Abstract
The expression of several muscle-specific genes is partially or completely regulated by MCAT elements, which bind members of the TEF family of transcription factors. TEF1 itself is unable to activate reporter plasmids bearing TEF1-binding sites, suggesting that additional bridging or co-activating factors are necessary to allow interaction of TEF1 with the transcriptional machinery. In addition, none of the known TEF genes are exclusively expressed in the cardiac or skeletal muscle lineage to account for the muscle-specific expression of MCAT-dependent genes. Here we describe that VITO-1, a new SID (scalloped interaction domain)-containing protein, binds to TEF1 in vitro and strongly stimulates transcription of a MCAT reporter plasmid together with TEF-1. Since VITO-1 is predominantly expressed in the skeletal muscle lineage, it might serve as an essential transcriptional intermediary factor to promote muscle-specific expression via MCAT cis-regulatory elements. Although VITO-1 alone is not sufficient to initiate myogenic conversion of 10T1/2 fibroblastic cells, it enhanced MyoD-mediated myogenic conversion. In addition, interference with VITO-1 expression by siRNA attenuated differentiation of C2C12 muscle cells and MyoD-dependent myogenesis in 10T1/2 cells. We conclude that VITO-1 is a crucial new cofactor of the muscle regulatory programme.
INTRODUCTION
Transcriptional control of skeletal muscle-specific gene expression is achieved by the combined action of various transcription factors that are either specifically expressed in the skeletal muscle lineage or show a more widespread expression pattern. Several groups of transcription factors such as the MyoD (1), MEF-2 (2) and TEF families of DNA-binding proteins (3) have been identified by different means as playing important roles in the development of muscle tissues and in the regulated expression of muscle-specific genes. Despite a considerable gain in knowledge concerning the biological role of these molecules, it remains a challenge to understand how these factors act on a molecular level.
It seems rather easy to anticipate how tissue-specific regulation of muscle cell-specific genes might be achieved by transcription factors that are themselves tissue-specific, such as the MyoD family of transcription factors (4–6). A variation on this theme is a combination of basal factors with tissue-specific transcription factors or an exclusive blend of transcription factors, which are not unique as individual molecules in a given tissue but exclusive with respect to their joint expression profiles. A seeming paradox to this rule is the finding that individual DNA-binding sites, which bind widely expressed transcription factors such as serum response factors (SRF) (7) and transcriptional enhancer factors (TEF), are able to confer muscle-specific expression of individual genes. Examples comprise the skeletal α-actin and cardiac Troponin T genes, which depend on SRF and TEF transcription factors, respectively. In such cases muscle-specific transcription might be achieved by the assembly of larger transcriptional complexes, which utilize DNA-binding proteins as docking stations for tissue-specific bridging or co-activating factors that might confer additional activities to DNA-bound proteins (8).
So far the TEF family of transcription factors encompasses four different members that have been cloned in different organisms such as human, mouse and chicken. All family members show complex expression patterns in various tissues. TEF-1 has attracted particular attention since its inactivation by a gene trap approach in the mouse revealed its critical importance for cardiac muscle development. TEF-1 homozygous null mice die between E10.5 and E11.5 from cardiac insufficiency (9). Redundancy between different TEF proteins seems possible since TEF proteins, which share a common TEA DNA-binding domain, all bind to the previously defined TEF consensus binding site (SV40 GT-IIC) despite several amino acid substitutions in the conserved DNA-binding domain (10,11). On the other hand, TEF proteins differ in their ability to bind cooperatively to tandemly repeated binding sites (11). It also seems probable that the amino acid substitution in the TEA domain might enable individual TEFs to bind to other sites and/or to interact with different sets of proteins.
TEF-1 was found to be entirely inactive after transfection into cell lines in which the endogenous protein is not present (12). Likewise, overexpression of TEF-1 and TEF-3 in cell lines producing endogenous TEF-1 or TEF-3 led to a strong repression of reporter plasmids that were dependent on MCAT-binding sites to which TEFs attach (13,14). It has been proposed that the failure of TEF-1 to transactivate reporter plasmids in transfection experiments is due to the limiting presence of co-activators or transcriptional intermediary factors that allow TEFs to interact with the general transcription machinery. However, no TEF-1 cofactors have been identified in vertebrates so far.
The TEA/ATTS domain, which mediates DNA binding of TEF proteins, is conserved during evolution (15). It has been identified in different proteins such as TEC1, AbaA (16) and scalloped (Sd) (17) found in yeast, Aspergillus nidulans and Drosophila melanogaster, respectively. The functional properties of the TEA/ATTS domain appear to be sufficient for the function of the protein since TEF-1, which has little similarity to the Drosophila scalloped protein outside the TEA/ATTS domain, is able to substitute for the loss of scalloped in Drososphila mutants (18). Both vertebrate TEF-1 and Drosophila scalloped show similar DNA-binding specificities in vitro (19). In Drososphila wing cells Sd interacts with vestigal (Vg) via its scalloped interaction domain (SID) (19,20). Both proteins form a wing-specific complex that acts as a selector for wing development (19,21). It has been postulated that Vg might act as a transcriptional activator that is recruited by Sd (22). In another study it was shown that Vg changed the DNA target selectivity of Sd, thereby demonstrating that the presence or absence of Vg determines the set of cis-regulatory elements bound and regulated by Sd (23).
Recently, we have identified a novel SID-containing protein in mouse and humans using a subtractive hybridization approach for genes expressed in skeletal muscle but not in other tissues (24), as well as in another screen for target genes of Lbx1 (unpublished data), which controls the fate of limb muscle precursor cells (25). This protein, which we named VITO-1 due to its homology to the Drosophila vestigial and the human TONDU proteins in the SID (54 and 40%, respectively) is expressed in the somitic myotome from E8.75 mouse embryos onwards and later on in skeletal muscle but not in the heart (24). VITO-1 is up-regulated in differentiated C2C12 myotubes, although some expression was detected in proliferating C2C12 myoblasts.
Here we report that VITO-1 binds to TEF1 in vitro and modifies its DNA-binding properties. It strongly stimulates transcription of a MCAT reporter plasmid together with TEF-1 but not with TEF-3. Furthermore, we show that VITO-1 enhances MyoD-mediated myogenic conversion in 10T1/2 cells. In addition, interference with VITO-1 expression by siRNA diminishes MyoD-dependent myogenesis in 10T1/2 cells and attenuates differentiation of C2C12 cells. We conclude that VITO-1 is an essential cofactor of the muscle regulatory programme.
MATERIALS AND METHODS
Construction of plasmids
For expression of VITO-1 in eukaryotic cells and for coupled in vitro transcription/translation the VITO-1 cDNA was amplified by PCR and inserted into pCS2+MT either before or after the myc tag epitope. Sequences for human (AJ578053) and mouse VITO-1 (AJ578054) cDNAs have been deposited in the EMBL Nucleotide Sequence Database. All constructs used in this report were derived from mouse cDNA. Deletion mutants of VITO-1 lacking the C-terminus or the SID domain were generated by PCR and inserted into pCS2+MT. Plasmids for expression of mouse TEF-1 and TEF-3 in vitro and in vivo were generously supplied by I. Davidson (Strasbourg, France). The TEF reporter plasmid is based on pTA-Luc (BD Bioscience) and contains either one or four copies of a TEF-1 consensus binding site (CATTCCA) inserted into the XhoI site. GAL4-VITO-1 fusion genes were constructed by insertion of different PCR-amplified VITO-1 cDNA fragments into the BamHI site of pGM4polyII (4) containing the GAL4 DNA-binding domain (VHD-1-SID-VHD-2, full-length VITO-1, amino acids 1–321; VHD-1-SID, amino acids 1–169; SID, amino acids 62–169; VHD-2; amino acids 169–321). siRNA against VITO-1 and Myogenin was synthesized from constructs based on pSuper (kindly supplied by R. Agami, Amsterdam, The Netherlands). In order to obtain pSuper-Vito-1, a 64 nt long double-stranded fragment with the sequence 5′-GAT CCC CGA CAT CAG CTC TGT GGT GGT TCA AGA GAC CAC CAC AGA GCT GAT GTC TTT TTG GAA A-3′ was cloned into the HindIII and EcoRI sites of pSUPER. pSuper-Myogenin was constructed similarly by inserting a 64 nt double-stranded fragment with the sequence 5′-GAT CCC CGT GAA TGA GGC CTT CGA GGT TCA AGA GAC CTC GAA GGC CTC ATT CAC TTT TTG GAA A-3′. The efficiency of siRNA-mediated knockdown of VITO-1 expression was controlled using a GFP–VITO-1 fusion construct. The GFP vector used in co-transfection experiments was pEGFP-C2 (Clontech). All constructs were sequenced using an ABI 310 DNA sequencer.
Electrophoretic mobility shift assays (EMSAs) and co-immunoprecipitation assays
Recombinant proteins used for EMSAs and co-immunoprecipitation assays were obtained by coupled in vitro transcription/translation using the TNT system (Promega) following the instructions of the manufacturer. EMSA reactions were performed as described previously (26) using equimolar amounts of 35S-labelled proteins in each binding reaction. The following binding sites were used: Bs01, 5′-CCG GCG ATC ATT CCA CAT GCC GG-3′; Bs02, 5′-CCG GCG ATC ATT CCC CAT GCC GG-3′; Bs03, 5′-CCG GCG ATC ATT CCT CAT GCC GG-3′. Bs01 is present as the SV40GTIIC binding site in the SV40 promoter/enhancer, the β-MyHC promoter of human, rat and rabbit and in the promoter of the α-MyHC gene of the rat. Bs02 is present in the promoters of the skeletal and cardiac TnC genes of the mouse. Bs03 is present in the promoters of the chicken cTnT gene, the α-MyHC gene of the rat, the skeletal α-actin gene of chicken and the ACh receptor β gene of the rat (for further details see 3). Complexes were separated on 5% polyacrylamide gels in 0.5× TBE buffer. Immunoprecipitation was performed as outlined previously using antibodies against the myc tag and the bHLH protein E2-2 and protein G–Sepharose (26). Bound proteins were separated by SDS–PAGE and visualized by fluorography using BiomaxMR film (Kodak).
Cell culture, transactivation and gene silencing assays
Luciferase reporter constructs were transactivated by transfecting 2 µg of expression plasmids coding for TEF-1 and VITO-1 along with 2 µg of the reporter plasmid and 0.2 µg of RSV-β-galactosidase plasmid as internal standard into C3H10T1/2, HEK293 and C2C12 cells. DNA transfection was performed by calcium phosphate precipitation. After transfection the medium was changed to DMEM containing 4% horse serum to induce terminal differentiation of C2C12 cells. Luciferase activity was determined 2–3 days after transfection by standard methods in 10–30% cellular extracts. The presented data were derived from at least three independently performed transfections for each individual experiment. The transactivation ability of VITO-1 was investigated by co-transfection of VITO-1-GAL4 DNA-binding domain fusion genes together with a GAL4 reporter plasmid into HEK293 and C2C12 cells (4). CAT activity was determined as described previously and compared to the activity evoked by the GAL4 DNA-binding domain without a transactivation domain (4). Potential interactions between VITO-1 and MEF2C were studied by co-transfection of VITO1, MEF2C and TEF-1 together with a CAT reporter plasmid carrying functional or mutated (mt) oligomerized MEF2-binding sites in front of a minimal TK promoter (plasmid pE102MEF2×2CAT) (27) into HEK293 cells, C2C12 myoblasts and C2C12 myotubes (28). All transfection assays were normalized to β-galactosidase activities.
To knockdown VITO-1 and Myogenin expression C2C12 cells were transfected using 2.5 µg pEGFP, 15 µg pSuper, 15 µg pSuper-Vito and 15 µg pSuper-Myogenin, respectively, per 6 cm dish. After 8 h the medium was changed to differentiation medium consisting of DMEM containing 2% horse serum. Expression of Myogenin and MyHC was scored 72 h after transfection as described (29). C3H10T1/2 mouse mesenchymal cells were converted to myoblasts using pEMSV-MyoD (30). Aliquots of 2 × 106 C3H10T1/2 were electroporated in 400 µl of medium containing 75% cytosalts and 25% OptiMEM at 475 V, 1 ms pulse length using four pulses. Appropriate plasmids were added at the following concentrations: 2.5 µg pEMSV-MyoD, 2.5 µg pHook2-LacZ, 15 µg pSuper, 15 µg pDsRED-N1-VITO1, 15 µg pSuper-VITO1 and 15 µg pSuper-Myogenin. Samples of 105 cells were seeded into 6 cm plates in DMEM with 10%FCS and analysed after 72 h by staining for MyHC. Transfection efficiencies were standardized to a co-transfected LacZ vector.
RESULTS
Association of VITO-1 with TEF-1 in vitro requires the SID
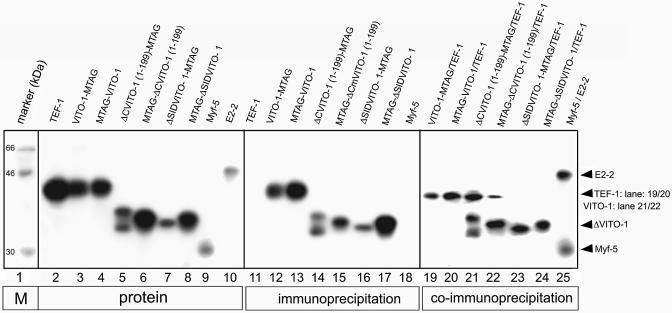
To start to decipher the functional interactions of VITO-1 with other molecules we decided to first analyse the putative interaction of VITO-1 with TEF-1 by co-immunoprecipitation. Three different fragments of the VITO-1 coding region were amplified by PCR and fused in-frame either downstream or upstream of the myc epitope of the pCS2+MT expression vector. Fragment 1 contained the complete coding region (amino acids 1–323) of VITO-1, yielding VITO-1-MTAG and MTAG-VITO-1. Fragment 2 ranged from amino acid 108 to 323, thus excluding the SID, yielding ΔSIDVITO-1-MTAG and MTAG-ΔSIDVITO-1, while fragment 3 ranged from amino acid 1–199, yielding ΔCVITO-1-MTAG and MTAG-ΔCVITO-1. The latter protein lacked the C-terminus but included the SID. The different VITO-1 protein fragments contained the myc epitope fused either to the N- (constructs MTAG-VITO-1, MTAG-ΔSIDVITO-1, MTAG-ΔCVITO-1) or the C-terminus (constructs VITO-1-MTAG, ΔSIDVITO-1-MTAG, ΔCVITO-1-MTAG) and could be immunoprecipitated with an anti-myc antibody as shown in Figure 1 (lanes 11–18). Unfortunately, the full-length VITO-1 protein migrated at approximately the same position in the SDS gel as TEF-1, thereby preventing simultaneous detection of both full-length proteins. We therefore incubated radioactively labelled TEF-1 with unlabelled VITO-1 proteins VITO-1-MTAG and MTAG-VITO-1, which carried myc epitopes at the C- and N-terminus, respectively. Hence, precipitation of labelled protein would indicate complex formation of TEF-1 and VITO-1. As shown in Figure 1, labelled TEF-1 was indeed brought down with unlabelled VITO-1-MTAG and MTAG-VITO-1 proteins (Fig. 1, lanes 19 and 20). Due to their reduced molecular weights we were able to use radioactively labelled N- and C-terminal deletions of VITO-1 together with labelled TEF-1 in the co-immunoprecipitation procedure. As expected, neither ΔSIDVITO-1-MTAG nor MTAG-ΔSIDVITO-1 proteins, which lacked the SID, were able to form complexes with TEF-1 (Fig. 1, lanes 23 and 24). The ΔCVITO-1-MTAG and MTAG-ΔCVITO-1 proteins, however, which retained the SID but lacked the C-terminal end of the proteins, readily co-immunoprecipitated with TEF-1 (Fig. 1, lanes 21 and 22).
Figure 1.
Interaction of VITO-1 with TEF-1 depends on the SID domain. SDS–gel electrophoresis of in vitro translated, [35S]methionine-labelled TEF-1 (lanes 2), VITO-1-MTAG (lane 3), MTAG-VITO-1 (lane 4), ΔCVITO-1(1–199)-MTAG (lane 5), MTAG-ΔC-VITO-1(1–199) (lane 6), ΔSID-VITO-1(108–323)-MTAG (lane 7), MTAG-ΔSID-VITO-1(108–323) (lane 8), Myf-5 (lane 9) and E2-2 (lane 10). Control immunoprecipitations with an anti-myc epitope antibody (lanes 11–17) and an anti-E2-2 antibody (lane 18). Co-immunoprecipitations of unlabelled VITO-1-MTAG together with labelled TEF-1 (lane 19), unlabelled MTAG-VITO-1 together with labelled TEF-1 (lane 20) and labelled TEF-1 (lanes 21–24) together with labelled ΔCVITO-1(1–199)-MTAG (lane 21), MTAG-ΔCVITO-1(1–199) (lane 22), ΔSID-VITO-1(108–323)-MTAG (lane 23) and MTAG-ΔSID-VITO-1(108–323) (lane 24). A control co-immunprecipation of E2-2- and Myf-5 with an antibody against E2-2 is shown in lane 25. The co-immunoprecipitation clearly demonstrates that VITO-1 and TEF-1 form a complex in the absence of DNA.
The relative amounts of precipitated VITO-1 and TEF-1 proteins were approximately equal based on densito metric scanning and the respective numbers of labelled [35S]methionine residues. It therefore seems likely that both proteins form complexes at equimolar ratios. To corroborate these findings control co-immunoprecipitation of the myogenic bHLH proteins Myf-5 and E2-2 were performed, which are known to form stoichometric complexes with each other. As shown in Figure 1, lane 25, similar amounts of Myf5 and E2-2 proteins compared to VITO-1 and TEF-1 were co-precipitated, confirming that the different proteins associated at similar ratios.
VITO-1 is required for efficient transactivation of MCAT reporter constructs by TEF-1 in HEK293 and C2C12 muscle cells
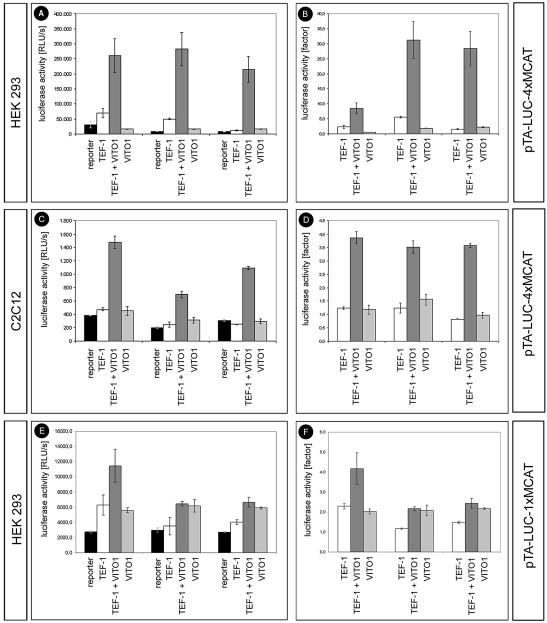
In order to establish a functional role for the interaction between VITO-1 and TEF-1, we performed a series of transfection experiments in HEK 293 and 10T1/2 cells as well as in C2C12 muscle cells using newly constructed pTA-LUC-MCAT reporter constructs. The reporter plasmids contained either one (pTA-LUC-1×MCAT) or four (pTA-LUC-4×MCAT) tandemly repeated copies of a MCAT-binding motif derived from the skeletal α-actin gene in front of a weak basal promoter driving Luciferase gene expression. As shown in Figure 2, neither the expression of TEF-1 nor VITO-1 alone was sufficient to significantly activate the pTA-LUC-MCAT reporter constructs in any of the analysed cell types. These results were in line with previous reports suggesting that the failure of TEF-1 to transactivate reporter plasmids in transfection experiments is due to the limiting presence of co-activators or transcriptional intermediary factors that allow TEFs to contact the transcriptional machinery (12).
Figure 2.
TEF-1-mediated transactivation of MCAT-dependent reporter constructs in HEK 293 (A, B, E and F) and C2C12 muscle cells (C and D) depends on VITO-1. HEK 293 cells and C2C12 cells were co-transfected with the reporter construct pTA-LUC-4×MCAT (A–D) and pTA-LUC-1×MCAT (E and F) and pCS2-based expression vectors for TEF-1 and VITO-1, either alone or in combination. The results of three different experimental series each consisting of five independent transfections are shown. Luciferase activities in relative light units (RLU) (A, C and E). Luciferase activities displayed as fold stimulation of pTA-LUC-4×MCAT (A–D) or pTA-LUC-1×MCAT (E and F) background activities. Neither VITO-1 nor TEF-1 alone are sufficient for efficient transactivation of pTA-LUC-4×MCAT or pTA-LUC-1×MCAT while a combination of both leads to an increase in luciferase activity.
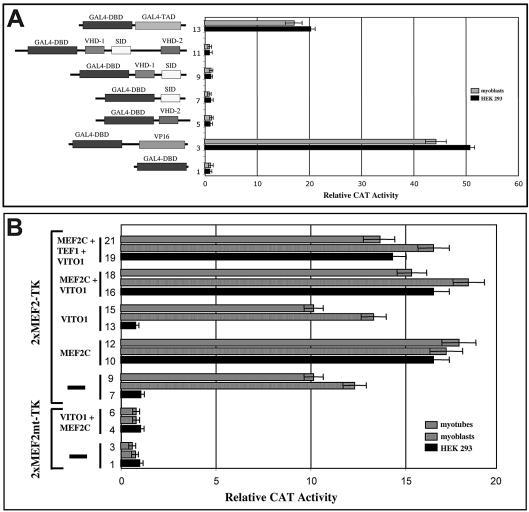
Interestingly, when both VITO-1 and TEF-1 were transfected together with the pTA-LUC-4×MCAT reporter construct, which harbours four MCAT-binding sites, a substantial increase in reporter gene activity was observed. As shown in Figure 2, combined expression of VITO-1 and TEF-1 resulted in an up to 35-fold increase in reporter gene activity in HEK293 cells and in an up to 4-fold increase in C2C12 muscle cells. The higher activation in HEK293 cells resulted from lower background activities and higher transfection efficiencies compared to C2C12 cells. Essentially the same results were obtained in 10T1/2 cells (data not shown). In contrast, the stimulation of reporter gene activity was about one order of magnitude lower in HEK293 cells when the pTA-LUC-1×MCAT reporter construct was used, which contains only a single MCAT-binding site. No induction of reporter gene activity was detected in C2C12 cells using pTA-LUC-1×MCAT (data not shown). From these results it was tempting to speculate that VITO-1 contributed a transactivation domain to the TEF–VITO complex, thereby enabling transcription. However, analysis of the VITO-1 protein sequence did not reveal an obvious transactivation domain. Likewise, fusion of full-length VITO-1 and various parts of the molecule to the yeast GAL4 DNA-binding domain (4) failed to stimulate activation of a GAL4-dependent reporter construct demonstrating the lack of an intrinsic transactivation domain in VITO-1 (Fig. 3A). From these data we conclude that VITO-1 might serve as an intermediary factor to release the action of a TEF co-repressor, as originally postulated by Chaudhary et al. (12). In contrast to the stimulation of the pTA-LUC-4×MCAT reporter construct that was achieved upon combined expression of VITO-1 with TEF-1, no stimulation of Luciferase activity was measured when VITO-1 was co-expressed with TEF-3, which is, like TEF-1, strongly expressed in skeletal muscle (data not shown). In fact, the background activity exerted by the pTA-LUC-4×MCAT reporter construct was repressed after co-transfection of TEF-3 and not stimulated as observed with TEF-1. Currently, we do not know whether this is due to the recruitment of different cellular factors by TEF-3, differences in the interaction between VITO-1 and TEF-3 or both.
Figure 3.
(A) VITO-1 is not a direct transcriptional activator. The complete coding region of VITO-1 containing vestigal homology region 1 (VHD-1), vestigal homology region 2 (VHD-2) and the scalloped interaction domain (SID) as well as various parts of the VITO-1 protein were fused to the GAL4 DNA-binding domain (DBD) and analysed on a GAL4 reporter plasmid. Wild-type GAL4 containing the GAL4 transactivation domain (TAD) and a GAL4–VP16 fusion protein were used as positive controls. No significant VITO-1-dependent transactivation activity was detected. (B) VITO-1 does not contribute to MEF2C-mediated activation of MEF2-dependent reporter genes. A functional MEF2 reporter construct (2×MEF2C-TK) and a reporter construct carrying mutated Mef2-binding sites (2×MEF2Cmt-TK) were transfected into HEK293 and C2C12 myoblasts and myotubes either alone (bars 1–3 and 7–9) or together with MEF2C (bars 4–6, 10–12 and 16–21). Addition of VITO-1 either alone (bars 16–18) or in combination with TEF1 (bars 19–21) did not increase activation of the active MEF2 reporter plasmid by MEF2C.
Previously, it had been described that TEF-1 interacts with MEF2 factors via their respective DNA-binding domains (31). We therefore asked whether VITO-1 might stimulate a MEF2 reporter construct containing a multimerized MEF2-binding site (28) together with TEF-1. As shown in Figure 3B, no significant activation of the MEF2 reporter plasmid was observed in several independent experiments in HEK293 cells and C2C12 myoblasts and myotubes, indicating that the VITO-1–TEF-1 complex and the MEF2 transcription factors exert their function by distinct cognate binding sites.
VITO-1 modulates binding of TEF1 to MCAT sequence motifs in vitro
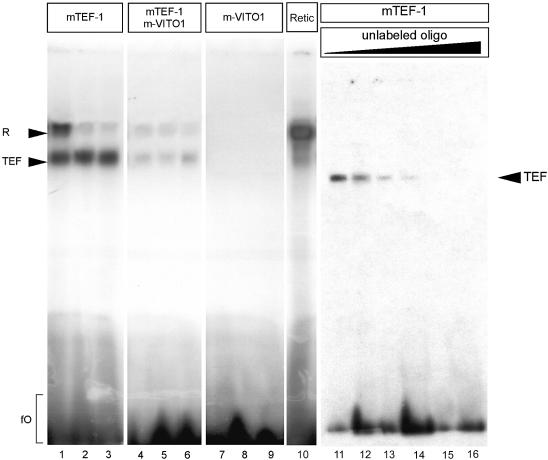
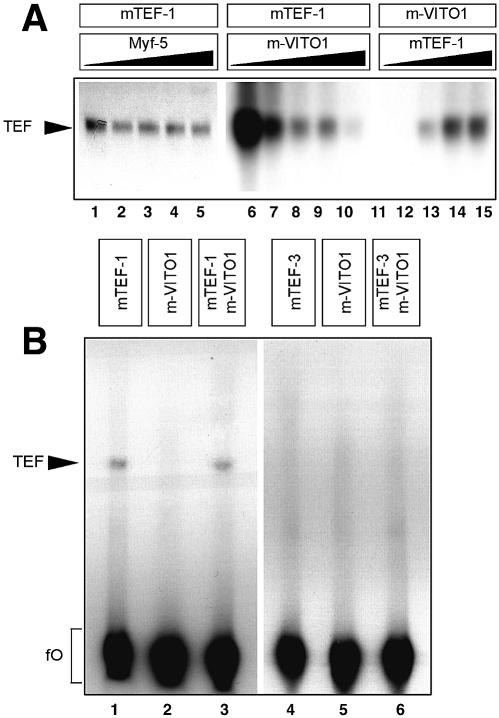
The ability of VITO-1 to bind to TEF-1 and to stimulate TEF-1- dependent reporter constructs prompted us to investigate whether the interaction of VITO-1 with TEF-1 modulates binding of TEF-1 to DNA-binding sites. We therefore analysed the DNA-binding properties of TEF1 with or without VITO-1 in electrophoretic mobility shift assays (EMSA). As shown in Figure 4, lanes 1–3, TEF-1 bound specifically to MCAT sites present in different muscle-specific promoters. However, when recombinant VITO-1 was added to the binding reactions the formation of TEF-1–MCAT complexes was significantly diminished (Fig. 4, lanes 4–6). This phenomenon was not due to a simple competition of VITO-1 and TEF1 for the same binding sites since VITO-1 alone was unable to form a stable complex with various MCAT-binding sites (Fig. 4, lanes 7–9). Since we sometimes observed an non-specific binding complex that also formed with unprimed reticulocyte lysate (Fig. 4, lane 10), the specificity of TEF-1 binding was verified by competition with increasing amounts of unlabelled binding sites (Fig. 4, lanes 11–16). To corroborate these results a number of additional competition and control experiments were performed. We examined whether addition of VITO-1 to other DNA binding reactions would decrease complex formation due to non-specific adverse effects mediated by VITO-1. There was no hint that VITO-1 affected DNA complex formation of the muscle transcription factors E2-2/Myf-4, E2-2 and MEF2C (data not shown). We also included additional reticulocyte lysate primed with Myf-5 to the TEF-1 binding reaction to rule out that increased concentrations of reticulocyte lysate affected DNA binding of TEF-1 (Fig. 5A, lanes 1–5). In contrast to unprimed reticulocyte lysate or unrelated DNA-binding proteins, addition of increasing amounts of VITO-1 to mTEF-1 EMSAs resulted in a continuous decrease in mTEF–MCAT complex formation (Fig. 5A, lanes 6–11). In a complementary experiment (Fig. 5A, lanes 12–15), addition of increasing amounts of mTEF-1 efficiently neutralized VITO-1-mediated inhibition of mTEF–MCAT complex formation and demonstrated the reversibility of the adverse effects of VITO-1 on TEF-1 DNA binding.
Figure 4.
DNA binding of TEF-1 and VITO-1 to different MCAT DNA motifs. Electrophoretic mobility shift assays (EMSA) with in vitro translated TEF-1 (lanes 1–3), TEF-1 and VITO-1 (lanes 4–6), VITO-1 (lanes 7–9) using oligo bs01 (lanes 1, 4, 7 and 10), oligo bs02 (lanes 2, 5 and 8) and oligo bs03 (lanes 3, 6 and 9). Equimolar concentrations of in vitro translated proteins were loaded as estimated from incorporation of 35S per methionine residue present in each protein and densitometric scanning. Between 4 and 8% of the volume of coupled in vitro transcription/translation reaction was used for an EMSA reaction if not indicated otherwise. An unstable, non-specific complex (R) that sometimes formed with bs01 and which was also present in unprimed reticulocyte lysate (lane 10) was clearly distinguishable from TEF-1-dependent complexes (TEF). Addition of VITO-1 led to a reduction in complex formation of TEF-1 with all three binding DNA fragments containing single MCAT motifs (lanes 4–6). No complex formation was noted when VITO-1 was used alone (lanes 7–9). To demonstrate specificity of TEF-1 DNA binding, increasing amounts of unlabelled Bs01 were incubated with the same labelled binding site (lanes 10–15). Lane 11, 0 ng; lane 12, 25 ng (10×); lane 13, 250 ng (100×); lane 14, 500 ng (200×); lane 15, 1250 ng (500×); lane 16, 2500 ng (1000×). FO, free or unbound oligonucleotides.
Figure 5.
(A) VITO-1 reduces binding of TEF-1 to single MCAT binding sites. Complex formation of TEF-1 with a single MCAT motif present on bs03 was challenged with increasing concentrations of the bHLH protein Myf-5 (lanes 1–5) and VITO-1 (lanes 6–10) and visualized by EMSA. No reduction in complex formation was discernable (lanes 1–5) after addition of increasing amounts of Myf-5 (lane1, 0%; lane 2, 5%; lane 3, 10%; lane 4, 20%; lane 5, 40% of a translation reaction) while supplementation with the same increasing concentrations of VITO-1 (up to 40% of a translation reaction) led to a clear reduction in TEF-1 binding (lanes 6–11). In a reverse experiment a constant concentration of VITO-1 was titrated against increasing concentrations of TEF-1 (up to 40% of a translation reaction) (lanes 12–15). TEF-1 complex formation was restored when the TEF-1 concentration exceeded the concentration of VITO-1. (B) Binding of TEF-1 to a multimerized MCAT binding site is resistant to modulation by VITO-1. An oligonucleotide containing four copies of the same tandemly repeated MCAT binding motif as used for transfection experiments was analysed for complex formation with TEF-1 (lane 1), VITO-1 (lane 2) and a combination of TEF-1 and VITO-1 (lane 3). Binding assays were primed with 10% of a VITO-1 and 20% of a TEF-1 translation assay to account for differences in the concentration of proteins as judged by densitometric scanning. In contrast to single MCAT binding sites, TEF-1 complexes forming on tandem repeated MCAT motifs were not disrupted by VITO-1. No complex formations on tandem repeated MCAT motifs were observed with TEF-3 under our experimental conditions (lanes 4–6). The unbound oligonucleotides are visible in the lower part (fO).
The modulation of TEF-1 DNA binding activities by VITO-1, which resulted in a reduction in complex formation with single MCAT sequence motifs, seemed to be in contrast to the strong stimulation of MCAT-dependent reporter constructs. An apparent difference in both assays was the use of single MCAT motifs in band shift experiments whereas in the efficient co-activation experiments a pTA-LUC-4×MCAT reporter was used that contained tandemly repeated copies of MCAT-binding sites. Since the multimerization of TEF-1-binding sites results in enhanced cooperative binding of TEF-1 (11,14,32), it appeared possible that TEF-1 molecules bound to multimerized MCAT motifs were resistant to the modulatory effects of VITO-1 compared to TEF-1 molecules bound to single MCAT sequences. In line with this hypothesis, we only observed a 10-fold lower stimulation of reporter gene activity when the pTA-LUC-1×MCAT reporter was used, which contains only a single MCAT-binding site. It should be mentioned, however, that in some experiments a weak induction of pTA-LUC-1×MCAT reporter activity was observed, which might be the result of a higher sensitivity of the reporter gene assays compared to the band shift assays or the presence of an additional activity in vivo that is not present in our in vitro binding reactions and which might stabilize TEF-1 DNA-binding complexes.
To solve the potential conflict between the transactivation and the binding data we used the multimerized DNA-binding site derived from the pTA-LUC-4×MCAT reporter in band shift experiments. As shown in Figure 5B, the complex formed by TEF-1 and the multimerized MCAT-binding site proved to be resistant to the modulatory action of VITO-1. Essentially, the mTEF–multimerized MCAT complex was unaffected by the presence or absence of extra amounts of VITO-1, demonstrating that VITO-1 destabilizes TEF-1 binding to single but not tandemly repeated MCAT sites.
VITO-1 enhances MyoD-mediated myogenic conversion of 10T1/2 cells
The proposed function of VITO-1 as a co-activator of TEF-1 implies that VITO-1 plays an important role in the control of the myogenic differentiation programme. We therefore asked whether VITO-1 is required for MyoD-mediated myogenic conversion of 10T1/2 cells (30). In order to achieve repression of VITO-1 in MyoD-transfected 10T1/2 cells we used the recently introduced pSUPER vector system that is based on the polymerase III H1-RNA gene promoter (33) to generate small interfering RNA (siRNA). Introduction of siRNA into cells has been demonstrated to suppress gene expression in several organisms (33). Since no antibodies against VITO-1 were available we proved the efficiency of VITO-1 suppression in 10T1/2 cells in a number of control experiments using a VITO-1-GFP fusion protein. Co-transfection of pSUPER-VITO-1 with a VITO-1-GFP expression vector resulted in a reproducible elimination of GFP expression in >90% of GFP-positive cells (data not shown).
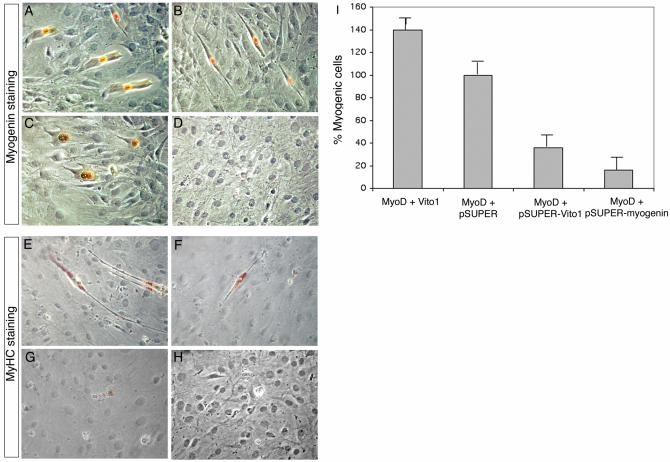
As shown in Figure 6, knockdown of VITO-1 expression in 10T1/2 cells by co-transfection of pSUPER-VITO-1 and MyoD led to a 64% reduction in muscle cells in 10T1/2 conversion assays. The requirement for VITO-1 for MyoD-mediated myogenic conversion was compared to the requirement for Myogenin, a factor that is known to be necessary for differentiation of muscle cells acting downstream of MyoD (34). As expected, inhibition of expression of Myogenin in converted 10T1/2 cells by co-transfection of pSUPER-Myogenin and MyoD resulted in robust inhibition of myogenic differentiation similar to the inhibition achieved with VITO-1 siRNA (Fig. 6). Although some differences in the degree of inhibition were noted (83% with pSUPER-Myogenin and MyoD compared to 64% with pSUPER-VITO-1 and MyoD), these differences were not statistically significant. Taken together our results clearly demonstrate that VITO-1 expression is instrumental in the expression of muscle-specific gene expression downstream of MyoD.
Figure 6.
MyoD-mediated myogenic conversion of C3H10T1/2 cells is modulated by VITO-1 expression levels. C3H10T1/2 cells were transfected with pEMSV-MyoD and pCS2-VITO-1 (A and E), pEMSV-MyoD and pSUPER (B and F), pEMSV-MyoD and pSUPER-VITO-1 (C and G), pEMSV-MyoD and pSUPER-Myogenin (D and H) and stained after 72 h with an antibody against Myogenin (A–D) or MyHC (E–H). Cumulative results of myogenic conversion experiments (I). The number of myogenic cells obtained after transfection of MyoD was set as 100%. Supplementary expression of VITO-1 led to an improvement in MyoD-mediated myogenic conversion while knockdown of VITO-1 or Myogenin expression decreases muscle cell formation. Transfection efficiencies were normalized against a co-transfected LacZ expression construct.
We also wanted to know whether supplementation of VITO-1 enhances recruitment of non-muscle cells into the myogenic fate. This hypothesis was based on the assumption that the presence of additional key regulatory factors might facilitate MyoD-mediated initiation of regulatory circuits that drive myogenic differentiation. Indeed, co-transfection of VITO-1 with MyoD into 10T1/2 cells increased the number of Myogenin- and MyHC-positive muscle cells by ∼40% (Fig. 6 and data not shown). In contrast, the expression of VITO-1 alone did not give rise to MyHC- or Myogenin-positive muscle cells. This demonstrates that VITO-1 is able to augment/enhance myogenesis but is not sufficient to initiate it (Fig. 6 and data not shown).
Down-regulation of VITO-1 expression by siRNA attenuates differentiation of C2C12 muscle cells
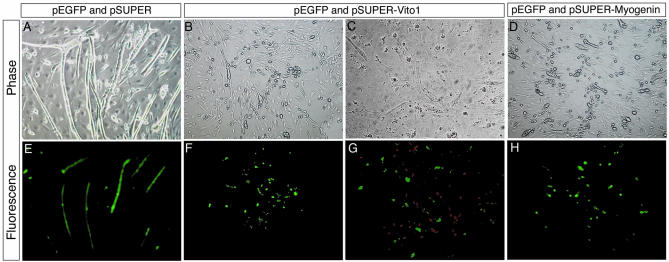
The requirement for VITO-1 for MyoD-mediated myogenic conversion of mesenchymal cells suggested an important role of VITO-1 in development of the myogenic lineage. To confirm the critical role of VITO-1 for formation of muscle cells we analysed whether knockdown of VITO-1 affected differentiation of C2C12 cells, which is a well-established system to analyse myogenic differentiation. The efficiency of the siRNA approach to change terminal differentiation of C2C12 muscle cells was analysed with reference to Myogenin, which is necessary for differentiation but not for determination of muscle cells. The siRNA vector pSUPER-VITO-1 was transiently transfected into C2C12 cells together with an EGFP-expressing plasmid to distinguish cells which expressed VITO-1 siRNA from those that had not taken up the plasmids.
The number of differentiated myotubes and the expression of Myogenin in EGFP-positive cells was accessed 48 h after transfection and removal of mitotic stimuli. As shown in Figure 7A and E, transfection of the pSUPER vector did not affect C2C12 myotube formation whereas a knockdown of VITO-1 and Myogenin efficiently blocked formation of terminally differentiated myotubes (Fig. 7B and F and Fig. 6D and H). Simultaneous detection of VITO-1 siRNA-expressing cells by EGFP fluorescence and anti-Myogenin antibody staining (red fluorescence) revealed that Myogenin was only translated in those C2C12 cells that did not express VITO-1 siRNA. The parallel and intermingled existence of cells, which appeared to be unable to undergo terminal differentiation, and of Myogenin-expressing cells might explain why the formation of myotubes was efficiently abrogated, despite the fact that only a subset of C2C12 cell expressed VITO-1 siRNA.
Figure 7.
Knockdown of VITO-1 expression attenuates differentiation of C2C12 muscle cells. C2C12 myoblasts were co-transfected with pSUPER and pEGFP-C2 (A and E), pSUPER-Vito-1 and pEGFP-C2 (B, C, F and G) and pSUPER-Myogenin and pEGFP-C2 (D and H). Cells were examined for EGFP fluorescence to visualize transfected cells (E–H) and stained with a Myogenin antibody (G) (detected with a rhodamine coupled secondary antibody) or inspected by phase contrast microscopy (A–D). Note that transfected cells expressing VITO-1 siRNA (green) are negative for Myogenin (red). Knockdown of VITO-1 and Myogenin expression led to an inhibition of myotube formation while differentiation of C2C12 cells occurred normally after transfection with pSUPER.
It should be mentioned that our initial attempts to generate cell lines stably expressing VITO-1 siRNA failed. During the selection process selected colonies became restricted in their growth, appeared unhealthy and finally died due to programmed cell death. Currently, we do not know whether this effect reflects a general toxicity of extended, high level expression of VITO-1 siRNA or a specific function of VITO-1 for myoblast maintenance.
DISCUSSION
In this study we have shown that VITO-1 is a crucial new cofactor of the muscle regulatory programme which interacts with TEF-1 to activate TEF-1 target genes. Using siRNA knockdown technology we have demonstrated that VITO-1 is essential for the expression of Myogenin in MyoD transfected 10T1/2 cells and for terminal differentiation (myotube formation) of C2C12 muscle cells.
Is VITO-1 one of the long sought co-regulatory factors of TEF1?
The failure of TEF-1 to activate transcription of target genes containing putative MCAT-binding sites suggested the existence of additional co-regulators that modulate the function of TEF-1. Davidson and colleagues have suggested that cells might contain a negatively acting factor(s) which inhibits transactivation of TEF-1 (35). They postulated that the relative lack of activity of the TEF-1 activation function does not result from the absence of positively acting factors but from the presence of a cell-specific negatively acting factor(s). Alternatively it was proposed that positively acting co-activators are required for TEF-1 transactivation (36). However, little evidence exists for the latter hypothesis.
So far our knowledge of TEF-1 interacting partners is rather limited. Only relatively weak interactions of TEF-1 with components of the TFIID complex were identified by chromatographic techniques using nuclear extracts (12). In addition, other groups reported that TEF-1 binds directly to the TATA-binding protein TBP (37) independently of an interaction with components of the TFIID complex. Studies using the viral protein SV40 TAg indicated that TEF-1 attaches to SV40 TAg and that this interaction requires the TEA domain of TEF-1 (38).
The original observation that transfected TEF-1 is inactive in transactivation experiments has been explained by the limiting presence of a bridging factors(s) that allows TEF-1 to interact with the transcriptional machinery (reviewed in 3). Thus, the failure of an excess concentration of TEF-1 in transactivation experiments to drive transcription from target genes might result from the competition of co-activators away from the promoter, a phenomenon known as squelching (39). It also seems possible that TEF-1 interacts with a negatively acting factor(s) which inhibits transactivation of TEF-1, as proposed by Davidson and colleagues (35), and that this repression is alleviated by other proteins that interact with TEF-1 and release a negatively acting factor(s). In our view the results presented in this study support the latter hypothesis. We have demonstrated that VITO-1 interacts with TEF-1 in vitro and strongly stimulates transcription of a MCAT reporter plasmid together with TEF-1. Since we obtained no evidence that VITO-1 contains a transactivating domain but TEF-1 is known to carry a multipartite activation domain (13,14,40) which interacts with cellular repressors (35), it seems logical to conclude that VITO-1 might elevate repression imposed on TEF-1 by such repressors.
Although we do not have definitive evidence as to how VITO-1 stimulates transctivation by TEF-1 and neutralizes putative repressors, it seems likely that this might happen via a conformational change instigated on the TEF-1 protein. We deduce this hypothesis from EMSA experiments with MCAT-binding sites, which revealed that VITO-1 disrupts binding of TEF-1 with singular but not with multimerized binding sites. Hence, the interaction of TEF-1 and VITO-1 might result in conformational changes in the TEF-1 protein simultaneously leading to a modification of the DNA-binding properties of TEF-1 and to interference with the binding of putative co-repressors. In addition, VITO-1 seems to restrict activation of genes that contain only a single MCAT-binding site while promoting activation of genes with repeated MCAT-binding sites or other elements that stabilize binding of TEF-1. From these results we conclude that the interaction of VITO-1 with TEF-1, which probably prevents interaction of a repressor with TEF-1, reduces the binding activity of TEF-1 to single but not to multimerized or otherwise ‘stabilized’ MCAT sequence motifs. As a consequence, VITO-1 will enable activation of certain promoters but repress others.
VITO-1 is necessary for the expression of Myogenin and for the terminal differentiation of C2C12 muscle cells
We and others have described in the past that the myogenic regulatory programme is established by a network of myogenic bHLH transcription factors, which can usurp the differentiation programme of many other pre-existing cell fates and reprogramme them to form muscle (reviewed in 1). An instrumental property of myogenic bHLH transcription factors to initiate and to maintain myogenesis seems to be their ability to set up autoregulatory loops and to stimulate the expression of other regulatory factors that might collaborate to regulate the expression of muscle structural genes such as myosins, which will build the contractile apparatus (34). Binding to and competition between a complex array of positive and negative regulators eventually determines the ability of myogeneic bHLH proteins to initiate the onset of myogenesis. VITO-1 seems to be an important component of this machinery that appears to be necessary to activate TEF-1 transcription factors. The block in VITO-1 expression, which we have achieved by siRNA knockdown, resulted in an inhibition of Myogenin expression and an obstruction of C2C12 myotube formation. At present it is hard to decide whether the inhibition of MyHC expression and myotube formation is solely due to the absence of Myogenin expression or due to a direct need for VITO-1 activity for the expression of muscle structural genes required to form a differentiated myotube. In vivo knockout studies will be necessary to extend and corroborate these findings. The Myogenin promoter does contain divergent MCAT motifs, although the functional significance of these sites is not known. In this context it is interesting to note that VITO-1 expression is significantly up-regulated during terminal differentiation of C2C12 cells. Although VITO-1 is already expressed in dividing myoblasts it is, like Myogenin, up-regulated in differentiated myotubes (24), suggesting a potential role of VITO-1 in terminal differentiation of muscle cells.
As mentioned above, myogenic factors of the MyoD family are able to reprogramme numerous cell types to adopt a muscle fate, but the efficiency of myogenic conversion varies considerably between different cell types (41). It is likely that the differential response depends on the molecular repertoire of various cell types and that supplementation of additional key regulatory factors facilitates MyoD-mediated myogenic differentiation. Therefore it did not come as a surprise that co-transfection of VITO-1 with MyoD into 10T1/2 cells considerably increased the number of muscle cells, further emphasizing the role of VITO-1 as an important cofactor of the muscle regulatory programme.
In general, there is emerging evidence that DNA-binding proteins interact with co-regulators which either stimulate or repress binding or function of the polymerase II transcription complex to specify cell fate during development (reviewed in 42). Cell-specific patterns of gene expression might depend on threshold effects determined at the level of transcription by integration of combinatorial information brought together by a series of independent interactions of various regulatory elements. In contrast to the traditional view, it seems possible that the polymerase II transcription complex might not be the primary target for integrating different activities but that cis-regulatory elements such as the MCAT motif serve as the substrate for the combinatorial integration of various regulatory molecules.
While this work was in preparation Stewart and colleagues (43) reported the cloning of Vestigial-like 2, which is identical to VITO-1. The study by Maeda et al. and our work are complementary, although some differences were noted. For example, Maeda et al. addressed the transcriptional activation of TEF-1 with TEF-1-Gal fusion proteins while we show activation of TEF-1 on its cognate binding sites. Likewise, Maeda et al. demonstrated a functional interaction of VITO-1 and TEF-1 but did not establish a physical interaction of the two proteins in vitro. On the other hand, we were unable to demonstrate a significant co-activation of MEF-2-dependent promoters by VITO-1 or a combination of TEF-1 and VITO-1 in co-transfection experiments. Despite numerous attempts, VITO-1 was not sufficient in our hands to boost transcription of reporter genes governed by MEF-2 regulatory elements in 10T1/2 and HEK 292 cells (Fig. 3B). At present it is difficult to address whether these differences were due to the use of different reporter plasmids, different expression levels achieved in transient transfections or other differences in the experimental design. Finally, we were able to demonstrate, by loss-of-function experiments, that VITO-1 is required for terminal differentiation of C2C12 cells and MyoD-mediated conversion of 10T1/2 cells.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to I. Davidson (Strasbourg) for kindly supplying various TEF constructs, to R. Agami (Amsterdam) for pSUPER and to Eva Bober for critically reading the manuscript. We greatly acknowledge the expert technical assistance of Sonja Krüger and Katja Zabel. This work was supported by the Deutsche Forschungsgemeinschaft, the ‘Fonds der Chemischen Industrie’ and the Wilhelm-Roux-Program for Research of the Martin-Luther-University.
DDBJ/EMBL/GenBank accession nos+ AJ578053 and AJ578054
REFERENCES
- 1.Neuhaus P. and Braun,T. (2002) Transcription factors in skeletal myogenesis of vertebrates. Results Probl. Cell Differ., 38, 109–126. [DOI] [PubMed] [Google Scholar]
- 2.Black B.L. and Olson,E.N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol., 14, 167–196. [DOI] [PubMed] [Google Scholar]
- 3.Larkin S.B. and Ordahl,C.P. (1999) Multiple layers of control in transcriptional regulation by MCAT elements and the TEF-1 protein family. In Harvey,R.P. and Rosenthal,N. (eds), Heart Development. Academic Press, San Diego, CA, pp. 307–329. [Google Scholar]
- 4.Braun T., Winter,B., Bober,E. and Arnold,H.H. (1990) Transcriptional activation domain of the muscle-specific gene-regulatory protein myf5. Nature, 346, 663–665. [DOI] [PubMed] [Google Scholar]
- 5.Braun T., Bober,E. and Arnold,H.H. (1992) Inhibition of muscle differentiation by the adenovirus E1a protein: repression of the transcriptional activating function of the HLH protein Myf-5. Genes Dev., 6, 888–902. [DOI] [PubMed] [Google Scholar]
- 6.Weintraub H., Davis,R., Tapscott,S., Thayer,M., Krause,M., Benezra,R., Blackwell,T.K., Turner,D., Rupp,R., Hollenberg,S. et al. (1991) The myoD gene family: nodal point during specification of the muscle cell lineage. Science, 251, 761–766. [DOI] [PubMed] [Google Scholar]
- 7.Carson J.A., Schwartz,R.J. and Booth,F.W. (1996) SRF and TEF-1 control of chicken skeletal alpha-actin gene during slow-muscle hypertrophy. Am. J. Physiol., 270, C1624–C1633. [DOI] [PubMed] [Google Scholar]
- 8.McKenna N.J. and O’Malley,B.W. (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell, 108, 465–474. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Friedrich,G.A. and Soriano,P. (1994) Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev., 8, 2293–2301. [DOI] [PubMed] [Google Scholar]
- 10.Azakie A., Larkin,S.B., Farrance,I.K., Grenningloh,G. and Ordahl,C.P. (1996) DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J. Biol. Chem., 271, 8260–8265. [DOI] [PubMed] [Google Scholar]
- 11.Jacquemin P., Hwang,J.J., Martial,J.A., Dolle,P. and Davidson,I. (1996) A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J. Biol. Chem., 271, 21775–21785. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary S., Brou,C., Valentin,M.E., Burton,N., Tora,L., Chambon,P. and Davidson,I. (1994) A cell-specific factor represses stimulation of transcription in vitro by transcriptional enhancer factor 1. Mol. Cell. Biol., 14, 5290–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart A.F., Larkin,S.B., Farrance,I.K., Mar,J.H., Hall,D.E. and Ordahl,C.P. (1994) Muscle-enriched TEF-1 isoforms bind M-CAT elements from muscle-specific promoters and differentially activate transcription. J. Biol. Chem., 269, 3147–3150. [PubMed] [Google Scholar]
- 14.Hwang J.J., Chambon,P. and Davidson,I. (1993) Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J., 12, 2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burglin T.R. (1991) The TEA domain: a novel, highly conserved DNA-binding motif. Cell, 66, 11–12. [DOI] [PubMed] [Google Scholar]
- 16.Andrianopoulos A. and Timberlake,W.E. (1994) The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol., 14, 2503–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell S., Inamdar,M., Rodrigues,V., Raghavan,V., Palazzolo,M. and Chovnick,A. (1992) The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev., 6, 367–379. [DOI] [PubMed] [Google Scholar]
- 18.Deshpande N., Chopra,A., Rangarajan,A., Shashidhara,L.S., Rodrigues,V. and Krishna,S. (1997) The human transcription enhancer factor-1, TEF-1, can substitute for Drosophila scalloped during wingblade development. J. Biol. Chem., 272, 10664–10668. [DOI] [PubMed] [Google Scholar]
- 19.Halder G., Polaczyk,P., Kraus,M.E., Hudson,A., Kim,J., Laughon,A. and Carroll,S. (1998) The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev., 12, 3900–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paumard-Rigal S., Zider,A., Vaudin,P. and Silber,J. (1998) Specific interactions between vestigial and scalloped are required to promote wing tissue proliferation in Drosophila melanogaster. Dev. Genes Evol., 208, 440–446. [DOI] [PubMed] [Google Scholar]
- 21.Simmonds A.J., Liu,X., Soanes,K.H., Krause,H.M., Irvine,K.D. and Bell,J.B. (1998) Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev., 12, 3815–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaudin P., Delanoue,R., Davidson,I., Silber,J. and Zider,A. (1999) TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development, 126, 4807–4816. [DOI] [PubMed] [Google Scholar]
- 23.Halder G. and Carroll,S.B. (2001) Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development, 128, 3295–3305. [DOI] [PubMed] [Google Scholar]
- 24.Mielcarek M., Günther,S., Kruger,M. and Braun,T. (2002) VITO-1, a novel vestigial related protein is predominantly expressed in the skeletal muscle lineage. Mech. Dev., 119S, S269–S274. [DOI] [PubMed] [Google Scholar]
- 25.Schafer K. and Braun,T. (1999) Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nature Genet., 23, 213–216. [DOI] [PubMed] [Google Scholar]
- 26.Braun T. and Arnold,H.H. (1991) The four human muscle regulatory helix–loop–helix proteins Myf3-Myf6 exhibit similar hetero-dimerization and DNA binding properties. Nucleic Acids Res., 19, 5645–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y.T., Breitbart,R.E., Smoot,L.B., Lee,Y., Mahdavi,V. and Nadal-Ginard,B. (1992) Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev., 6, 1783–1798. [DOI] [PubMed] [Google Scholar]
- 28.Molkentin J.D., Black,B.L., Martin,J.F. and Olson,E.N. (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell, 83, 1125–1136. [DOI] [PubMed] [Google Scholar]
- 29.Kruger M., Mennerich,D., Fees,S., Schafer,R., Mundlos,S. and Braun,T. (2001) Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development, 128, 743–752. [DOI] [PubMed] [Google Scholar]
- 30.Braun T., Buschhausen,D.G., Bober,E., Tannich,E. and Arnold,H.H. (1989) A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J., 8, 701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda T., Gupta,M.P. and Stewart,A.F. (2002) TEF-1 and MEF2 transcription factors interact to regulate muscle-specific promoters. Biochem. Biophys. Res. Commun., 294, 791–797. [DOI] [PubMed] [Google Scholar]
- 32.Jacquemin P., Martial,J.A. and Davidson,I. (1997) Human TEF-5 is preferentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-B gene enhancer. J. Biol. Chem., 272, 12928–12937. [DOI] [PubMed] [Google Scholar]
- 33.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 34.Braun T., Bober,E., Buschhausen-Denker,G., Kohtz,S., Grzeschik,K.H., Arnold,H.H. and Kotz,S. (1989) Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J., 8, 3617–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhary S., Tora,L. and Davidson,I. (1995) Characterization of a HeLa cell factor which negatively regulates transcriptional activation in vitro by transcriptional enhancer factor-1 (TEF-1). J. Biol. Chem., 270, 3631–3637. [DOI] [PubMed] [Google Scholar]
- 36.Xiao J.H., Davidson,I., Matthes,H., Garnier,J.M. and Chambon,P. (1991) Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell, 65, 551–568. [DOI] [PubMed] [Google Scholar]
- 37.Jiang S.W. and Eberhardt,N.L. (1996) TEF-1 transrepression in BeWo cells is mediated through interactions with the TATA-binding protein, TBP. J. Biol. Chem., 271, 9510–9518. [DOI] [PubMed] [Google Scholar]
- 38.Berger L.C., Smith,D.B., Davidson,I., Hwang,J.J., Fanning,E. and Wildeman,A.G. (1996) Interaction between T antigen and TEA domain of the factor TEF-1 derepresses simian virus 40 late promoter in vitro: identification of T-antigen domains important for transcription control. J. Virol., 70, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ptashne M. (1988) How eukaryotic transcriptional activators work. Nature, 335, 683–689. [DOI] [PubMed] [Google Scholar]
- 40.Yockey C.E., Smith,G., Izumo,S. and Shimizu,N. (1996) cDNA cloning and characterization of murine transcriptional enhancer factor-1-related protein 1, a transcription factor that binds to the M-CAT motif. J. Biol. Chem., 271, 3727–3736. [DOI] [PubMed] [Google Scholar]
- 41.Davis R.L., Weintraub,H. and Lassar,A.B. (1987) Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell, 51, 987–1000. [DOI] [PubMed] [Google Scholar]
- 42.Mannervik M., Nibu,Y., Zhang,H. and Levine,M. (1999) Transcriptional coregulators in development. Science, 284, 606–609. [DOI] [PubMed] [Google Scholar]
- 43.Maeda T., Chapman,D.L. and Stewart,A.F. (2002) Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem., 277, 48889–48898. [DOI] [PubMed] [Google Scholar]