Abstract
Nutritional and genetic factors influence aging and life expectancy. The reduction of food intake without malnutrition, referred to caloric restriction (CR), has been shown to increase lifespan in a wide variety of species. The nematode Caenorhabditis elegans (C. elegans) is one of the principle models with which to study the biology of aging and search for anti-aging compounds. In this study, we validated and optimized a high-throughput liquid culture system to monitor C. elegans lifespan with minimized mechanical stress. We used alive and ultraviolet (UV)-killed Escherichia coli (E. coli) OP50 at 108 or 109 colony-forming units (cfu)/ml to feed Bristol N2 wild-type (WT) and mutant worms of a well-characterized insulin/insulin-like growth factor signaling (ILS) pathway: the insulin receptor homolog daf-2 (e1370), phosphatidylinositol 3-kinase age-1 (hx546), and transcriptional factor FOXO homolog daf-16 (mu86 and mgDf50). Compared with alive E. coli at 109 cfu/ml, supplementations of alive E. coli at 108 cfu/ml or UV-killed E. coli at 109 cfu/ml dramatically prolonged lifespan in WT and age-1 mutants, and to a lesser extent, in daf-2 and daf-16 mutants, suggesting that signaling pathways in CR and ILS do not overlap fully. Feeding 108 cfu/ml UV-killed E. coli, which led to maximally saturated longevity in WT and daf-2 mutant, can prolonged lifespan in age-1, but not daf-16, mutants. This approach will be useful for investigating the biology of aging, physiological responses and gene functions under CR conditions and also for screening pharmacologic compounds to extend lifespan or affect other biologic processes.
Keywords: C. elegans, aging, lifespan, caloric restriction
Aging eventually affects every living thing with an intrinsic and progressive decline in function. For decades, researchers have been exploring aging mechanisms and trying to discover methods to prolong lifespan and reduce age-related diseases [1]. Lifespan is influenced by many factors including nutrients and genetic determinants. Among them, caloric restriction (CR) is one of the most robust and reproducible interventions known to extend lifespan and delay the onset of age-related phenotypes in a wide range of species [2–5]. The efficacy of CR in primates has not yet been fully resolved, but a conclusive settlement is eagerly awaited [6,7].
The nematode Caenorhabditis elegans (C. elegans) has proved an excellent and convenient model of 2- to 3-week lifespan for the study of dietary and genetic determinants of longevity and aging [1,8]. A good example of the genetic factors in play is the insulin/insulin-like growth factor signaling (ILS) pathway [9]. The lifespan of C. elegans reportedly doubles owing to mutations in the age-1 or daf-2 genes, orthologs of the mammalian phosphatidiylinositol 3-kinase and insulin/insulin-like growth factor-1 receptor, respectively [10,11]. CR can further lengthen lifespan of long-lived age-1 and daf-2 mutants [12–14]. In addition, the FOXO transcription factor daf-16 has been discovered to be the primary transcription factor required for the profound lifespan extension observed in daf-2 mutants [15]. Thus, daf-16 null mutants have shortened lifespans.
To accomplish a fixed amount of feeding for CR assays, ultraviolet (UV)-killed Escherichia coli (E. coli) instead of proliferating alive E. coli is provided to C. elegans. The feeding of UV-killed E. coli reportedly increases C. elegans lifespan 20% relative to that of animals maintained on alive E. coli. Toxins produced by the alive bacteria and bacterial invasion explain the vulnerability [16].
Despite accumulating results, experimental systems for monitoring lifespan of C. elegans are not fixed; in some cases, assays have been carried out in liquid media, whereas other studies have been used nematode growth agar medium (NGM) plates. Whereas NGM agar plates are the standard and common method for the cultivation of C. elegans, they have several disadvantages, including losing or killing worms unintentionally during their transfer and through desiccation after they crawl up the wall of the plate. On the contrary, a liquid culture system overcomes these disadvantages and may be more useful and convenient for testing a variety of compounds, fixed concentrations of feeding bacteria, or combinations thereof through addition to the culture media using high-throughput experiments.
In this study, we implemented a C. elegans liquid culture system to optimize lifespan extension using Bristol N2 wild-type (WT) and genetic mutant worms fed alive or UV-killed E.coli in 96-well plates. This approach will be an excellent model for high-throughput drug and chemical screenings to identify positive candidates for lifespan prolongation. This system will also be useful for studying physiological responses and gene functions in aging processes and under CR conditions.
MATERIALS AND METHODS
C. elegans strain
N2 Bristol (WT), long lived mutants of daf-2 (e1370) and age-1 (hx546) and short lived mutants of daf-16 (mu86) and daf 16 (mgDf50) were used [3,17,18].
Culture conditions
C. elegans strains were maintained at 20°C on NGM with alive E. coli OP50 as the food source as described elsewhere [19]. Liquid culture conditions of C. elegans with 96-well plates were used [20] with a minor modification. Briefly, worms were maintained in S complete media containing ampicillin (50 μg/ml) (Wako Pure Chemical Industries, Japan) and amphotericin B (0.1 μg/ml) (Sigma Aldrich, USA). To prevent hatching, we included 50 μg/ml 5-fluoro-2′-deoxyuridine (5-FUdR) in the media. All experiments were performed at 20°C.
Preparation of E. coli
To assess the lifespan of worms on NGM plates, we used M9 medium before synchronization to prepare E. coli OP50. For the liquid culture system, ampicillin-resistant E. coli OP50 was used to avoid bacterial contamination. Colony-forming units (cfu) per milliliter of the E. coli were determined after overnight cultivation at 37°C on agar plates. After being counted, alive E. coli were irradiated with 254-nm UV light using a UV Stratalinker model 2400 (Stratagene, USA) at 9,999 J/cm2 for 30 minutes. The complete killing of E. coli was confirmed by spreading UV-exposed E. coli on agar plates and incubating overnight at 37°C.
Lifespan determination
Worms were synchronized via hypochlorite bleaching, hatched overnight, and subsequently cultured on NGM plates. The day of synchronization was regarded as day 0. Age-synchronized worms were monitored until all were dead, which was declared when the worms did not move after repeated stimuli. For measuring lifespan on NGM plates, WT C. elegans were transferred to the NGM plates containing 100 μg/ml 5-FUdR and fed alive or UV-killed E. coli after 7 hours of synchronization. Worms were transferred to new plates using a worm picker, and their viability was simultaneously checked every other day. Animals that crawled away from the plate or showed internal hatching were excluded. Experiments were performed twice independently.
For the liquid culture system, worms hatched overnight were transferred to NGM plates (10-cm diameter) after synchronization. At the young adult stage on day 3, worms were transferred to 96-well plates with indicated concentrations of alive or UV-killed E. coli. Each well contained 5–15 worms in 150 μl S complete medium containing 50 μg/ml ampicillin and 0.1 μg/ml amphotericin B. After the wells were shaken for 3 minutes, worm survival was checked every other day under an inverted microscope (Axiovert 35, Zeiss, Germany) at 50x or 100x magnification. To confirm whether worms were alive or dead, we activated their movement via exposure to strong lights in some cases. Culture media containing the indicated concentrations of alive or UV-killed E. coli were exchanged 4 times per week.
Statistical analysis
Kaplan-Meier curves were generated using survival duration in days for each worm. We compared curves for the 2 groups with the log-rank test. All values are expressed as means ± standard error (SE). Differences in the mean values between 2 groups were assessed using the 2-tailed Student’s t-test. Differences in the mean values among more than 2 groups were determined with analysis of variance. A value of p < 0.05 was considered statistically significant.
RESULTS
Most of the CR studies reported to date have been performed using agar plates to assess the lifespan of C. elegans. We examined first whether feeding with UV-killed E. coli (109 cfu/ml) could affect the lifespan of Bristol N2 WT worms on NGM agar plates compared with lifespan achieved with alive E. coli (109 cfu/ml) feeding. The mean, median and maximal lifespans of WT were significantly longer in the UV-killed E. coli group (20, 18 and 40 days) compared with those in the alive E. coli group (16, 15 and 24 days, respectively) (Fig. 1). This difference is compatible to that of previous reports [21].
Figure 1.
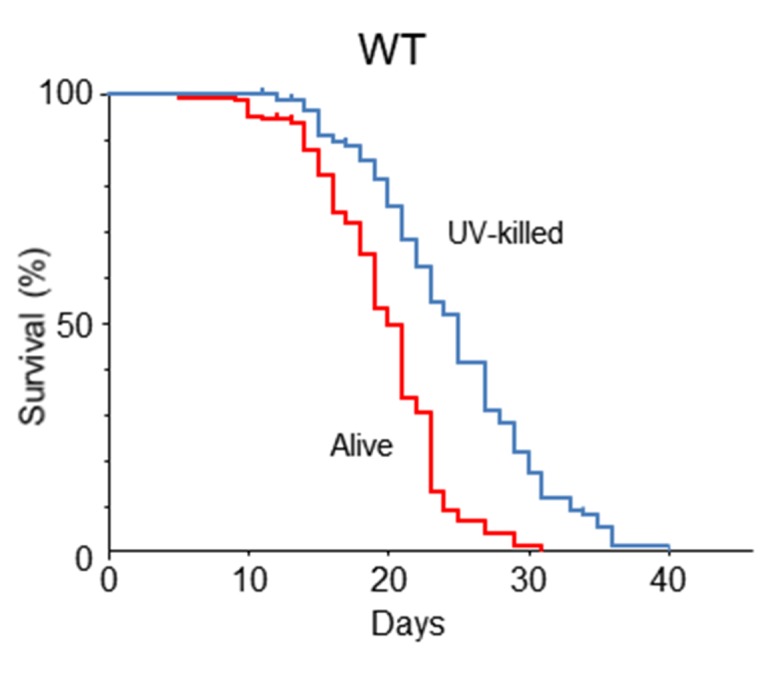
C. elegans survival in an agar-based system. Kaplan-Meier survival curves of N2 Bristol wild-type (WT) C. elegans fed alive E. coli (109 cfu/ml) and UV-killed E. coli (109 cfu/ml) on solid nematode growth medium. n = 68 (alive E. coli), n = 70 (UV-killed E. coli), Log-rank test, p < 0.0001. Values represent the mean ± SE of the sum of 2 independent experiments.
In this study, we validated and optimized a liquid culture system for C. elegans for the observation of lifespan extension using Bristol N2 WT and genetic mutant worms of the insulin receptor homolog daf-2 (e1370), phosphatidylinositol 3-kinase age-1 (hx546), and transcriptional factor FOXO homolog daf-16 (mu86 and mgDf50) through the feeding of alive or UV-killed E.coli in 96-well plates (Fig. 2). The daf-2 and age-1 mutants have longer lifespans, whereas daf-16 mutants are models for shorter lifespans, compared with WT worms under non-CR conditions.
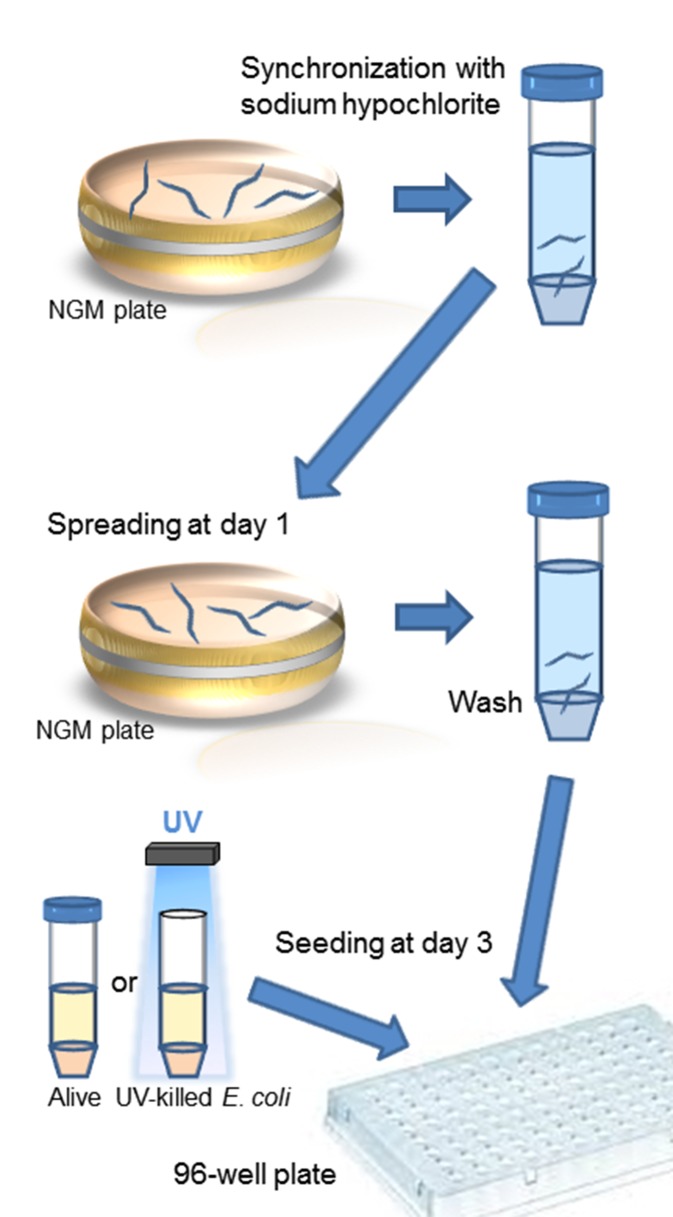
Figure 2.
Schematic representation of our liquid culture protocol for monitoring C. elegans survival. NGM, nematode growth medium.
In pilot studies, 3 concentrations of alive or UV-killed E. coli (109, 108 or 107 cfu/ml) were fed to the worms in 96-well plates. We found that feeding a 107 cfu/ml concentration of alive or UV-killed E. coli did not extend lifespan compared with feeding 108 cfu/ml E. coli, which instead shortened the lifespan of daf-16 mutants (data not shown), suggesting that feeding 107 cfu/ml E. coli could be considered a malnutrition condition in our experimental system. Therefore, we used 109 and 108 cfu/ml of E. coli for further studies.
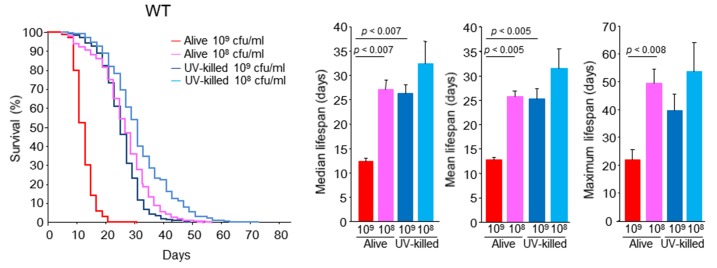
In WT C. elegans, feeding 108 cfu/ml alive E. coli significantly extended the mean, median and maximum lifespans of C. elegans 101.6%, 119.5%, and 125%, respectively, of that achieved feeding 109 cfu/ml alive E. coli (Fig. 3). The results clearly demonstrated that CR conditions prolonged WT C. elegans lifespan and slowed the aging of worms in this liquid culture system. Feeding with non-proliferating and UV-killed E. coli is also one of the CR methods used to examine C. elegans longevity [22]; fixed and stable concentrations of E. coli are provided to the worms. Feeding 109 cfu/ml UV-killed E. coli significantly prolonged the mean and median lifespans of WT C. elegans to the levels seen under the CR condition with 108 cfu/ml alive E. coli (Fig. 3).
Figure 3.
N2 Bristol WTC. elegans survival in the liquid culture system. Kaplan-Meier survival curves and median, mean, and maximal lifespans of WT C. elegans fed alive E. coli (109 or 108 cfu/ml) and UV-killed E. coli (109 or 108 cfu/ml). n = 396 (109 cfu/ml alive E. coli), n = 300 (109 cfu/ml UV-killed E. coli), n = 384 (108 cfu/ml alive E. coli), n = 300 (108 cfu/ml UV-killed E. coli), Log-rank test, p < 0.0001 between alive E. coli at 109 and 108 cfu/ml, between UV-killed E. coli at 109 and 108 cfu/ml, between 109 cfu/ml alive E. coli and 109 cfu/ml UV-killed E. coli, and between 108 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli 108. Values represent the mean ± SE of the sum of 3 independent experiments.
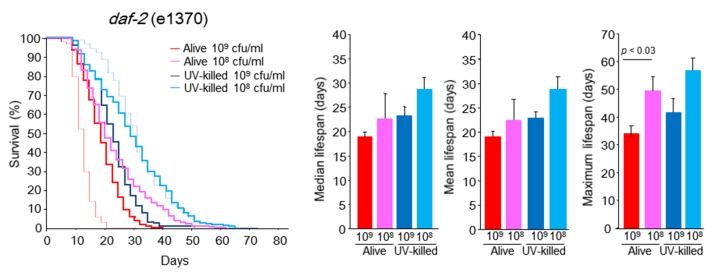
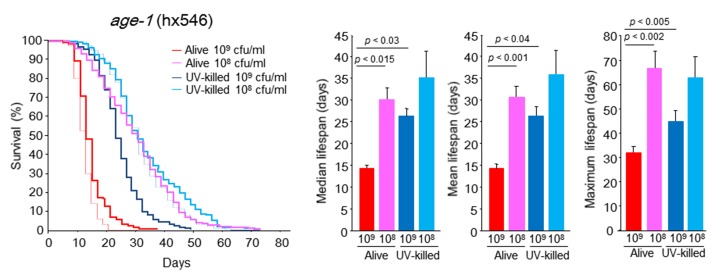
Next, we undertook experiments with longer survival worm models of daf-2 (e1370) and age-1 (hx546) mutants. The daf-2 mutants fed 108 cfu/ml alive or UV-killed E. coli showed survival curves that were significantly shifted to the right, but the mean and median lifespan values were not significantly longer than that of worms given 109 cfu/ml alive E. coli (Fig. 4). The daf-2 mutants fed 109 cfu/ml alive E. coli survived longer than WT worms did under this culture condition (Figs. 3 and 4). When age-1 mutants were subjected to 108 cfu/ml alive E. coli, a significant increase in survival was noted, and their mean, median, and maximal lifespans were significantly expanded by 107.5%, 121.6% and 106.3%, respectively, compared with those under the feeding condition of 109 cfu/ml alive E. coli (Fig. 5). In case of feeding UV-killed E. coli, we also observed significantly longer survival of age-1 mutants at the concentration of 108 cfu/ml compared with 109 cfu/ml (Fig. 5). Even in the longer survival models of daf-2 and age-1 mutants, 108 cfu/ml UV-killed E. coli yielded maximally saturated and prolonged survival curves similar to those seen in WT worms (Figs. 4 and 5). Non-overlapping signaling pathways between daf-2 and age-1 mutants may be involved in survival under the feeding condition of 109 cfu/ml alive E. coli.
Figure 4.
daf-2 (e1370) C. elegans survival in the liquid culture system. Kaplan-Meier survival curves and median, mean, and maximal lifespan of daf-2 (e1370) C. elegans fed alive E. coli (109 or 108 cfu/ml) and UV-killed E. coli (109 or 108 cfu/ml). Dashed red and blue lines indicate Kaplan-Meier survival curves of WT C. elegans fed 109 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli, respectively. n = 350 (109 cfu/ml alive E. coli), n = 300 (109 cfu/ml UV-killed E. coli), n = 350 (108 cfu/ml alive E. coli), n = 300 (108 cfu/ml UV-killed E. coli), Log-rank test, p < 0.0001 between alive E. coli at 109 and 108 cfu/ml, between UV-killed E. coli at 109 and 108 cfu/ml, between 109 cfu/ml alive E. coli and 109 cfu/ml UV-killed E. coli, and between 108 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli. Values represent the mean ± SE of the sum of 3 independent experiments.
Figure 5.
age-1 (hx546) C. elegans survival in the liquid culture system. Kaplan-Meier survival curves and median, mean, and maximal lifespan of age-1 (hx546) C. elegans fed alive E. coli (109 or 108 cfu/ml) and UV-killed E. coli (109 or 108 cfu/ml). Dashed red and blue lines indicate Kaplan-Meier survival curves of WT C. elegans fed 109 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli, respectively. n = 400 (109 cfu/ml alive E. coli), n = 300 (109 cfu/ml UV-killed E. coli), n = 400 (108 cfu/ml alive E. coli), n = 300 (108 cfu/ml UV-killed E. coli), Log-rank test, p < 0.0001 between alive E. coli at 109 and 108 cfu/ml, between UV-killed E. coli at 109 and 108 cfu/ml, between 109 cfu/ml alive E. coli and 109 cfu/ml UV-killed E. coli, and between 108 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli. Values represent the mean ± SE of the sum of 3 independent experiments.
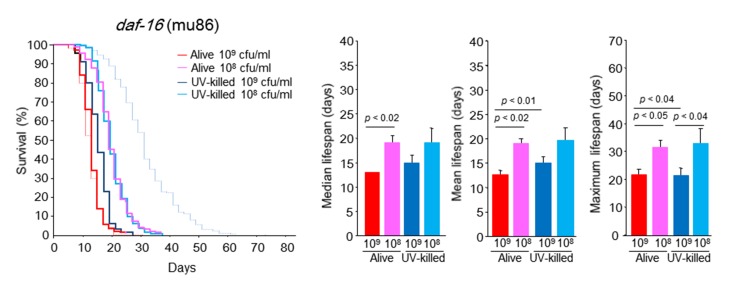
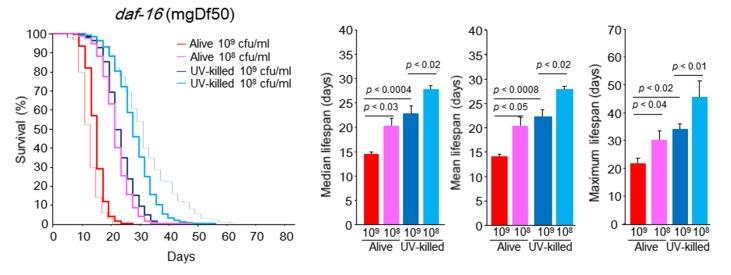
When the short lifespan models of mutant worms of daf-16 (mu86) and daf-16 (mgDf50) were fed 109 cfu/ml alive E. coli, their survival curves were very similar to those of WT worms fed 109 cfu/ml alive E. coli (Figs 3, 6 and 7). This result led us to suspect that DAF-16 activity would be inhibited in WT animals fed 109 cfu/ml alive E. coli. The CR-induced lifespan extension of daf-16 mutants occurred to lesser degree than that in WT worms (Figs. 6 and 7). However, in both the daf-16 (mu86) and daf-16 (mgDf50) mutants fed 108 cfu/ml alive E. coli, the mean, median and maximal lifespans were significantly 50%, 46.2% and 43.2% longer and 42.7%, 42.0% and 40.9% longer, respectively, than that of the groups fed alive E. coli at 109 cfu/ml (Figs. 6 and 7). The feeding of UV-killed E. coli improved C. elegans longevity but the increase did not reach the level seen in WT worms fed 108 cfu/ml UV-killed E. coli (Figs. 3, 6 and 7).
Figure 6.
daf-16 (mu86) C. elegans survival in the liquid culture system. Kaplan-Meier survival curves and median, mean, and maximal lifespan of daf-16 (mu86) C. elegans fed alive E. coli (109 or 108 cfu/ml) and UV-killed E. coli (109 or 108 cfu/ml). Dashed red and blue lines indicate Kaplan-Meier survival curves of WT C. elegans fed 109 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli, respectively. n = 400 (109 cfu/ml alive E. coli), n = 300 (109 cfu/ml UV-killed E. coli), n = 400 (108 cfu/ml alive E. coli), n = 300 (108 cfu/ml UV-killed E. coli), Log-rank test, p < 0.0001 between alive E. coli at 109 and 108 cfu/ml, between UV-killed E. coli at 109 and 108 cfu/ml, and between 109 cfu/ml alive E. coli at 109 and 109 cfu/ml UV-killed E. coli. Values represent the mean ± SE of the sum of 3 independent experiments.
Figure 7.
daf-16 (mgDf50) C. elegans survival in the liquid culture system. Kaplan-Meier survival curves and median, mean, and maximal lifespan of daf-16 (mgDf50) C. elegans fed alive E. coli (109 or 108 cfu/ml) and UV-killed E. coli (109 or 108 cfu/ml). Dashed red and blue lines indicate Kaplan-Meier survival curves of WT C. elegans fed 109 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli, respectively. n = 400 (109 cfu/ml alive E. coli), n = 300 (109 cfu/ml UV-killed E. coli), n = 400 (108 cfu/ml alive E. coli), n = 300 (108 cfu/ml UV-killed E. coli), Log-rank test, p < 0.0001 between alive E. coli at 109 and 108 cfu/ml, between UV-killed E. coli at 109 and 108 cfu/ml, between 109 cfu/ml alive E. coli and 109 cfu/ml UV-killed E. coli, and between 108 cfu/ml alive E. coli and 108 cfu/ml UV-killed E. coli. Values represent the mean ± SE of the sum of 3 independent experiments.
DISCUSSION
CR is one of the well-characterized strategies for retarding senescence, slowing aging, reducing mortality, and prolonging lifespan. In C. elegans, the CR condition of 5 × 108 bacteria/ml alive E. coli on agar plates has been reported to increase mean lifespan by 13.2% compared with that of the non-CR condition of 5 × 1010 bacteria/ml alive E. coli ad libitum[23]. Both dilution of food bacteria and deprivation of the bacteria are known to extend the lifespan of WT C. elegans[24]. In mice, a 40% decrease in food intake reportedly extends lifespan up to 43% [25]. Numerous hypotheses and pathways underlie CR-mediated longevity: reduction of metabolic rate and ILS pathways, activation of AMP-activated protein kinase, glyoxylate shunt, SKN-1, and daf-16 dependence [26–28].
In this study, we reproduced and observed significant prolongation of lifespan in C. elegans of daf-2 (e1370) and age-1 (hx546) mutants and shortened lifespan in daf-16 (mu86) and daf-16 (mgDf50) mutants using our liquid culture system, compared with that in WT worms at the same concentration of E. coli feeding (Figs. 3–7). The liquid culture system has advantages of minimized mechanical stress to C. elegans and a convenient method for testing compounds. CR of E. coli feeding from 109 to 108 cfu/ml also significantly extended lifespan in WT and all C. elegans mutants examined with the system (Figs. 1 and 3–7). We thus conclude that this liquid culture system will be useful for identifying positive candidates for lifespan prolongation via high-throughput drug and chemical screening. As an example of pharmacological intervention, treatment of metformin, a biguanide drug, significantly extended the lifespan of WT worms fed with 109 cfu/ml alive E. coli (data not shown). The beneficial effect of metformin has been reported previously [29]. We are confident that this liquid system will be commonly implemented in the near future. It will be powerful to be able to screen chemicals or compounds that prolong the lifespans of daf-2 mutants by feeding 108 cfu/ml UV-killed E. coli, which showed the maximal longest lifespan, and those of daf-16 mutants by feeding 109 cfu/ml alive E. coli, which showed the shortest lifespan in this study. We hope that novel pathways or unique genes will be identified in these screenings using our system.
In conclusion, we used for the first time a liquid culture system to optimize lifespan extension in C. elegans with Bristol WT and genetic mutant worms by feeding alive or UV-killed E. coli. This approach was evaluated as a useful tool in the high-throughput screening of drugs, chemicals and natural compounds for lifespan prolongation and in the study of physiological responses and gene functions under CR conditions and in aging processes.
Acknowledgments
We thank Ms Yuko Niimura for her assistance and Dr. Sumino Yanase (Daito Bunka University School of Sports and Health Science) for kindly providing daf-2 (e1370), age-1 (hx546), daf-16 (mu86), and daf 16 (mgDf50) worms. Editage reviewed the manuscript before submission. This study was supported by a Grant-in-Aid for Scientific Research (24590375) from the Japan Society for the Promotion of Sciences.
Footnotes
Disclosure Statement
The authors declare no competing interests.
References
- [1].Vanfleteren JR, Braeckman BP. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- [2].Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- [3].Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- [4].Barbieri M, Bonafè M, Franceschi C, Paolisso G. Insulin/IGF-1-signaling pathway: An evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab. 2003;285:1064–1071. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- [5].Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- [6].Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, et al. Caloric restriction delays disease onset and mortality in rhesus monkey. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Finch CE, Ruvkun G. The genetics of aging. Ann Rev Genomics Hum Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- [9].Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin- like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- [10].Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A Caenorhabditis elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- [11].Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signaling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- [14].Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, et al. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in Caenorhabditis elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- [16].Hansen E, Buecher EJ, Yarwood EA. Development and maturation of Caenorhabditis briggsae in response to growth factor. Nematologica. 1964;10:623–30. [Google Scholar]
- [17].Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Apfeld J, Kenyon C. Cell nonautonomy of Caenorhabditis elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- [19].Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Solis GM, Petrascheck M. Measuring Caenorhabditis elegans lifespan in 96 well plates. J Vis Exp. 2011;18:e49. doi: 10.3791/2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in Caenorhabditis elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Evason K, Huang C, Yamben I, Covey DF, Kornfeld K. Anticonvulsant medications extend worm lifespan. Science. 2005;307:258–262. doi: 10.1126/science.1105299. [DOI] [PubMed] [Google Scholar]
- [23].Honjoh S, Yamamoto T, Uno M, Nashida E. Signaling through RHEB-1 mediates intermittent fasting-induced longevity in Caenorhabditis elegans. Nature. 2008;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- [24].Sutphin GL, Kaeberlein M. Dietary restriction by bacterial deprivation increase lifespan in wild-derived nematodes. Exp Gerontol. 2008;43:130–135. doi: 10.1016/j.exger.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sohal RS, Ku HH, Agarwal S, Forster MJ, Lal H. Oxidative damage, mitochondrial oxidant generation and antioxidant defenses during aging and in response to food restriction in the mouse. Mech Ageing Dev. 1994;74:121–133. doi: 10.1016/0047-6374(94)90104-x. [DOI] [PubMed] [Google Scholar]
- [26].Johnson TE, Friedman DB, Foltz N, Fitzpatrick PA, Shoemaker JE. Genetic variants and mutations of C. elegans provide tools for dissecting the aging processes. In: Harrison DE, editor. Genetic Effects on Aging Caldwell. Vol. 2. New Jersey: Telford press; 1990. pp. 101–126. [Google Scholar]
- [27].Fuchs S, Bundy JG, Davies SK, Viney JM, Swire JS, Leroi AM. A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 2010;8:14. doi: 10.1186/1741-7007-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in Caenorhabditis elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- [29].Onken B, Driscoll M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans healthspan via AMPK, LKB1, and SKN-1. PLoS ONE. 2010;5:e8758. doi: 10.1371/journal.pone.0008758. [DOI] [PMC free article] [PubMed] [Google Scholar]