase–structural consequences and functional implications. Eur J Biochem. 1997;248:329–337. doi: 10.1111/j.1432-1033.1997.00329.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, Zhao J, Potter JD. Phosphorylation of both serine residues in cardiac troponin I is required to decrease the Ca2+ affinity of cardiac troponin C. J Biol Chem. 1995;270:30773–30780. doi: 10.1074/jbc.270.51.30773. [DOI] [PubMed] [Google Scholar]
- 40.Ward DG, Brewer SM, Gallon CE, Gao Y, Levine BA, Trayer IP. NMR and mutagenesis studies on the phosphorylation region of human cardiac troponin I. Biochemistry. 2004;43:5772–5781. doi: 10.1021/bi036310m. [DOI] [PubMed] [Google Scholar]
- 41.Hoffman RM, Blumenschein TM, Sykes BD. An interplay between protein disorder and structure confers the Ca2+ regulation of striated muscle. J Mol Biol. 2006;361:625–633. doi: 10.1016/j.jmb.2006.06.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.