Abstract
Two forms of human tryptophanyl-tRNA synthetase (TrpRS) are produced in vivo through alternative mRNA splicing. The two forms, full-length TrpRS and mini TrpRS, are catalytically active, but are distinguished by the striking anti-proliferative and anti-angiogenic activity specific to mini TrpRS. Here we describe two new splice variants of human TrpRS mRNA. Their production was strongly regulated by γ-interferon (IFN-γ), an anti-proliferative cytokine known to stimulate the expression of other anti-angiogenic factors. A new IFN-γ-sensitive promoter was demonstrated to drive production of these splice variants. In human endothelial cells, both the newly discovered and a previously reported promoter were shown to respond specifically to IFN-γ and not to other cytokines such as tumor necrosis factor-α, transforming growth factor-β, interleukin-4 or erythropoietin. In addition, both promoters were stimulated by the ‘downstream’ interferon regulatory factor 1 that, in turn, is known to be regulated by the ‘upstream’ signal transducer and activator of transcription 1α subunit. Thus, the tandem promoters provide a dual system to regulate expression and alternative splicing of human TrpRS in vivo.
INTRODUCTION
Aminoacyl-tRNA synthetases are essential enzymes that covalently link amino acids to their cognate tRNAs (1,2). The 20 enzymes are divided into two groups of 10 each (3–6). Class I enzymes share a common active site architecture which includes a Rossmann fold nucleotide-binding domain (7). The active sites of class II enzymes are made up of a seven-stranded antiparallel β-sheet flanked by α-helices (6,8). During their long evolution, many tRNA synthetases acquired extra domains in addition to the core active site structures. Initially, these extra domains added specificity to the aminoacylation reactions. Later additions and sequence adaptations, like those seen in mammalian synthetases, contribute to the additional cellular functions seen in several enzymes (9,10). Typically the added functions developed alongside the activity of tRNA synthetases in translation.
A striking example of functional expansion is human tyrosyl-tRNA synthetase (TyrRS), which is processed by the extracellular protease leukocyte elastase to release two fragments, one with IL-8-like cytokine and pro-angiogenic activity (mini TyrRS) and the other with EMAP II-like cytokine activity (C-domain) (11,12). Significantly, the unprocessed full-length enzyme lacks the cytokine activities. The closely related human enzyme tryptophanyl-tRNA synthetase (TrpRS) developed anti-angiogenic activity in the naturally occurring variant called mini TrpRS but not in the full-length, native protein (13,14). Several recent reports revealed that other mammalian synthetases have unique cellular activities beyond translation. For example, glutaminyl-tRNA synthetase inhibits apoptosis (15) and histidyl-tRNA synthetase stimulates inflammation signal transduction pathways (16). While these particular specializations and functional expansions show the remarkably versatile activities associated with particular tRNA synthetases, others have evolved secondary functions related to RNA processing such as human rRNA maturation, which requires methionyl-tRNA synthetase (17), mitochondrial tyrosyl-tRNA synthetase-dependent splicing of group I introns in the fungi Neurospora and Podospora (18,19) and splicing of the fourth intron (bI4) of the Saccharomyces cerevisiae cob gene by mitochondrial leucyl-tRNA synthetase (20).
Among tRNA synthetases with expanded functions, human TrpRS was one of the first studied. Early work established that TrpRS underwent limited proteolysis and was secreted from bovine pancreas (21,22). In separate studies, a truncated variant of human TrpRS (mini TrpRS) was demonstrated to be produced by alternative splicing in vivo (23,24). This variant, along with the full-length enzyme, was strongly up-regulated by IFN-γ (23,25) (Fig. 1A). The rationale and biological significance of IFN-γ regulation and truncated TrpRS forms were unknown.
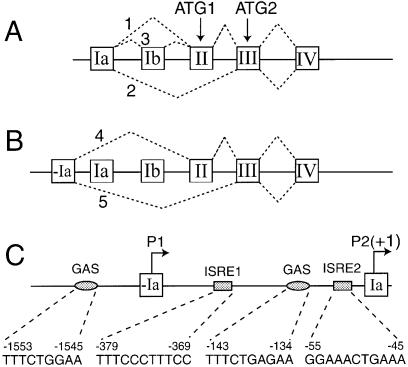
Figure 1.
Schematic presentation of genomic DNA organization and alternative splicing of the human TrpRS gene. Boxes and lines represent exons and introns, respectively. Discontinuous lines indicate major spliced forms of TrpRS mRNA. Translation initiated from ATG1 produces full-length TrpRS protein, the translation product from ATG2 corresponds to mini TrpRS protein. (A) Three alternative splicing forms of mRNA containing exon Ia (23). (B) Two newly discovered alternative splicing forms of mRNA containing exon –Ia. (C) Schematic presentation of the 5′ regulatory region of human TrpRS. The major transcription start site within exon Ia is referred to as +1. The potential IFN-related regulatory elements and their nucleotide sequences and positions are indicated, several of which were previously described (23,36). The position of exon –Ia is –1458 to –912. Two promoters are indicated as P1 and P2. GAS, γ-activated sequence; ISRE, interferon-stimulated response element.
Recent work showed that three related variants of human TrpRS, mini TrpRS, T1-TrpRS and T2-TrpRS, have potent anti-angiogenic activity (13,14). Thus, mini TrpRS blocked vascular endothelial growth factor (VEGF)-induced proliferation and migration of human umbilical vein endothelial cells (HUVECs) (13) and inhibited angiogenesis in chick chorioallantoic membranes, in a mouse matrigel model and in a mouse model of post-natal blood vessel development in the retina (13,14). Retinal vessel development is well correlated with VEGF expression and direct suppression of VEGF signaling effectively inhibited neovascularization (26,27). While the mechanistic details of mini TrpRS inhibition are not yet known, VEGF-mediated angiogenesis was inhibited by mini TrpRS. In the retina, T2-TrpRS potently inhibited blood vessel development and was localized to pre-existing blood vessels (14). These results collectively suggest that T2-TrpRS acts directly on existing blood vessels to inhibit further development. The discovery that these fragments of TrpRS have anti-angiogenic activity provided a biological rationale for their production.
IFN-γ is a pleiotropic cytokine that induces an anti-proliferative cellular state and regulates expression of over 200 genes, some of which are involved in host defense or inflammation (28). Other IFN-γ-regulated genes, like interferon-inducible protein-10 (IP-10), monokine induced by γ-interferon (MIG) and mini TrpRS, are anti-angiogenic factors (29,30). Although some lymphoid cell lines do not exhibit TrpRS regulation by IFN-γ, expression of TrpRS is specifically up-regulated by IFN-γ in epithelial cells and fibroblasts (31–33). Given a biological rationale for production of fragments of TrpRS and the role of IFN-γ in regulating expression of anti-angiogenic factors, we were particularly interested in examining the regulation of TrpRS and mini TrpRS in endothelial cells. The idea was to provide a framework for understanding how the remarkable activities of this human tRNA synthetase are generated and controlled. This framework, in turn, gave us the basis to establish a working model explaining how multiple factors control expression of this tRNA synthetase.
MATERIALS AND METHODS
Cell culture
HUVECs were obtained from Clonetics (Walkersville, MD) and maintained in EGM®-2 BulletKit® medium (Clonetics) in an atmosphere of 5% CO2 in air at 37°C according to the instructions of the supplier. HeLa cells were obtained from Clontech (Palo Alto, CA) and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% inactivated fetal bovine serum and antibiotics. HUVECs were treated with IFN-γ (Roche, Mannheim, Germany) at 50–500 U/ml, tumor necrosis factor-α (TNF-α) (Roche) at 500–1000 U/ml, transforming growth factor-β (TGF-β) (Roche) at 0.1–2 nM, interleukin-4 (IL-4) (R&D Systems, Minneapolis, MN) at 10–100 U/ml, erythropoietin (Epo) (Sigma, Saint Louis, MI) at 4 U/ml or human interferon-α (IFN-α) (Sigma) at 500 U/ml for 0–48 h incubation.
Reverse transcription–PCR (RT–PCR)
After treatment with IFN-γ or other cytokines, HUVECs (1 × 106 cells) were harvested at the times indicated, washed twice with ice-cold phosphate-buffered saline (PBS) and collected by centrifugation. Total RNA was isolated from HUVECs using the RNeasy Midi kit (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. Total RNA (1 µg) in 15 µl was converted to complementary DNA (cDNA) using M-MLV reverse transcriptase and an anchored oligo(dT) primer set following the manufacturer’s protocol (Qiagen). A 2 µl aliquot of the resulting cDNA was used as template for PCR amplification with specific primers: human TrpRS, pJL50 (forward, exon –Ia, 5′-GAGCGCTGACTGGCCCGGCTGGG-3′) and pJL51 (reverse, exon III, 5′-ATGAGCTTATCGTAGTCTATGCC-3′) or pKLEA (forward, exon Ia, 5′-AGCTCAACTGCCCAGCGTGACC-3′) and pKLEB (reverse, exon III, 5′-CAGTCAGCCTTGTAATCCTCCCCC-3′); human glyceraldehyde 3-phosphate dehydrogenase (GAPDH), pKLEC (forward, 5′-GGTCGGAGT CAACGGATTT-3′) and pKLED (reverse, 5′-CCAGCATCGCCCCACTTGA-3′). The amplifications were performed by initial denaturation (94°C for 1 min), 30 cycles of denaturation, annealing and extension (94°C for 1 min, 60°C for 45 s and 72°C for 45 s), and final extension (72°C for 10 min). The transcript of GAPDH was also amplified by RT–PCR from the same cDNA template and was used as an internal control. The resulting PCR products were resolved by 1.2% agarose gel electrophoresis and cloned into a pCR®4-TOPO vector (Invitrogen, Carlsbad, CA). The identity of each DNA band was confirmed by DNA sequencing.
Northern blot analysis
Total RNA (10 µg) was separated by electrophoresis in a 1% agarose/2.2 M formaldehyde gel, capillary transferred to a Zeta-Probe GT Genomic positively charged nylon membrane (Bio-Rad) with 10× SSC. The membrane was UV cross-linked using a HL-2000 HybriLinker (Upvon, Upland, CA). Oligonucleotide probes specific for the different TrpRS transcript isoforms were radioactively labeled with [α-32P] dCTP (3000 Ci/mmol; Amersham Pharmarcia Biotech Inc., Piscataway, NJ) by random priming (Promega, Madison, WI) and used as probes to hybridize with the total RNA samples. The blots were hybridized at 55–65°C for 20 h in hybridization solution as described (34). Detection was performed with a PhosphorImager screen. Membranes were stripped and re-probed with a 32P-labeled oligonucleotide probe specific for GAPDH to evaluate consistency of sample loading.
Plasmids and cloning
The putative promoters for human TrpRS were amplified by PCR from HUVEC genomic DNA with the following oligonucleotides pairs: pES01 (forward, 5′-cggaagatctCTTCTAAAGCCAGCCAGCCAGC-3′) and pES02 (reverse, 5′-ggaattcTGCCCAGCCGGGCCAGTCAGCG-3′) for amplification of putative promoter P1; pES03 (forward, 5′-cggaagatctCACTGCACTGTGCCGCCTCGG-3′) and pES04 (reverse, 5′-ggaattcGGAAGACACTGCAGAGGTGGCC-3′) for promoter P2; pES01 and pES04 for putative tandem promoters P1 + P2 (P12). The nucleotide sequences in lowercase and underlined BglII and EcoRI restriction sites were added for cloning PCR products between the BglII and EcoRI restriction sites of a luciferase reporter vector phRL-null (Promega) to give pP1-Luc, pP2-Luc and pP12-Luc, respectively. Mutant forms of γ-activated sequence (GAS) or interferon-stimulated response element (ISRE) consensus sequences within the promoter constructs were created by site-directed mutagenesis with a QuickChange XL kit (Stratagene, La Jolla, CA) as follows: ISRE1 (–379 to –369, 5′-GTTTCCCTTTC-3′→5′-GTTTCCAGCGCC-3′), ISRE2 (–55 to –45, 5′-GGAAACTGAAA-3′→5′-GAAACCTCGAG-3′), ISRE3 (–1421 to –1408, 5′-CAAACACAAAAC-3′→5′-CTTGCACAAAAC-3′). Mutated nucleotides are underlined. Plasmids carrying human IRF-1 and IRF-2 in vector pcDNA (Invitrogen) were gifts from Drs Mark Perrella and Richard Riese at Brigham and Women’s Hospital (Boston, MA). Endotoxin-free plasmid DNA was prepared with an EndoFree Plasmid Maxi Kit (Qiagen).
Transient transfection luciferase assays
Transient transfections of 60–70% confluent HUVEC or HeLa cells were performed with SuperFect transfection reagent (Qiagen) and each of the luciferase reporter vectors (5 or 10 µg) according to the manufacturer’s instructions (Qiagen). After transfection, cells were incubated with fresh medium with or without IFN-γ (100 U/ml) for 24 h. At the designated times, treated cells were rinsed with PBS and lysed for quantification of luciferase. Luciferase activity was measured following the manufacture’s protocol (Promega) using the Analyst AD Assay Development System (Molecular Devices, Sunnyvale, CA). Luciferase activity was initially calculated as relative luminescence units (RLU)/µg protein. Relative luciferase activity was expressed as (sample – vector control)/vector control.
RESULTS
Multiple 5′-untranslated regions (UTRs) of human TrpRS mRNA
Through extensive nucleotide BLAST searches of the human Expressed Sequence Tag (EST) database at the National Center for Biotechnology Information we deduced alternative splice variants of TrpRS from a variety of tissues/cell lines, including lung, prostate tumor, testis, brain, lymphoma, eye and skin. Unexpectedly, multiple 5′-UTRs of TrpRS mRNA were identified. Among the five distinct sequences described here, two were novel and three were previously identified (Fig. 1A) (23,25). The two newly identified variants showed excellent sequence identity downstream of exon II or exon III in TrpRS cDNA, but had unique sequences at their 5′ ends. Comparison of these nucleotide sequences to the genomic DNA sequence of human TrpRS showed that the 5′-sequence of the two novel mRNAs corresponded to a previously unidentified exon upstream of exon Ia. The existence of the upstream exon (exon –Ia) strongly suggested that the five different TrpRS mRNAs were generated by two different promoters and that transcription events initiated from exon –Ia produced distinct TrpRS mRNAs due to alternative splicing (Fig. 1B).
Analysis of human TrpRS promoters
Analysis of the 2000 bp DNA sequence upstream of the human TrpRS gene by the program MatInspector (35) revealed several potential interferon regulatory elements: two GAS at –143 to –134 (5′-TTTCTGAGAA-3′) and –1553 to –1545 (5′-TTTCTGGAA-3′); two ISRE at –55 to –45 (5′-GGAAACTGAAA-3′) and –379 to –369 (5′-TTTCCCTTTCC-3′) (Fig. 1C). [The transcription response element positions are indicated relative to the transcription start site within exon Ia (as +1) (23).] These response elements were located in two separate regions of the upstream 2000 bp of the TrpRS gene. One GAS element was located upstream of exon –Ia between positions –1545 and –1553 within the region of the newly identified promoter (named promoter P1 because it is in front of the previously identified promoter P2). The other GAS element (23) and the two ISRE sequences (36) were located within the intervening sequence between exon –Ia and exon Ia within promoter P2. This analysis suggested that both promoters (P1 and P2) would be responsive to IFN-γ stimulation.
Regulation of TrpRS in HUVECs
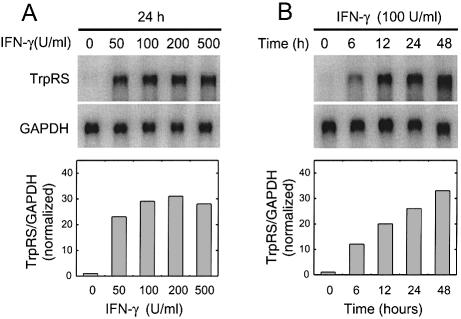
Several human cell types up-regulate TrpRS and mini TrpRS expression in response to IFN-γ (23,31–33). However, studies on TrpRS expression in HUVECs have not been previously reported. Because the proliferation and migration of HUVECs are inhibited by mini TrpRS, whether TrpRS expression was regulated by IFN-γ in these cells was of obvious interest. The level of TrpRS mRNAs was measured by northern blot hybridization in HUVECs treated with IFN-γ (50–500 U/ml IFN-γ for 24 h). A dramatic increase in TrpRS mRNA was observed following IFN-γ treatment (Fig. 2A), with maximum induction of ∼30-fold higher mRNA levels at 24 h than in HUVECs without IFN-γ stimulation. The induction of TrpRS mRNA by IFN-γ was also time-dependent, with a 10-fold increase in mRNA observed after 6 h IFN-γ incubation (Fig. 2B). The level of TrpRS mRNA continued to increase from 6 to 24 h, after which it reached a stable plateau after 48 h of treatment. The amount of TrpRS protein increased up to 10-fold after IFN-γ stimulation (data not shown). Expression of another synthetase, TyrRS (which is not regulated by IFN-γ), was not changed during this time. TrpRS expression was also examined in HUVECs following treatment with IFN-α, TNF-α, TGF-β, IL-4 and Epo, because earlier reports suggested that these factors modulated TrpRS expression in other cell types (32,37–39). However, none of these factors affected TrpRS expression in HUVECs, whether alone or in combination with IFN-γ (data not shown). Collectively, these data demonstrate that human TrpRS expression in HUVECs was strongly and specifically stimulated by IFN-γ in a dose- and time-dependent fashion.
Figure 2.
IFN-γ up-regulates TrpRS in HUVECs. (A) Concentration- and (B) time-dependent up-regulation of TrpRS mRNA in HUVECs stimulated with IFN-γ. Total RNA was isolated from the cells stimulated with various amount of IFN-γ for different lengths of time and the levels of mRNAs for TrpRS and GAPDH were analyzed by northern blot hybridization. TrpRS mRNA levels were normalized to GAPDH mRNA levels to control for variations in loading between samples.
Alternative splice variants of TrpRS in HUVECs
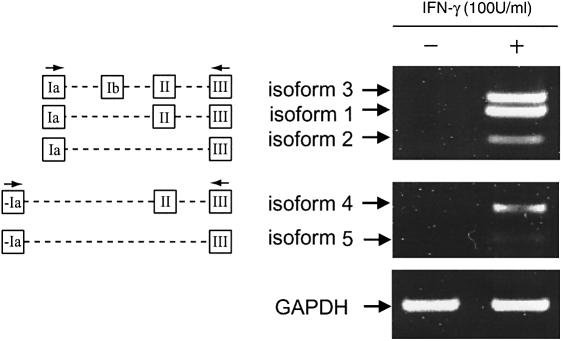
Alternative splicing of TrpRS mRNA in HUVECs was examined by RT–PCR analysis. Two sets of oligonucleotide primers were used to PCR amplify cDNAs that corresponded to the various TrpRS mRNAs predicted from the EST analysis described above. The first set of primers corresponded to exon Ia and exon III and the second set of primers corresponded to exon –Ia and exon III (Fig. 3). Amplification with the first set of primers gave three cDNA bands (isoforms 1–3) from HUVECs. The identities of the cDNA bands were determined by DNA sequencing and these three isoforms corresponded to those previously identified from HeLa cells (23,25). To show the differences in TrpRS mRNA isoform production before and after IFN-γ treatment, we used cDNA (prepared as described in Materials and Methods) that was diluted 1:100 in RT–PCRs. With this dilution, DNA bands representing different isoforms are barely visible before IFN-γ treatment, while the level of all TrpRS mRNAs was increased following IFN-γ stimulation (Fig. 3). Amplification with the second set of primers (–Ia and III) gave two cDNA bands (isoforms 4 and 5) in both HeLa cells (data not shown) and HUVECs (Fig. 3). The amount of isoforms 4 and 5 increased following IFN-γ stimulation (Fig. 3). Among the five TrpRS variants, isoforms 2 and 5 were relatively less than the other bands, which is consistent with the amounts of the corresponding protein products (data not shown) (24). As an internal control, GAPDH mRNA was PCR amplified from the same samples and the level was invariant.
Figure 3.
RT–PCR analyses of IFN-γ up-regulated TrpRS isoforms in HUVECs. Cells were treated with or without IFN-γ (100 U/ml) for 24 h. Total RNAs were isolated and converted to cDNA. PCR was performed with cDNA (prepared as described in Materials and Methods and diluted 1:100) and analyzed by 1.2% agarose gel electrophoresis. Under these conditions, the differences between the levels of TrpRS mRNA isoforms could be seen. GAPDH is an internal control.
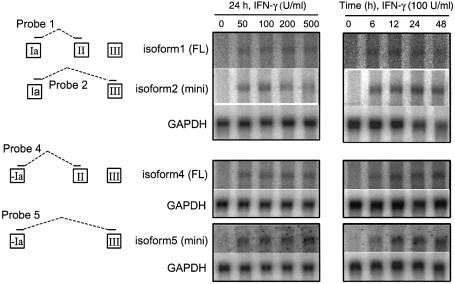
Northern blot hybridization analyses were also carried out to evaluate the expression of different TrpRS mRNA isoforms. Four oligonucleotide probes were designed to hybridize to the distinct isoforms of TrpRS mRNA (Fig. 4). Before IFN-γ treatment, the mRNA level of each variant was low, while after IFN-γ stimulation the amount of all variants increased in a dose- and time-dependent manner (Fig. 4). Although it is difficult to determine the relative ratio between the TrpRS mRNA isoforms, it is clear that all the isoforms were strongly stimulated by IFN-γ. These data were consistent with the results obtained from RT–PCR analysis. Together, these studies showed that two newly identified TrpRS mRNA isoforms (4 and 5) were produced in HUVECs and HeLa cells in addition to the three previously reported forms (1, 2 and 3). The new isoforms have a unique 5′ exon, but they share the characteristic IFN-γ regulation of the previously known TrpRS isoforms.
Figure 4.
Northern blot analyses of IFN-γ up-regulated TrpRS isoforms expressed in HUVECs. Total RNA was isolated from the cells stimulated with various amounts of IFN-γ for different lengths of time and the levels of mRNAs for TrpRS were analyzed by northern blot hybridization. GAPDH is an internal control for even loading. Specific probes for different mRNA isoforms are illustrated to the left in the figure.
Two inducible promoters regulate TrpRS expression in HUVECs
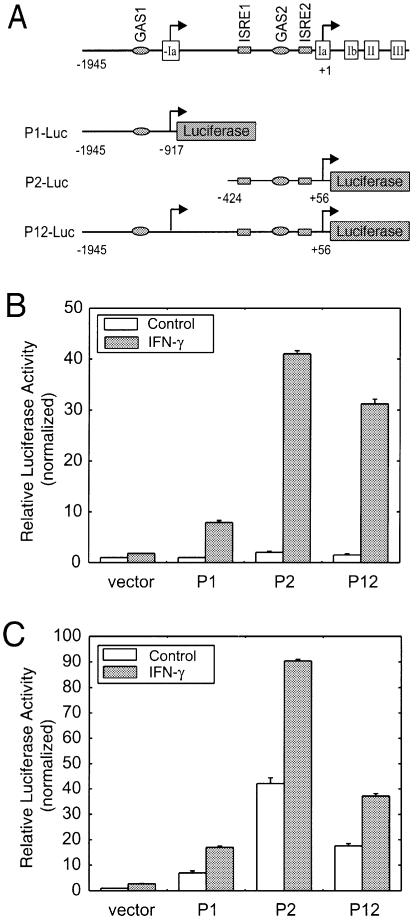
We next sought to dissect the activity of the two TrpRS promoters using a promoter-driven luciferase assay. A series of luciferase reporter vectors was constructed containing different portions of the DNA region upstream of base +1 and extending 56 bp into exon Ia followed by the luciferase gene for direct readout of promoter activity (Fig. 5A). Each construct was transiently transfected into HeLa cells and HUVECs and the relative luciferase activity was determined in the presence or absence of IFN-γ. In HeLa cells, the basal level of luciferase expression from promoter P1, P2 or the combined promoter P12 was low. In contrast, IFN-γ strongly stimulated luciferase expression from all three constructs, with the P2-Luc construct giving the most robust expression (Fig. 5B). Similar results were obtained in HUVECs, where IFN-γ treatment strongly induced luciferase expression (Fig. 5C). This result was obtained from three independent experiments. Interestingly, in HUVECs, the basal expression of luciferase from promoter constructs P1-Luc, P2-Luc and P12-Luc was higher than that in HeLa cells. This result is consistent with the previous observation that TrpRS gene expression is higher in HUVECs compared to most other human tissues and cells (40). We also noticed that the promoter activity of P12 is lower than that of promoter P2 alone in HUVECs, which is also observed in HeLa cells (23). In summary, these data demonstrate that promoters P1 and P2 were functional promoters responsive to IFN-γ stimulation.
Figure 5.
Transient transfection assays. (A) Schematic diagram showing different luciferase reporter constructs. Open boxes represent exons. The DNA sequence of exon –Ia is included in the P12-Luc construct. The putative IFN-related regulatory elements are indicated. GAS, γ-activated sequence; ISRE, interferon-stimulated response element. (B) HeLa cells transfected with TrpRS promoter constructs and incubated with or without IFN-γ (100 U/ml) for 24 h. (C) HUVECs transfected with TrpRS promoter constructs and incubated with or without IFN-γ (100 U/ml) for 24 h. Data shown here are representative of three independent measurements. Results are expressed as fold induction relative to the activity of the empty reporter vector control and represent the mean ± SE (n = 3).
IRF-1 activates TrpRS promoter activity
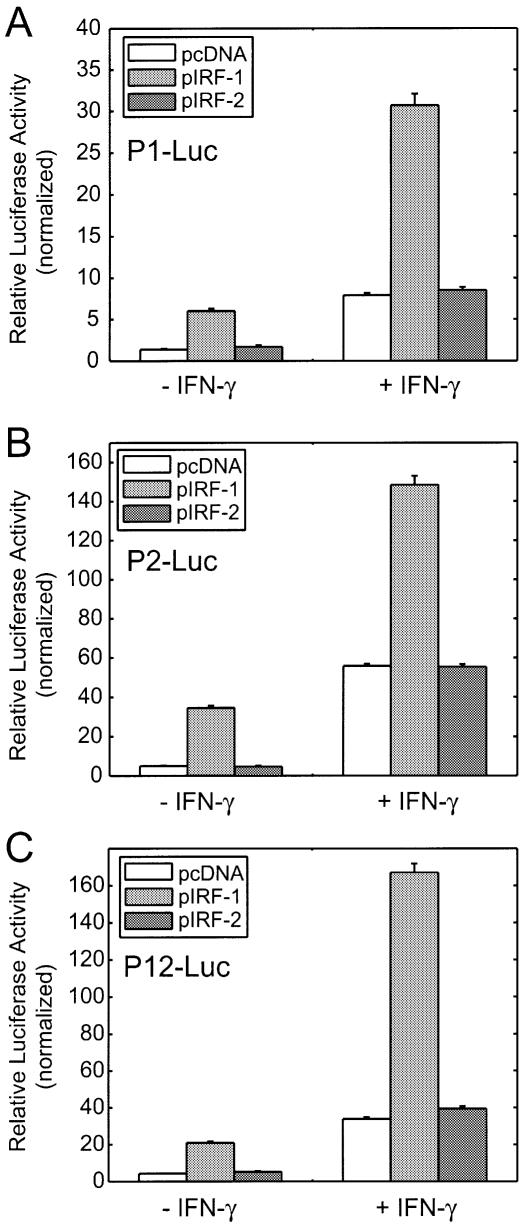
Because GAS sites are well defined and known to be activated by signal transducer and activator of transcription 1α subunit (STAT1), we focused on new features within the TrpRS promoter such as the potential ISRE motifs and their possible activator(s). Interferon regulatory factors (IRFs) belong to a growing family of transcription factors. Several recent reports showed that IRF-1, which itself is up-regulated by IFN-γ, stimulates IFN-γ-induced target gene expression by binding to ISRE consensus sites (41,42). IRF-2 is expressed constitutively and acts mostly as a repressor by competing with IRF-1 for the same binding site(s) (41,43). There were two potential ISRE consensus sequences identified by the program MatInspector within promoter P2 that were potential binding sites for IRF-1. We examined whether overexpression of IRF-1 would activate TrpRS promoter activity independent of IFN-γ stimulation. An IRF-1-containing expression vector was co-transfected into HeLa cells with each of the three promoter constructs. Overexpression of IRF-1 led to dramatic increases in P1, P2 and P12 promoter activity compared with cells transfected with empty vector in the absence or presence of IFN-γ (Fig. 6). Endogenous TrpRS expression was also stimulated by IRF-1 overexpression in western blot analyses (data not shown). In contrast, co-transfection of IRF-2 had no effect on promoter activity. Because MatInspector did not identify a consensus sequence for ISRE in this promoter, it was surprising to find that IRF-1 stimulated expression from promoter P1 (construct P1-Luc). However, upon closer inspection a nucleotide segment (–1421 to –1408, 5′-CAAACACAAAAC-3′) was found that may serve as an ISRE and thus explain the effect of IRF-1 on P1-Luc.
Figure 6.
Overexpression of IRF-1 activated TrpRS promoters. HeLa cells were transiently co-transfected with indicated promoter constructs along with IRF-1 or IRF-2 vector for 24 h in the absence or presence of IFN-γ (100 U/ml). (A) P1-Luc; (B) P2-Luc; (C) P12-Luc. Data shown here are representative of three independent measurements. Results are expressed as fold induction relative to the activity of the empty reporter vector control and represent the mean ± SE (n = 3).
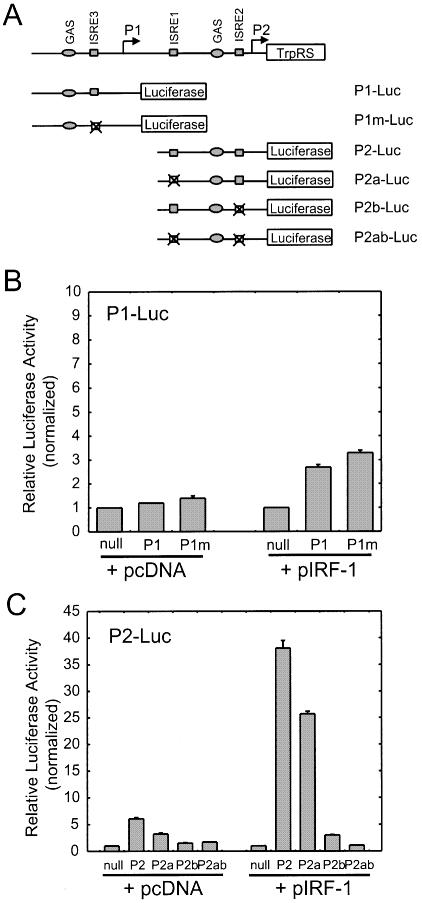
To find out which of the putative ISREs is involved in IRF-1 stimulation, we mutated each ISRE motif within promoters P1 and P2 (Fig. 7A). Each construct was co-transfected with an IRF-1-containing vector into HeLa cells and the luciferase activity was measured. The putative ISRE motif within promoter P1 (ISRE3) was mutated, but the promoter activity was only marginally changed compared to that of wild-type P1 (Fig. 7B). This result suggested that promoter P1 may contain an unidentified nucleotide element to mediate IRF-1 stimulation or that the IRF-1 stimulation was regulated by a mechanism not requiring ISRE3. As shown in Figure 7C, mutation of ISRE1, associated with promoter P2, had a modest effect on luciferase activity, which decreased to ∼70% of wild-type P2. In contrast, when ISRE2 was mutated, the P2 promoter activity was strongly inhibited. When both ISREs were mutated (P2ab), there was no measurable luciferase activity. These results suggested that ISRE2 within promoter P2 was the primary site for IRF-1 binding and stimulation of TrpRS expression, while ISRE1 was needed for maximal expression.
Figure 7.
Transient transfection assays. HeLa cells were transiently co-transfected with the indicated promoter constructs along with IRF-1 for 24 h. (A) Schematic diagram showing different luciferase reporter constructs. Open boxes represent exons. The putative IFN-related regulatory elements GAS and ISRE are indicated; (B) P1-Luc; (C) P2-Luc. Data shown here are representative of three independent measurements. Results are expressed as fold induction relative to the activity of the empty reporter vector control and represent the mean ± SE (n = 3).
DISCUSSION
TrpRS is overexpressed in guinea pigs during delayed-type hypersensitivity reactions (44), in Drosophila during development of salivary gland (45) and in aged human epidermis (46). Elevated TrpRS expression was also found in IFN-γ-treated bladder transitional cells (47). The increase in TrpRS levels during specific developmental events and in specific diseases may be a response related to its role in cell signaling. While the canonical function of human TrpRS is aminoacylation of tRNATrp for protein synthesis, regulation of angiogenesis is an extended function provided by an alternative splicing variant (13,14). The regulation of expression of human TrpRS by IFN-γ is perhaps related to this extended function. Supporting this idea, IFN-γ was shown here to stimulate the production of the anti-angiogenic, N-terminally truncated variant.
Although previous data by others described TNF-α, TGF-β and Epo stimulation of TrpRS expression in certain cell types (38,39), in our studies those effects were not observed in HUVECs. None of these cytokines increased TrpRS expression whether alone or in combination. In contrast, TrpRS expression in HUVECs was specifically and robustly stimulated by IFN-γ. Previous reports that IL-4 and TGF-β had a negative effect on IFN-γ-induced TrpRS expression (37) led us to test these cytokines on HUVECs. However, neither IL-4 nor TGF-β had an effect on IFN-γ-stimulated TrpRS expression. Thus, TrpRS expression in HUVECs was uniquely stimulated by IFN-γ.
The new promoter (reported here) controlling expression was identified within a 2000 bp region of the gene. [This promoter is upstream and in tandem with the previously identified promoter (23).] Overall, the 2000 bp region contains four IFN-γ regulatory sequences and all elements needed to direct basal level TrpRS expression. Promoter P1 contains one GAS element, while promoter 2 contains one GAS element and two ISRE elements. Both promoters respond to IFN-γ stimulation. The five TrpRS mRNA variants seen in vivo [two new TrpRS mRNA isoforms identified in this study and three isoforms reported previously (23,25)] result in expression of two protein forms, TrpRS and mini TrpRS. Expression of all five TrpRS mRNA isoforms was stimulated by IFN-γ (Figs 3 and 4). That two promoters control the gene expression of TrpRS and mini TrpRS reveals that expression is complex and warrants further investigation.
Interferon signaling occurs by binding of IFN-γ to a heterodimeric receptor that is linked to Janus kinases (JAKs) 1 and 2. Thus activated, JAK1/2 in turn activates STAT1, which is a transcription factor that translocates to the nucleus and activates several IFN-γ-responsive genes through binding to GAS elements. Among the primary IFN-γ response genes are some transcription factors, including IRF-1, which itself binds to ISRE elements to modulate gene expression (41,42,48). The two TrpRS promoters examined here contained two GAS sites and two putative ISRE motifs. Previous work suggested that IFN-γ stimulation of expression of human TrpRS involved STAT1 and the GAS site of promoter P2. However, IFN-γ-sensitive expression of TrpRS could not be completely ascribed to the STAT1 transcription factor acting through the GAS site of the P2 promoter (23). In this work, transcription factor IRF-1 and the new P1 promoter were demonstrated to be regulators in the IFN-γ-sensitive expression of TrpRS, which thereby suggests a way to explain more broadly the IFN-γ-related regulation of TrpRS expression.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Mark Perrella and Richard Riese at Brigham and Women’s Hospital in Boston for the generous gifts of vectors carrying human IRF-1 and IRF-2. We also thank Bonnie M. Slike and Francella Otero for kind assistance in culturing cells and Dr Jun Liu for helping with the luciferase activity determinations. This work was supported by grant CA92577 from the National Cancer Institute and by a fellowship from the National Foundation for Cancer Research.
REFERENCES
- 1.Schimmel P. (1987) Aminoacyl tRNA synthetases: general scheme of structure–function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem., 56, 125–158. [DOI] [PubMed] [Google Scholar]
- 2.Moras D. (1992) Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem. Sci., 17, 159–164. [DOI] [PubMed] [Google Scholar]
- 3.Webster T., Tsai,H., Kula,M., Mackie,G.A. and Schimmel,P. (1984) Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science, 226, 1315–1317. [DOI] [PubMed] [Google Scholar]
- 4.Ludmerer S.W. and Schimmel,P. (1987) Gene for yeast glutamine tRNA synthetase encodes a large amino-terminal extension and provides a strong confirmation of the signature sequence for a group of the aminoacyl-tRNA synthetases. J. Biol. Chem., 262, 10801–10806. [PubMed] [Google Scholar]
- 5.Eriani G., Delarue,M., Poch,O., Gangloff,J. and Moras,D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature, 347, 203–206. [DOI] [PubMed] [Google Scholar]
- 6.Cusack S., Berthet-Colominas,C., Härtlein,M., Nassar,N. and Leberman,R. (1990) A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature, 347, 249–255. [DOI] [PubMed] [Google Scholar]
- 7.Rao S.T. and Rossmann,M.G. (1973) Comparison of super-secondary structures in proteins. J. Mol. Biol., 76, 241–256. [DOI] [PubMed] [Google Scholar]
- 8.Ruff M., Krishnaswamy,S., Boeglin,M., Poterszman,A., Mitschler,A., Podjarny,A., Rees,B., Thierry,J.C. and Moras,D. (1991) Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- 9.Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999) Aminoacyl-tRNA synthetases: a family of expanding functions. EMBO J., 18, 4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francklyn C., Perona,J.J., Puetz,J. and Hou,Y.M. (2002) Aminoacyl-tRNA synthetases: versatile players in the changing theater of translation. RNA, 8, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakasugi K. and Schimmel,P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science, 284, 147–151. [DOI] [PubMed] [Google Scholar]
- 12.Wakasugi K., Slike,B.M., Hood,J., Ewalt,K.L., Cheresh,D.A. and Schimmel,P. (2002) Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J. Biol. Chem., 277, 20124–20126. [DOI] [PubMed] [Google Scholar]
- 13.Wakasugi K., Slike,B.M., Hood,J., Otani,A., Ewalt,K.L., Friedlander,M., Cheresh,D.A. and Schimmel,P. (2002) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl Acad. Sci. USA, 99, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otani A., Slike,B.M., Dorrell,M.I., Hood,J., Kinder,K., Ewalt,K.L., Cheresh,D., Schimmel,P. and Friedlander,M. (2002) A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl Acad. Sci. USA, 99, 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko Y.G., Kim,E.Y., Kim,T., Park,H., Park,H.S., Choi,E.J. and Kim,S. (2001) Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J. Biol. Chem., 276, 6030–6036. [DOI] [PubMed] [Google Scholar]
- 16.Howard O.M., Dong,H.F., Yang,D., Raben,N., Nagaraju,K., Rosen,A., Casciola-Rosen,L., Härtlein,M., Kron,M., Yiadom,K. et al. (2002) Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J. Exp. Med., 196, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko Y.G., Kang,Y.S., Kim,E.K., Park,S.G. and Kim,S. (2000) Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol., 149, 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akins R.A. and Lambowitz,A.M. (1987) A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell, 50, 331–345. [DOI] [PubMed] [Google Scholar]
- 19.Kämper U., Kuck,U., Cherniack,A.D. and Lambowitz,A.M. (1992) The mitochondrial tyrosyl-tRNA synthetase of Podospora anserina is a bifunctional enzyme active in protein synthesis and RNA splicing. Mol. Cell. Biol., 12, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rho S.B. and Martinis,S.A. (2000) The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA, 6, 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheinker V.S., Beresten,S.F., Degtyarev,S.K. and Kisselev,L.L. (1979) The effect of tRNA and tryptophanyl adenylate on limited proteolysis of beef pancreas tryptophanyl-tRNA synthetase. Nucleic Acids Res., 7, 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Favorova O.O., Zargarova,T.A., Rukosuyev,V.S., Beresten,S.F. and Kisselev,L.L. (1989) Molecular and cellular studies of tryptophanyl-tRNA synthetases using monoclonal antibodies. Remarkable variations in the content of tryptophanyl-tRNA synthetase in the pancreas of different mammals. Eur. J. Biochem., 184, 583–588. [DOI] [PubMed] [Google Scholar]
- 23.Tolstrup A.B., Bejder,A., Fleckner,J. and Justesen,J. (1995) Transcriptional regulation of the interferon-gamma-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J. Biol. Chem., 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 24.Shaw A.C., Røssel Larsen,M., Roepstorff,P., Justesen,J., Christiansen,G. and Birkelund,S. (1999) Mapping and identification of interferon gamma-regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis, 20, 984–993. [DOI] [PubMed] [Google Scholar]
- 25.Turpaev K.T., Zakhariev,V.M., Sokolova,I.V., Narovlyansky,A.N., Amchenkova,A.M., Justesen,J. and Frolova,L.Y. (1996) Alternative processing of the tryptophanyl-tRNA synthetase mRNA from interferon-treated human cells. Eur. J. Biochem., 240, 732–737. [DOI] [PubMed] [Google Scholar]
- 26.Aiello L.P., Pierce,E.A., Foley,E.D., Takagi,H., Chen,H., Riddle,L., Ferrara,N., King,G.L. and Smith,L.E. (1995) Suppression of retinal neovascularization in vivo by inhibition of vascular endothelial growth factor (VEGF) using soluble VEGF-receptor chimeric proteins. Proc. Natl Acad. Sci. USA, 92, 10457–10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih S.C., Robinson,G.S., Perruzzi,C.A., Calvo,A., Desai,K., Green,J.E., Ali,I.U., Smith,L.E. and Senger,D.R. (2002) Molecular profiling of angiogenesis markers. Am. J. Pathol., 161, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 29.Farber J.M. (1993) HuMig: a new human member of the chemokine family of cytokines. Biochem. Biophys. Res. Commun., 192, 223–230. [DOI] [PubMed] [Google Scholar]
- 30.Strieter R.M., Kunkel,S.L., Arenberg,D.A., Burdick,M.D. and Polverini,P.J. (1995) Interferon gamma-inducible protein 10 (IP-10), a member of the C-X-C chemokine family, is an inhibitor of angiogenesis. Biochem. Biophys. Res. Commun., 210, 51–57. [DOI] [PubMed] [Google Scholar]
- 31.Bange F.C., Flohr,T., Buwitt,U. and Böttger,E.C. (1992) An interferon-induced protein with release factor activity is a tryptophanyl-tRNA synthetase. FEBS Lett., 300, 162–166. [DOI] [PubMed] [Google Scholar]
- 32.Fleckner J., Martensen,P.M., Tolstrup,A.B., Kjeldgaard,N.O. and Justesen,J. (1995) Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine, 7, 70–77. [DOI] [PubMed] [Google Scholar]
- 33.Barceló-Batllori S., André,M., Servis,C., Lévy,N., Takikawa,O., Michetti,P., Reymond,M. and Felley-Bosco,E. (2002) Proteomic analysis of cytokine induced proteins in human intestinal epithelial cells: implications for inflammatory bowel diseases. Proteomics, 2, 551–560. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J., Fritsch,E.F. and Maniatis,T. (2000) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 35.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frolova L.Y., Grigorieva,A.Y., Sudomoina,M.A. and Kisselev,L.L. (1993) The human gene encoding tryptophanyl-tRNA synthetase: interferon-response elements and exon-intron organization. Gene, 128, 237–245. [DOI] [PubMed] [Google Scholar]
- 37.Yuan W., Collado-Hidalgo,A., Yufit,T., Taylor,M. and Varga,J. (1998) Modulation of cellular tryptophan metabolism in human fibroblasts by transforming growth factor-beta: selective inhibition of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA synthetase gene expression. J. Cell. Physiol., 177, 174–186. [DOI] [PubMed] [Google Scholar]
- 38.Födinger M., Fritsche-Polanz,R., Buchmayer,H., Skoupy,S., Sengoelge,G., Hörl,W.H. and Sunder-Plassmann,G. (2000) Erythropoietin-inducible immediate-early genes in human vascular endothelial cells. J. Invest. Med., 48, 137–149. [PubMed] [Google Scholar]
- 39.Matsunaga T., Ishida,T., Takekawa,M., Nishimura,S., Adachi,M. and Imai,K. (2002) Analysis of gene expression during maturation of immature dendritic cells derived from peripheral blood monocytes. Scand. J. Immunol., 56, 593–601. [DOI] [PubMed] [Google Scholar]
- 40.Ewalt K.L. and Schimmel,P. (2002) Activation of angiogenic signaling pathways by two human tRNA synthetases. Biochemistry, 41, 13344–13349. [DOI] [PubMed] [Google Scholar]
- 41.Blanco J.C., Contursi,C., Salkowski,C.A., DeWitt,D.L., Ozato,K. and Vogel,S.N. (2000) Interferon regulatory factor (IRF)-1 and IRF-2 regulate interferon gamma-dependent cyclooxygenase 2 expression. J. Exp. Med., 191, 2131–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storm van's Gravesande K., Layne,M.D., Ye,Q., Le,L., Baron,R.M., Perrella,M.A., Santambrogio,L., Silverman,E.S. and Riese,R.J. (2002) IFN regulatory factor-1 regulates IFN-gamma-dependent cathepsin S expression. J. Immunol., 168, 4488–4494. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto H., Lamphier,M.S., Fujita,T., Taniguchi,T. and Harada,H. (1994) The oncogenic transcription factor IRF-2 possesses a transcriptional repression and a latent activation domain. Oncogene, 9, 1423–1428. [PubMed] [Google Scholar]
- 44.Yang D., Nakada-Tsukui,K., Ohtani,M., Goto,R., Yoshimura,T., Kobayashi,Y. and Watanabe,N. (2001) Identification and cloning of genes associated with the guinea pig skin delayed-type hypersensitivity reaction. J. Biochem. (Tokyo), 129, 561–568. [DOI] [PubMed] [Google Scholar]
- 45.Seshaiah P. and Andrew,D.J. (1999) WRS-85D: a tryptophanyl-tRNA synthetase expressed to high levels in the developing Drosophila salivary gland. Mol. Biol. Cell, 10, 1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gromov P., Skovgaard,G.L., Palsdottir,H., Gromova,I., Østergaard,M. and Celis,J.E. (2003) Protein profiling of the human epidermis from the elderly reveals up-regulation of a signature of interferon-gamma-induced polypeptides that includes manganese-superoxide dismutase and the p85-beta subunit of phosphatidylinositol 3-kinase. Mol. Cell. Proteomics, 2, 70–84. [DOI] [PubMed] [Google Scholar]
- 47.Aboagye-Mathiesen G., Ebbesen,P., von der Maase,H. and Celis,J.E. (1999) Interferon gamma regulates a unique set of proteins in fresh human bladder transitional cell carcinomas. Electrophoresis, 20, 344–348. [DOI] [PubMed] [Google Scholar]
- 48.Kumatori A., Yang,D., Suzuki,S. and Nakamura,M. (2002) Cooperation of STAT-1 and IRF-1 in interferon-gamma-induced transcription of the gp91(phox) gene. J. Biol. Chem., 277, 9103–9111. [DOI] [PubMed] [Google Scholar]