Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) has been involved in endothelial cell dysfunction associated with various pathophysiological conditions. The intrinsic mechanism of PARP-1-mediated endothelial cell dysfunction could be related to PARP-1 overactivation, NAD+ consumption and ATP depletion. An alternative way could involve transcription regulation. By using high-density microarrays, we examined early tumor necrosis factor α (TNF-α)-stimulated gene expression profiles in PARP-1+/+ and PARP-1–/– murine heart endothelial cells. TNF-α modulated a significant number of genes in both cell types. We have identified a set of genes whose expression in response to TNF-α is modulated by PARP-1, whereas the expression of others is PARP-1-independent. Up-regulation of several genes involved in the inflammatory response is hampered in the absence of PARP-1. Moreover, NF-κB-dependent transcriptional activation is partially inhibited in PARP-1–/– compared to PARP-1+/+ cells. However, we found that PARP-1 might also silence transcription of several NF-κB target genes. Overall, our results show that PARP-1 is regulating the expression of genes by the endothelial cells both in a positive and a negative fashion, with the final effects depending on the gene. Individual studies of these genes are now necessary to clarify the intrinsic mechanism by which PARP-1 is controlling transcription and thereby finding out different therapeutic approaches involving PARP-1.
INTRODUCTION
Endothelial cell dysfunction is an important early-recurring phenomenon in virtually all forms of ischemia and reperfusion injury (1,2) and plays a critical role in uncontrolled inflammatory conditions such as sepsis and multiorgan dysfunction syndrome (3). Pro-inflammatory mediators released following insult initiate signaling pathways to the nucleus in the endothelial cells, mediated by NF-κB and other stress-responsive transcription factors which reprogram gene expression (4). The overall result is a dysregulation of endothelial function, leading to fluid leakage, transmigration of leukocytes across the endothelium, thrombosis, end organ damage, multiple organ dysfunction and often death (5). The cytokine tumor necrosis factor α (TNF-α) is one of the most important mediators of the inflammatory process, and a large number of genes have been identified that are responsive to TNF-α (6). Such genes include cytokines, transcription factors, adhesion molecules and structural proteins. TNF-α-dependent gene expression is mainly mediated by the transcription factor NF-κB. Under basal conditions, NF-κB is found in an inactive cytoplasmic form bound to the inhibitor IκB (7). Upon TNF-α signaling, IκB undergoes post-translational modification (phosphorylation and polyubiquitination) that leads to its degradation and dissociation from NF-κB. The released NF-κB is then translocated to the nucleus where it activates the transcription of genes carrying NF-κB binding sites in their promoters.
Recent evidence suggests that the enzyme poly(ADP-ribose) polymerase 1 (PARP-1) is involved in the endothelial dysfunction observed in various pathophysiological conditions such as reperfusion (8), endotoxic shock (9), diabetes (10) and aging (11). PARP-1 (EC 2.4.2.30) is a highly conserved nuclear zinc-finger DNA-binding protein (113 kDa). PARP-1 specifically detects DNA-strand breaks or nicks generated by different genotoxic agents (12) and, using NAD+ as a substrate, synthesizes and transfers ADP-ribose onto glutamic acid residues of acceptor proteins, including itself (automodification), histones, transcription factors and DNA repair proteins (13). In addition, it has been suggested that signals other than DNA lesions, including steroid hormones, stress and infection, may also activate PARP molecules at specific chromosome sites (14). Despite intense recent interest in the biochemical properties and signaling function of PARP-1, its physiological function remains under debate.
An important tool for the analysis of this issue has been the development of PARP-1 knockout mice (PARP-1–/–) (15–17). Although PARP-1-deficient mice are viable, they accumulate chromosomal abnormalities and are defective in DNA damage repair. Moreover, PARP-1–/– mice are protected against a variety of experimentally induced disorders with a clear inflammatory component (18). The underlying mechanism of endothelial cell dysfunction mediated by PARP-1 could be attributed to PARP-1 overactivation, with resulting NAD+ consumption and ATP depletion (10,19). However, an alternative way in which PARP-1 may influence endothelial cell function could be through its transcription regulation function (20,21). Thus, it has been demonstrated that PARP-1 is necessary for the induction of NF-κB-dependent gene expression after exposure to LPS, TNF-α or hydrogen peroxide (22,23). However, since translocation of NF-κB to the nucleus occurred in PARP-1–/– cells as it did in the wild-type (23), a new level of control of NF-κB must occur after its translocation to the nucleus, in which PARP-1 seems to play a crucial role (24). Hassa et al. (25) have suggested that PARP-1 is an essential and novel transcription coactivator for κB-dependent gene expression. Recently, it has also been shown that PARP-1 is required for the activation of other inflammation-related transcription factors such as AP-1, SP-1, Oct-1, YY-1 and STAT-1 (26,27). Thus, a large body of evidence has implicated PARP-1 in the regulation of transcriptional activity of eukaryotic genes (21). Indeed, PARP-1 might play a critical role as a signaling molecule, which controls the expression of multiple genes involved in the inflammatory response (18).
In the present study we use oligonucleotide microarrays analysis to elucidate the role of PARP-1 in the early gene expression of endothelial cell in response to TNF-α. This approach allows a comprehensive analysis of global gene expression in both PARP-1+/+ and PARP-1–/– cells under different activation conditions. Our results show that the expression of some genes by endothelial cells in response to TNF-α is modulated by PARP-1 in both a positive and a negative fashion, whereas the expression of other genes is PARP-1-independent. Likewise, the expression of several NF-κB-dependent genes is dramatically inhibited in the absence of PARP-1 while the expression of other NF-κB-dependent genes is up-regulated in PARP-1-deficient cells suggesting multiple regulatory processes that should be studied in an individual way.
MATERIALS AND METHODS
Antibodies and chemicals
The following monoclonal antibodies were used: rat anti-mouse CD102 (3C4), CD31 (MEC 13.3), CD105 (MJ7/18), CD62E (10E9.6), CD106 (MVCAM.A) and CD54 (3E2), all from BD Pharmingen (San Diego, CA). Fluorescein isothiocyanate-conjugated goat anti-rat IgG (H+L) antibody from Caltag (Burlingame, CA). Horseradish peroxidase-conjugated goat anti-rat IgG from Sigma (St Louis, MO). Proteasome inhibitor MG-132 (Z-Leu-Leu-Leu-al) was purchased from Sigma.
Cell isolation and culture
Murine heart endothelial cells (MHEC) were isolated from female PARP-1 knockout (PARP-1–/–) and their wild-type (PARP-1+/+) littermates (strain 129/Sv X C57BL/6) mice (9–10 weeks old) (kindly provided by Dr de Murcia, Strasbourg, France) (16), by collagenase treatment and cell sorting of ICAM2-positive cells. Briefly, the hearts were washed extensively with cold phosphate-buffered saline (PBS) (Biowhittaker, Verviers, Belgium). Diced tissue was incubated in PBS supplemented with 0.5 mg/ml of collagenase (Boehringer, Mannheim, Germany) for 1 h at 37°C. After washing twice in PBS, cells were incubated for a further 10 min in 0.25% trypsin/0.04% EDTA solution (Life Technologies, Inc., Grand Island, NY), washed twice in PBS supplement with 2.5% heat inactivated fetal calf serum (FCS) (Life Technologies), and incubated for 30 min at 4°C with a rat anti-mouse CD31 and a rat anti-mouse CD102 monoclonal antibodies. After washing twice with cold PBS, cells were incubated with sheep anti-rat Ig-conjugated microbeads (107 beads/ml) (Dynal, Oslo, Norway) for 15 min at 4°C. The magnetically labeled cells were collected with a Dynal magnetic particle concentrator. Positively selected cells attached to the Dynabeads were washed five times in PBS. After detachment, cells were resuspended in RPMI 1640 medium (Life Technologies) supplemented with 20% FCS, 2 mM l-glutamine (Life Technologies), 1% penicillin/streptomycin (Life Technologies), 1 mM sodium pyruvate (Sigma), 20 mM HEPES (Sigma), 1% non-essential amino acids (Sigma), 50 mM 2-mercaptoethanol (Sigma), 100 µg/ml endothelial cell growth supplement (Beckton Dickinson, Mountain View, CA), 12 U/ml heparin (Rovi Laboratories, Madrid, Spain) and plated onto flasks coated with gelatin (100 µg/ml) (Sigma). Cells were split upon confluence by tripsinization (0.25% trypsin/0.04% EDTA), washed and incubated with rat anti-mouse CD102 mAb for 30 min at 4°C. After washing twice with cold PBS, cells were incubated in the same buffer containing fluorescein-conjugated goat anti-rat IgG (H+L) antibody. Following 30 min incubation at 4°C in the dark, the samples were again washed twice with cold PBS, and resuspended in 500 µl of RPMI 1640 medium. Positive cells were collected by fluorescence activated cell sorting (FACS) using a MoFlo® cell sorter (Cytomation Inc., Fort Collins, CO) equipped with an Argon-ion blue laser (excitation 488 nm) and a Red Diode Laser (excitation 635 nm). Forward and side light scatter and specific fluorescence were used to establish sort regions by using SummitTM software (Cytomation) in a 1–2 drop single cell mode. Cells were plated, and subcultured as described above. In all cases, >99% of the cells stained positively for cell surface CD102, displayed typical cobblestone morphology and were positive for other typical endothelial cell markers as CD105, CD31, CD54 and CD106 by flow cytometry analysis in a FACS cytofluorimeter (Becton Dickinson) using Cell Quest software (Becton Dickinson) (data not shown).
Microarray
Primary MHECs derived from PARP-1+/+ and PARP-1–/– mice were grown under identical conditions to 80–90% confluency. After treatment with murine recombinant TNF-α (20 ng/ml) (Sigma) for 2 h, total RNA was isolated from cells by using a Rneasy Total RNA Isolation kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. Total RNA (8 µg) was subjected to reverse transcription with Superscript (Life Technologies), using a T77-(dT)24 primer containing a T7 RNA polymerase promoter site. Biotinylated complementary RNA was made from 1 µg of cDNA and then fragmented to ∼50–100 nt, following Affymetrix’s instructions. Fifteen micrograms of the in vitro transcripts with appropriate controls and spikes were hybridized for 16 h at 45°C with constant rotation at 60 r.p.m. to an Affymetrix U74Av2 microarray (Affymetrix, Santa Clara, CA), which contain probes for 12 488 known genes and expressed sequences tags (EST). Chips were washed and stained by using the EukGE-WS2v4 protocol on an Affymetrix fluidics station. The stain included streptavidin–phycoerythrin (10 µg/ml) (Molecular Probes, Eugene, OR) and biotinylated goat anti-streptavidin (3 µg/ml) (Vector Laboratories, Burlingame, CA). Chips were scanned with Agilent Gene Array Scanner and visualized and analyzed using Affymetrix software (Affymetrix Microarray Suite 5.0; Affymetrix Data Mining Tool 3.0; Affymetrix MicroDB 3.0). Expression values of transcripts were normalized, according to the total intensity on the chip. Only those differences in RNA abundance that were reproducible in independent experiments with different batches of cells and represented a change of 2-fold or greater were considered.
Quantitative RT–PCR
Total RNA was prepared from resting or stimulated MHEC using Rneasy Total RNA Isolation kit. cDNA was synthesized using oligo-(dT)16 primer and the GeneAmp RNA-PCR kit (Roche, Foster City, CA). Quantitative real-time PCR was performed in a Light-Cycler (Roche) using a Light Cycler-FastStart DNA Master SYBR Green I kit (Roche). Specific primers to the different genes analyzed are listed in Table 1. A threshold was set in the linear part of the amplification curve (fluorescence = f[cycle number]), and the number of cycles needed to reach it was calculated for every gene. Melting curves and agarose gel electrophoresis established the purity of the amplified band. Each gene was normalized to the housekeeping gene β-actin before fold change was calculated. Results are expressed as the relative fold increase or decrease of the stimulated over the resting cells.
Table 1. Primer sets used for quantitative real-time PCR analysis.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Cxcl2 | TATCGATTCGCTAATTCACTG | AAATGATGTGAACATGCACAC |
| Cxcl1 | TGCTAAAAGGTGTCCCCAAG | GCAGAACTGAACTACCATCG |
| Cxcl5 | TCATGCAGAAACCTGTGTAG | ATACGTAGTGGCCCAAATAG |
| Ccl7 | CAAAGAAGGGCATGGAAGTCTG | ATCCCTTAGGACCGTGATCAAC |
| Ccl5 | TGAAGATCTCTGCAGCTGCCC | GATTGGAGCACTTGCTGCTGG |
| Cx3cl1 | AGTGACTGCTGAAGCAAGGAGC | TCGACAAAGGGTTGGCTTCCTC |
| Cxcl10 | AGTTCTAAGTTTACCTGAGCTC | ACAAGTTCTTCCATATACAATGC |
| Tnfaip3 | CTGCATGTATTTTGGGACTCC | ATGGAATCTCTGGTTCTGAGC |
| Gadd45b | GTTCAGAAGATGCAGGCGGTG | AGCAGAACGACTGGATCAGGG |
| Slfn2 | GGAAAATGCAAAGGCTGCTGGG | GATTGCTCCACCTCCCGAATTC |
| Areg | CCACATCCCCAGCGGTTCCAG | GGATCCAGACAGACCTCCTTC |
| Csf2 | CAACTCCGGAAACGGACTGTG | GCTGTGCCACATCTCTTGGTC |
| Itga5 | AGGAAGTCTGCAGCTAAGAGC | GGTCTGGCTCCATTCTCTTTC |
| Gem | CTCACTGCATACTTATATCCC | AGCTCCTAAGTCTACAATCTC |
| Actin | TCCCTGGAGAAGAGCTACGA | AGGAAGGAAGGCTGGAAGAG |
Transient transfection and luciferase activity
For transient-transfection experiments, MHEC were plated into 24-well plates (2 × 104 cells/well) the day before transfection. Cells were transfected by the liposome-mediated gene transfer method as previously described (28). Briefly, 1.5 µl of Lipofectamine (Life Technologies) was mixed in 48.5 µl of Opti-MEM 1 (Life Technologies) with 0.475 µg of a firefly luciferase reporter plasmid under the control of a 3 × κB consensus site from the HIV enhancer (29) or the firefly luciferase pGL3-Control vector (Promega, Madison, WI), and 0.025 µg (ratio 20:1) of the Renilla luciferase expression vector pRLCMV (Promega) as an internal control to normalize the values obtained with the firefly luciferase construct. Mixture was incubated for 30 min at room temperature to allow the formation of DNA–lipid complexes. The mixture was then diluted in 200 µl of Opti-MEM 1 and added to the cell cultures. After 6 h in culture, 1 ml of growth medium was added to the transfection mixture. Next day, medium was replaced by a fresh one. Cells were cultured for 72 h, and lysed with passive lysis buffer (Promega). Firefly and Renilla luciferase activity was measured by using the Dual luciferase assay kit (Promega), as specified by the manufacturer, to discriminate the activity of the two types of luciferases, in a Optocomp I luminometer (MGM Instruments, Inc., Hamden, CT).
ELISA
To determine the amount of secreted chemokines, cells were seeded in 96-well plates at subclonfluent density overnight. The enzyme-linked immunosorbent assays (ELISA) for MIP-2 and Cxcl10 were carried out using supernatants according to the manufacturer’s protocol (Quantikine® mouse MIP-2 and Cxcl10 immunoassay, R&D Systems, Minneapolis, MN). Ccl7 production was measured by a sandwich ELISA in which 500 ng of goat polyclonal antibody to mouse Ccl7 (Abcam, Cambridge, UK) was coated into a microtiter plate overnight at 4°C. The plates were blocked with PBS containing 1% bovine serum albumin (BSA) for 1 h at 22°C. Then, cell-free supernatants from untreated or TNF-α-treated endothelial cells were added and incubated for 1 h at 22°C. After washing, biotinylated goat polyclonal antibody to mouse Ccl7 (R&D Systems) was added for 1 h at 22°C. For developing the ELISA, after washing, horseradish peroxidase-conjugated streptavidin (Promega) was added at the final step and incubated for 30 min at 22°C. Color was developed using ABTS (Sigma) substrate, and the absorbance was measured at 405 nm. Recombinant mouse Ccl7 protein (R&D Systems) was used as standard for quantification.
E-selectin and VCAM-1 surface expression on MHEC was quantified as previously described (30). Briefly, MHEC were seeded in 96-well plates and culture until confluence. Cells were then washed in PBS and fixed in 0.2% glutaraldehyde (Sigma) for 40 min at room temperature. Plates were saturated overnight in PBS containing 1% BSA. After three washes, the cells were incubated with rat anti-mouse E-selectin or VCAM-1 monoclonal antibodies at 37°C for 1 h. After three more washes, the cells were incubated with horseradish peroxidase-conjugated goat anti-rat IgG for 1 h at 37°C. The reaction was developed using ABTS substrate, and the absorbance was measured at 405 nm.
Adhesion assay
For cell adhesion, Concanavalin A-stimulated murine splenocytes cultured in the presence of 500 U/ml of IL-2 (gift of Hoffmann-La Roche, Nutley, NJ) for 3 days were labeled with the fluorescent dye BCECF-AM (Molecular probes, The Netherlands), and added in RPMI medium to a monolayer of resting or TNF-α-activated endothelial cells from both genotypes. After incubation for 20 min at 37°C, unbound cells were removed by three washes with RPMI medium, and adhered cells were quantified using a fluorescence analyzer (BMG, Durham, NC). In all experiments a control plate using titrated numbers of labeled splenocytes was set up to establish a linear relationship between cell number and the mean fluorescence.
RESULTS
Transcriptional programs in PARP-1+/+ versus PARP-1–/– endothelial cells in response to TNF-α
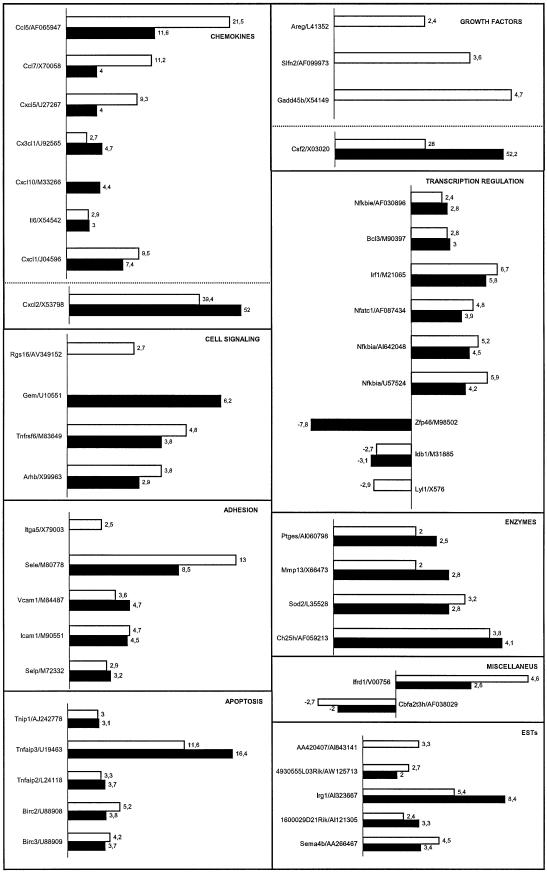
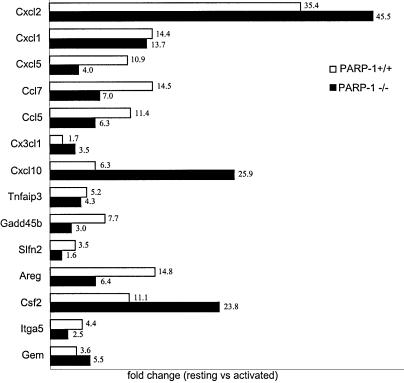
The gene expression profiles induced by TNF-α in primary MHEC derived from PARP-1-wild-type and PARP-1-deficient mice were analyzed using oligonucleotide microarray technology. Primary cells (passage 3) grown under identical conditions to ∼80% confluency were stimulated with 20 ng/ml recombinant mouse TNF-α for 2 h. Total RNA prepared from resting or TNF-α-stimulated cells were further purified, and labeled cRNA was prepared for DNA microarray analyses using Affymetrix oligonucleotide chips (murine genome U74A) containing probes for 12 488 known mouse genes or ESTs. Only those changes in RNA abundance that were reproducible in independent experiments using different batches of endothelial cells were considered. Genes that appeared to be differentially expressed by a factor >2-fold between untreated and TNF-α-stimulated cells are represented in Figure 1, classified into a number of functional categories. Those include chemokines, adhesion molecules, growth factors, cell signaling molecules, transcription factors, apoptosis related molecules and metabolic mediators. In addition to these differentially expressed known full-length genes, we also identified a number of differentially expressed ESTs (Fig. 1). The identification of full cDNA encoding for these unknown genes as well as their biological relevance will be addressed in additional studies. TNF-α modulated a significant number of early-response genes (0.34%) in PARP-1-expressing endothelial cells, up-regulating 40 and concomitantly down-regulating three transcripts, while in PARP-1-deficient endothelial cells 39 of genes (0.32%) were modulated, up-regulating 36 genes and down-regulating three transcripts (Fig. 1). Expression pattern of these genes was similar under basic conditions in both cell types. Expression of 28 early-response genes to TNF-α was similar in PARP-1+/+ and PARP-1–/– endothelial cells (fold change <1.5). However, we found 18 early-response genes to TNF-α whose expression seems to be PARP-1-dependent (Table 2). To validate the array data, we also evaluated expression of several of these genes by quantitative real-time PCR. Although there were differences in the fold-change absolute values detected by the two methods, in every case, PCR results correlated well with the differential gene expression data produced using Affymetrix GeneChips (Fig. 2).
Figure 1.
Differential gene expression patterns in PARP-1+/+ and PARP-1–/– MHECs in response to TNF-α. Gene expression profiles were generated using an Affymetrix U74Av2 microarray and analyzed using GeneChip software. The x-axis represents fold-change of activated versus resting cells for both PARP-1+/+ (white bars) and PARP-1–/– (black bars) cells. TNF-α-responsive genes have been clustered according to function, and the corresponding GenBank accession numbers are indicated. Genes with no change >2-fold between untreated and TNF-α-stimulated in PARP-1+/+ cells (Cxcl10, Gem and Zfp46) or in PARP-1–/– cells (Rgs16, Itga5, Areg, Slfn2, Gadd45b, Lyl1 and AA420407) are not indicated. Results represent the mean value of two independent experiments using different batches of heart endothelial cells.
Table 2. Silencing and enhancing transcription of early-response genes to TNF-α by PARP-1 in MHECsa.
| Positive regulation | Negative regulation | ||
|---|---|---|---|
| Gene | Fold change | Gene | Fold change |
| Ccl5 | 1.8 | Cx3cl1 | 1.7 |
| Cxcl5 | 2.3 | Cxcl10 | 4.4 |
| Ccl7 | 2.8 | Csf2 | 1.9 |
| E-selectin | 1.5 | Gem | 6.2 |
| Itga5 | 2.5 | Lyl1 | 2.9 |
| Gadd45b | 4.7 | Irg1 | 1.6 |
| Slfn2 | 3.6 | ||
| Areg | 2.4 | ||
| Rgs 16 | 2.7 | ||
| Ifrd1 | 1.8 | ||
| EST (AA420407) | 3.3 | ||
| Zfp46 | 7.8 | ||
aGenes were classified as PARP-1-dependent genes when their expression in response to TNF-α was changed by a factor of at least 1.5 between PARP-1+/+ and PARP-1–/– cells in the microarrays analysis. Fold change expressed the ratio of gene expression in PARP-1+/+ over PARP-1–/– cells (positive regulation column) or the ratio of gene expression in PARP-1–/– over PARP-1+/+ cells (negative regulation column).
Figure 2.
Real-time PCR of select genes identified as differentially expressed by GeneChip analysis. Samples were normalized according to actin expression level. The x-axis represents fold-change of activated versus resting cells for both PARP-1+/+ (white bars) and PARP-1–/– (black bars) cells. Results represent the mean value of two independent experiments using different batches of heart endothelial cells.
Role of PARP-1 in endothelial cells gene expression of molecules involved in inflammation after TNF-α treatment
A subset of genes (∼40%) with altered expression by MHEC in response to TNF-α encode for chemokines, growth factors and adhesion molecules which are major regulators of mononuclear and polymorphonuclear cell trafficking across the endothelium in various forms of inflammation. All genes in the chemokines group were upregulated (or unmodified) after stimulation in both PARP-1-wild-type and PARP-1-deficient endothelial cells (Fig. 1). Transcription of Ccl5, Ccl7 and Cxcl5 genes is markedly induced in PARP-1+/+ endothelial cells in response to TNF-α but their upregulation was inhibited in PARP-1–/– endothelial cells (Fig. 1). However, macrophage inflammatory protein 2 (MIP-2 or Cxcl2), Cxcl1 and IL6 genes were similarly up-regulated in both cell types. Cx3cl1 chemokine gene was slightly up-regulated in PARP-1+/+ and higher in PARP-1–/–, while Cxcl10 was up-regulated only in PARP-1-deficient cells in response to TNF-α. Genes included in the growth factors group were also upregulated after stimulation in PARP-1-wild-type cells, while only one was up-regulated in PARP-1-deficient endothelial cells (Fig. 1). The up-regulation of macrophage colony stimulating factor 2 gene (Csf2) in PARP-1–/– cells was 2-fold higher than observed in wild-type cells (Fig. 1). Differences in the expression of several genes included in both functional groups were verified by quantitative real-time PCR (Fig. 2).
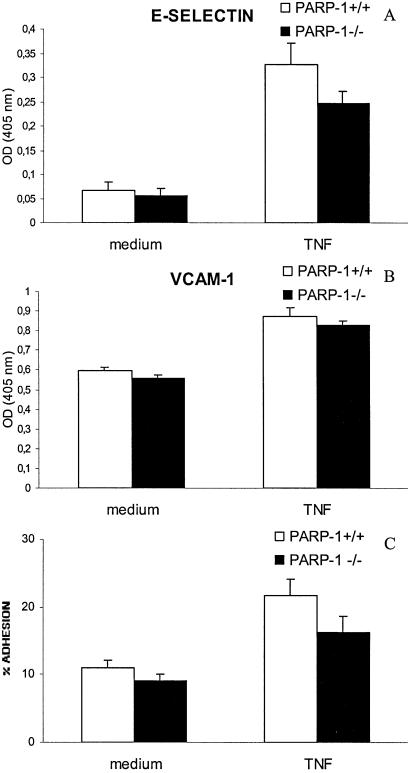
A fundamental and early event in inflammation is adhesion of leukocytes to the endothelium. This is mediated by binding of leukocytes to endothelial cells adhesion molecules such as E-selectin (Sele), vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1). Expression of E-selectin gene was highly up-regulated in PARP-1+/+ cells and to a lesser extent in PARP-1–/– cells in response to 2 h TNF-α treatment. On the other hand, similar level of up-regulation was found for ICAM-1, VCAM-1 and Selp genes in both PARP-1-wild-type and PARP-1-deficient endothelial cells at this early stage of activation. The fibronectin 2 receptor gene (Itga5) was up-regulated in PARP-1+/+ but not in PARP-1–/– cells (Fig. 1). Expression of several adhesion molecules genes (E-selectin and VCAM-1) in response to TNF-α was also verified at protein level by ELISA in both cell types (Fig. 3A and B). Moreover, lymphocyte adhesion to a monolayer of TNF-α-activated endothelial cells was higher in PARP-1+/+ than PARP-1–/– endothelial cells (Fig. 3C).
Figure 3.
Cell surface expression of E-selectin (A) and VCAM-1 (B) in both PARP-1+/+ (white bars) and PARP-1–/– (black bars) endothelial cells in response to 6 h TNF-α treatment (20 ng/ml). ELISAs were performed on fixed cells. Absorbencies measured at 405 nm are plotted in the y-axis. A representative of three experiments is shown. (C) Cell adhesion of activated murine splenocytes to TNF-α-activated PARP-1+/+ (white bars) and PARP-1–/– (black bars) endothelial cells. Error bars indicate standard error.
Silencing and enhancing transcription of NF-κB-target genes by PARP-1 in MHECs
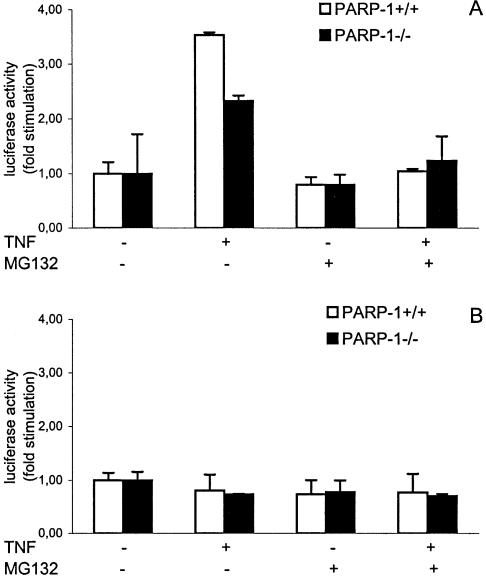
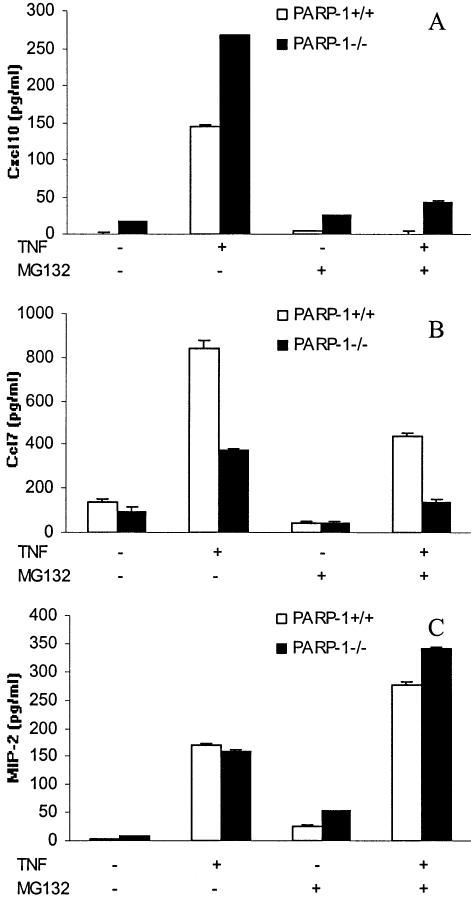
NF-κB pathway plays a critical role in the response to TNF-α by endothelial cells (31). TNF-α signaling led to proteolytic degradation of IκBα by the 26S proteasome complex, which is necessary for NF-κB nuclear translocation and therefore NF-κB-dependent transcription activation (32). The transcriptional status of NF-κB was examined in transiently transfected heart endothelial cells of both PARP-1+/+ and PARP-1–/– genotypes stimulated with TNF-α (for 6 h) using a luciferase reporter plasmid under the control of a 3 × κB consensus site from the HIV enhancer (29). Our results show that NF-κB-dependent transcriptional activation was partially inhibited in PARP-1-deficient MHEC in response to TNF-α compared to PARP-1+/+ cells (Fig. 4A). To demonstrate the specificity of TNF-α, we performed a control experiment using the pGL3-control vector, a basic luciferase reporter vector under the control of a SV40 promoter (Fig. 4B). To study a possible correlation between NF-κB target genes and a positive or a negative effect of PARP-1 in their regulation, we next examined the effect of proteasome inhibition on expression of several genes by TNF-α-stimulated MHEC by real-time PCR. IκBα degradation as a prerequisite for NF-κB activation could be blocked by incubation of the cells with proteasome inhibitors such as MG132 (32,33). As expected, pre-incubation of MHEC with MG132 inhibited the NF-κB-dependent transcriptional activation induced by TNF-α in both PARP- 1+/+ and PARP-1–/– cells (Fig. 4A). Preincubation of MHEC with MG132 blocked the TNF-α-mediated up-regulation of different genes, suggesting to us that NF-κB is critical for the transcription of these genes by MHEC (Fig. 5). Although NF-κB-dependent transcriptional activation is hampered in PARP-1–/– MHEC we found examples of NF-κB-target genes (Figs 5 and 6) whose expression is silencing by PARP-1 (Cxcl10, Csf2) (Figs 1 and 6A and Table 2) and NF-κB-target genes whose expression is enhancing by PARP-1 (Ccl7, Cxcl5) (Figs 1 and 6B and Table 2).
Figure 4.
Defective NF-κB-dependent transcriptional activation on TNF-α-stimulated PARP-1–/– heart endothelial cells. Cells were transiently co-transfected with a firefly luciferase reporter plasmid under the control of a 3 × κB promoter element (A) or the pGL3-control vector, a basic firefly luciferase reporter vector under the control of a SV40 promoter (B) and the Renilla luciferase expression vector pRLCMV. After 72 h, cells were pre-treated with MG-132 for 1 h and subsequently treated for 6 h with TNF-α (20 ng/ml) as indicated. Results were normalized as indicated in Materials and Methods. The ratio obtained for untreated cells was arbitrarily set to 1. A representative of three experiments is shown. Error bars indicate standard error.
Figure 5.
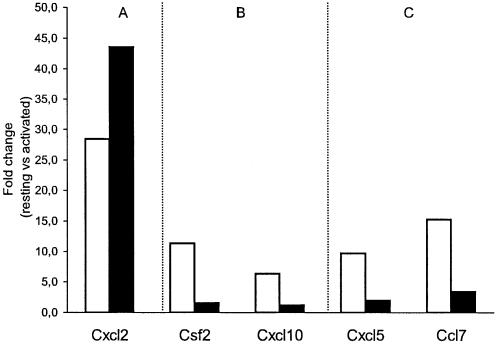
Effect of proteasome inhibition on gene expression of selected genes, (A) PARP-1-independent gene, (B) genes negatively regulated by PARP-1 and (C) genes positively regulated by PARP-1, in response to TNF-α. MHECs were pre-treated for 1 h with MG-132 and then exposed to TNF-α for 2 h. Expression analysis was carried out by real-time PCR as described in Figure 2. The y-axis represents fold-change of activated versus resting PARP-1+/+ cells in the absence of MG132 (white bars) or after pre-incubation with MG132 (black bars).
Figure 6.
TNF-α-induced chemokine production in both PARP-1+/+ (white bars) and PARP-1–/– (black bars) endothelial cells. MHECs were stimulated for 24 h (A and C) or 6 h (B) with 20 ng/ml TNF-α, in the presence or absence of MG132. The levels of Cxcl10 (A), Ccl7 (B) and MIP-2 (C) chemokines were measured by ELISA. A representative of three experiments is shown. Error bars indicate standard error.
Expression of MIP-2 by MHEC in response to TNF-α was similar in PARP-1+/+ and PARP-1–/– cells (Figs 1 and 6C). The up-regulation of this gene in MHECs seems to be NF-κB-independent as their up-regulation was not inhibited by using the proteasome inhibitor MG132 at both mRNA and protein level (Figs 5 and 6C). The results obtained for the MIP-2 gene in our MHEC confirms a recent observation of Hipp et al. (32), which has demonstrated high expression of human IL-8 (the human homolog to mouse MIP-2) (34) by human arterial endothelial cells despite complete suppression of NF-κB activity by proteasome inhibition.
DISCUSSION
In the present study, we used a large-scale gene expression analysis to study the role played by PARP-1 in MHECs early gene expression in reponse to TNF-α. Previously, it has been demonstrated that TNF-α-induced activation of the stress/inflammation transcription factors NF-κB, AP-1, SP-1, Oct-1, YY-1 and Stat-1 in macrophages, fibroblasts and glia cells requires PARP-1 (23,26). Our microarray analysis revealed early TNF-α-response genes in MHEC with various biological functions and include chemokines, growth factors, cell signaling molecules, adhesion molecules, transcription factors, apoptotic molecules and metabolic mediators. Eighteen of these genes (40%) seem to be PARP-1-dependent genes, whereas expression of the other 28 genes did not show differences between PARP-1+/+ and PARP-1–/– MHEC in response to TNF-α (PARP-1-independent genes) at this early time point after activation.
Many TNF-α-induced genes by MHEC represent potential mediators of the inflammatory response. Moreover, a number of them have been previously characterized as NF-κB targets, including MIP-2, Ccl5, Cxcl1, Cxcl10, IL-6, E-selectin, Csf (35) and Gadd45b (36). Recent reports have shown that NF-κB activation depends on PARP-1, suggesting a critical role of this protein as an essential transcriptional coactivator for κB-dependent gene expression (10,23,25). Our results showed that NF-κB-dependent transcriptional activation is hampered in PARP-1–/– MHEC in response to TNF-α. Similar results have been previously reported by Garcia-Soriano et al. after high-glucose stimulation of PARP-1+/+ and PARP-1–/– endothelial cells (10) and by Oliver et al. (23) in TNF-α-activated fibroblast from both genotypes. Likewise, we have also found that the upregulation of the majority of genes involved in the inflammatory response after TNF-α treatment seems to be PARP-1-dependent genes. Furthermore, expression of some of these genes is inhibited in the absence of PARP-1, confirming the results of previous workers that PARP-1 is an important molecule in the regulation of inflammation (18,37). However, a few genes included in this functional group, such as Cxcl10 and Csf2, are higher responders to TNF-α in the absence of PARP-1. A similar effect was observed in other genes belonging to different functional categories such as Gem and Irg1 genes. In fact, PARP-1–/– mice do not show the same phenotype as mice lacking NF-κB family members (38,39) indicated that only a subset of κB-dependent genes are modulated by PARP-1 and that the requirement of PARP-1 for κB-dependent gene expression may be dependent on the tissue and development stage-specific expression of PARP-1. On the other hand, using chromatin immunoprecipitation, Saccani et al. have shown that after an acute stimulation two distinct waves of NF-κB recruitment to target promoters occur: a fast recruitment to constitutively and immediately accessible promoters and a late recruitment to promoters requiring stimulus-dependent modifications in chromatin structure to make NF-κB sites accessible. This new regulatory level implies a mechanism of specificity in NF-κB-dependent transcriptional responses based on the ability of individual stimuli to make late recruitment promoters accessible to NF-κB before its rapid extrusion from the nucleus (40). PARP-1 could be involved in the second wave of NF-κB dependent transcription through its ability to modify chromatin-associated proteins.
Many of pathways in response to TNF-α have proved to be cell type-specific, requiring that observations made in other cell types be confirmed or ruled out in endothelial cells (41). Thus, we next studied whether these genes are indeed NF-κB targets in MHECs. TNF-α signaling involved IκBα degradation and subsequent NF-κB nuclear translocation. IκBα degradation could be blocked by incubation of the cells with proteasome inhibitors such as MG-132 (32,33). Our data showed that pre-incubation of MHEC with this inhibitor dramatically blocks the NF-κB-dependent transcriptional activity. By using this approach, we found that with the exception of MIP-2, expression of all genes analyzed was affected by proteasome inhibition, suggesting that they are NF-κB targets in MHECs. Although NF-κB-dependent transcriptional activity is hampered in PARP-1–/– MHEC, our results show that PARP-1 is regulating the expression of NF-κB-dependent genes both in a positive and a negative fashion, with the final effects depending on the gene. This result suggests different regulatory mechanisms controlling the transcription of these genes. In fact, PARP-1 might modulate gene expression through different mechanisms: (i) physical interactions with other proteins, especially transcription factors; (ii) direct binding to the gene-regulating sequences; and (iii) transient post-translational modifications of nuclear proteins by poly(ADP-ribosyl)ation. Recently, Soldatenkov et al. have demonstrated transcriptional repression of PARP gene expression by binding of PARP to its own promoter sequences (42). PARP has also been shown to bind the IL-6/glucocorticoid-responsive element of Reg gene, forming the active transcriptional DNA/protein complex for Reg gene expression (43). Thus, PARP-1 appears to have dual functions in the regulation of transcription working as a silencing or enhancing transcription factor.
Expression of MIP-2 at both mRNA and protein levels was not blocked by proteasome inhibitor treatment suggesting that its expression in MHEC was NF-κB independent. Our results on MIP-2 gene expression by MHEC confirm the data obtained by Hipp et al. (32) on the expression of human IL-8, the human homolog to murine MIP-2 (34), suggesting that NF-κB is dispensable for MIP-2 activation in murine endothelial cells as it is for IL-8 expression in human endothelial cells. Recently, Haskó et al. (37) have reported that the production of MIP-2 in response to lipopolysaccharide was abolished in PARP-1–/– fibroblasts. In here, we found similar expression in both cell types. This difference could be attributed to the fact that the factors involved in MIP-2 transcription regulation are different in fibroblasts and endothelial cells. In fact, we found that proteasome inhibition increases MIP-2 expression at both RNA and protein levels in MHEC. Hipp et al. (32) have also demonstrated that proteasome inhibition induced IL-8 expression in endothelial cells, and these inhibitors suppressed NF-κB but increased AP-1 activity.
In summary, by using DNA microarray technology, we have identified a set of genes in MHEC whose early expression in response to TNF-α is modulated by PARP-1, but with the final effect depending on the gene. Individual studies in each of these genes are now necessary to clarify the intrinsic mechanisms by which PARP-1 is controlling transcription and thereby finding out different therapeutic approaches involving PARP-1.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr de Murcia for kindly provided the PARP-1–/– mice, Dr Encarna Fermiñan for assistance with microarray analysis and Drs Cristina Rada, Pedro Aparicio and Marisa Galbis for critical reading of the manuscript. A.C. is a recipient of a fellowship from Fundación Séneca. J.Y. is an Investigator from the Ramon y Cajal Program (Spanish Ministerio de Ciencia y Tecnologia). This work was supported by the Instituto de Salud Carlos III Grants PI021138 and C03/02.
REFERENCES
- 1.Granger D.N. (1999) Ischemia-reperfusion: mechanisms of microvascular dysfunction and the influence of risk factors for cardiovascular disease. Microcirculation, 6, 167–178. [PubMed] [Google Scholar]
- 2.Cutrin J.C., Perrelli,M.G., Cavalieri,B., Peralta,C., Rosell Catafau,J. and Poli,G. (2002) Microvascular dysfunction induced by reperfusion injury and protective effect of ischemia preconditioning. Free Radic. Biol. Med., 33, 1200–1208. [DOI] [PubMed] [Google Scholar]
- 3.Aird W.C. (2003) The role of the endothelium in severe sepsis and the multiple organ dysfunction syndrome. Blood, 101, 3765–3777. [DOI] [PubMed] [Google Scholar]
- 4.Hawiger J. (2001) Innate immunity and inflammation: a transcriptional paradigm. Immunol. Res., 23, 99–109. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler A.P. and Bernard,G.R. (1999) Treating patients with severe sepsis. N. Engl. J. Med., 340, 207–214. [DOI] [PubMed] [Google Scholar]
- 6.Gordon H.M., Kucera,G., Salvo,R. and Boss,J.M. (1992) Tumor necrosis factor induces genes involved in inflammation, cellular and tissue repair and metabolism in murine fibroblasts. J. Immunol., 148, 4021–4027. [PubMed] [Google Scholar]
- 7.Karin M. and Ben-Neriah,Y. (2000) Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol., 18, 621–663. [DOI] [PubMed] [Google Scholar]
- 8.Szabó G., Bährle,S., Stumpf,N., Sonnenberg,K, Szabó,E., Pacher,P., Csont,T., Schulz,R., Dengler,T.J., Liaudet,L., Jagtap,P.G., Southan,G.J., Vahl,C.F., Hagl,S. and Szabó,C. (2002) Poly(ADP-ribose) polymerase inhibition reduces reperfusion injury after heart transplantation. Circ. Res., 90, 100–106. [DOI] [PubMed] [Google Scholar]
- 9.Szabó C., Cuzzocrea,S., Zingarelli,B., O’Connor,M. and Salzman,A.L. (1997) Endothelial dysfunction in a rat model of endotoxic shock. Importance of the activation of poly(ADP-ribose) synthetase by peroxynitrite. J. Clin. Invest., 100, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GarciaSoriano F., Virág,L., Jagtap,P., Szabó,E., Mabley,J.G., Liaudet,L., Marton,A., Hoyt,D.G., Murthy,G., Salzman,A.L., Southan,G.J. and Szabó,C. (2001) Diabetic endothelial dysfunction: the role of poly(ADP-ribose) polymerase activation. Nat. Med., 7, 108–113. [DOI] [PubMed] [Google Scholar]
- 11.Pacher P., Mabley,J.G., Soriano,F.G., Liaudet,L., Komjati,K. and Szabo,C. (2002) Endothelial dysfunction in aging animals: the role of poly(ADP-ribose) polymerase activation. Br. J. Pharmacol., 135, 1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Murcia G. and Menissier de Murcia,J. (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci., 19, 172–176. [DOI] [PubMed] [Google Scholar]
- 13.de Murcia G. and Shall,S. (2000) From DNA damage and stress signaling to cell death: poly (ADP-ribosylation) reactions. Oxford University Press, Oxford, UK. [Google Scholar]
- 14.Tulin A. and Spradling,A. (2003) Chromatin loosening by Poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science, 299, 560–562. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z.Q., Auer,B., Stingl,L., Berghammer,H., Haidacher,D., Schweiger,M. and Wagner,E.F. (1995) Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev., 9, 509–520. [DOI] [PubMed] [Google Scholar]
- 16.Menissier de Murcia J., Niedergang,C., Trucco,C., Ricoul,M., Dutrillaux,B., Mark,M., Oliver,F.J., Masson,M., Dierich,A., LeMeur,M., Walztinger,C., Chambon,P. and de Murcia,G. (1997) Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA, 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masutani M., Nozaki,T., Nishiyama,E., Shimokawa,T., Tachi,Y., Suzuki,H., Nakagama,H., Wakabayashi,K. and Sugimura,T. (1999) Function of poly(ADP-ribose) polymerase in response to DNA damage: Gene-disruption study in mice. Mol. Cell Biochem., 193, 149–152. [PubMed] [Google Scholar]
- 18.Yelamos J. and Oliver,F.J. (2002) Role of poly(ADP-ribose) polymerase-1 (PARP-1) in the inflammatory response. Inmunologia, 21, 219–227. [Google Scholar]
- 19.Szabo C. and Dawson,V. (1998) Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol. Sci., 19, 287–298. [DOI] [PubMed] [Google Scholar]
- 20.Chiarugi A. (2002) Poly(ADP-ribose) polymerase: killer or conspirator? The ‘suicide hypothesis’ revisited. Trends Pharmacol. Sci., 23, 122–129. [DOI] [PubMed] [Google Scholar]
- 21.Kraus W.L. and Lis,J.T. (2003) PARP goes transcription. Cell, 113, 677–683. [DOI] [PubMed] [Google Scholar]
- 22.Hasssa P.O. and Hottiger,M.O. (1999) A role of poly(ADP-ribose) polymerase in NF-κB transcriptional activation. Biol. Chem., 380, 953–959. [DOI] [PubMed] [Google Scholar]
- 23.Oliver F.J., Menissier de Murcia,J., Nací,C., Decker,P., Andriantsitohaina,R., Muller,S., De la Rubia,G., Stoclet,J.C. and de Murcia,G. (1999) Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly(ADP-ribose) polimerase-1 deficient mice. EMBO J., 18, 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassa P.O. and Hottiger,M.O. (2002) The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol. Life Sci., 59, 1534–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassa P.O., Covic,M., Hasan,S., Imhof,R. and Hottiger,M.O. (2001) The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J. Biol. Chem., 276, 45588–45597. [DOI] [PubMed] [Google Scholar]
- 26.Ha H.C., Hester,L.D. and Snyder,S.H. (2002) Poly(ADP-ribose) polymerase-1 dependence of stress-induced transcription factors and associated gene expression in glia. Proc. Natl Acad. Sci. USA, 99, 3270–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreone T.L., O’Connor,M., Denenberg,A., Hake,P.W. and Zingarelli,B. (2003) Poly(ADP-ribose) Polymerase-1 regulates activation of activator protein-1 in murine fibroblasts. J. Immunol., 170, 2113–2120. [DOI] [PubMed] [Google Scholar]
- 28.Carrillo A., Chamorro,S., Rodríguez-Gago,M., Alvarez,B., Molina,M.J., Rodríguez-Barbosa,J.I., Sánchez,A., Ramírez,P., Muñoz,A., Domínguez,J., Parrilla,P. and Yélamos,J. (2002) Isolation and characterization of immortalized porcine aortic endothelial cell lines. Vet. Immunol. Immunopathol., 89, 91–98. [DOI] [PubMed] [Google Scholar]
- 29.Bachelerie F., Alcami,J., Arenzana,S.F. and Virelizier,J.L. (1991) HIV enhancer activity perpetuated by NF-κB induction on infection of monocytes. Nature, 350, 709–712. [DOI] [PubMed] [Google Scholar]
- 30.Charreau B., Coupel,S., Goret,F., Pourcel,C. and Soulillou,J.P. (2000) Association of glucocorticoids and cyclosporin A or rapamycin prevents E-selectin and IL-8 expression during LPS- and TNFα-mediated endothelial cell activation. Transplantation, 69, 945–953. [DOI] [PubMed] [Google Scholar]
- 31.Brown K, Gerstberger,S., Carlson,L., Franzoso,G. and Siebenlist,U. (1995) Control of I kappa B-alpha proteolysis by site-specific, signal induced phosphorylation. Science, 267, 1485–1488. [DOI] [PubMed] [Google Scholar]
- 32.Hipp M.S., Urbich,C., Mayer,P., Wischhusen,J., Weller,M., Kracht,M. and Spyridopoulos,L. (2002) Proteasome inhibition leads to NF-κB-independent IL8 transactivation in human endothelial cells through induction of AP-1. Eur. J. Immmunol., 32, 2208–2217. [DOI] [PubMed] [Google Scholar]
- 33.Kalogeris T.J., Laroux,F.S., Cockrell,A., Ichikawa,H., Okayama,N., Phifer,T.J., Alexander,J.S. and Grisham,M.B. (1999) Effect of selective proteasome inhibitors on TNF-induced activation of primary and transformed endothelial cells. Am. J. Physiol., 276, 856–864. [DOI] [PubMed] [Google Scholar]
- 34.Gerard C., Frossard,J.L., Bhatia,M., Saluja,A., Gerard,N.P., Lu,B. and Steer,M. (1997) Targeted disruption of the beta-chemokine receptor CCR1 protects against pancreatitis-associated lung injury. J. Clin. Invest., 100, 2022–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pahl H.L. (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene, 18, 6853–6866. [DOI] [PubMed] [Google Scholar]
- 36.Jin R., De Smaele,E., Zazzeroni,F., Nguyen,D.U., Papa,S., Jones,J., Cox,C., Gelinas,C. and Franzoso,G. (2002) Regulation of the gadd45beta promoter by NF-kappaB. DNA Cell Biol., 21, 491–503. [DOI] [PubMed] [Google Scholar]
- 37.Haskó G., Mabley,J.G., Németh,Z.H., Pacher,P., Deitch,E.A. and Szabó,C. (2002) Poly(ADP-ribose) Polymerase is a regulator of chemokine production: relevance for the pathogenesis of shock and inflammation. Mol. Med., 8, 283–289. [PMC free article] [PubMed] [Google Scholar]
- 38.Sha W.C., Liou,H.C., Tuomanen,E.I. and Baltimore,D. (1995) Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell, 80, 321–330. [DOI] [PubMed] [Google Scholar]
- 39.Beg A.A., Sha,W.C., Bronson,R.T., Ghosh,S. and Baltimore,D. (1995) Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature, 376, 167–170. [DOI] [PubMed] [Google Scholar]
- 40.Saccani S., Pantano,S. and Natoli,G. (2001) Two waves of nuclear factor kappaB recruitment to target promoters. J. Exp. Med., 193, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madge L.A. and Pober,J.S. (2001) TNF signaling in vascular endothelial cells. Exp. Mol. Pathol., 70, 317–325. [DOI] [PubMed] [Google Scholar]
- 42.Soldatenkow V.A., Chasovskikh,S., Potaman,V.N., Trofimova,I., Smulson,M.E. and Dritschilo,A. (2002) Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J. Biol. Chem., 277, 665–670. [DOI] [PubMed] [Google Scholar]
- 43.Akiyama T., Takasawa,S., Nata,K., Kobayashi,S., Abe,M., Shervani,N.J., Ikeda,T., Nakagawa,K., Unno,M., Matsuno,S. and Okamoto,H. (2001) Activation of Reg gene, a gene for insuline-producing β-cell regeneration: Poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc. Natl Acad. Sci. USA, 98, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]