Abstract
The wealth of transcript information that has been made publicly available in recent years requires the development of high-throughput functional genomics and proteomics approaches for its analysis. Such approaches need suitable data integration procedures and a high level of automation in order to gain maximum benefit from the results generated. We have designed an automatic pipeline to analyse annotated open reading frames (ORFs) stemming from full-length cDNAs produced mainly by the German cDNA Consortium. The ORFs are cloned into expression vectors for use in large-scale assays such as the determination of subcellular protein localization or kinase reaction specificity. Additionally, all identified ORFs undergo exhaustive bioinformatic analysis such as similarity searches, protein domain architecture determination and prediction of physicochemical characteristics and secondary structure, using a wide variety of bioinformatic methods in combination with the most up-to-date public databases (e.g. PRINTS, BLOCKS, INTERPRO, PROSITE SWISSPROT). Data from experimental results and from the bioinformatic analysis are integrated and stored in a relational database (MS SQL-Server), which makes it possible for researchers to find answers to biological questions easily, thereby speeding up the selection of targets for further analysis. The designed pipeline constitutes a new automatic approach to obtaining and administrating relevant biological data from high-throughput investigations of cDNAs in order to systematically identify and characterize novel genes, as well as to comprehensively describe the function of the encoded proteins.
INTRODUCTION
Large-scale cDNA sequencing projects have led to an enormous amount of publicly available transcript information, which has proven to be fundamental for research efforts related to understanding both human biology and disease. Due to the large quantity of generated cDNAs, several high-throughput functional genomics and proteomics approaches are presently in use or being developed. Such approaches require, besides suitable data integration procedures, a high level of automation at the level of theoretical protein analysis to gain maximum benefit from the results generated.
In this respect, the exponentially growing number of available algorithms, tools and methods with different approaches and standards constitutes a bottleneck which is aggravated by the necessity of sound knowledge of the concepts underlying the different computing methods and how to combine them correctly. A further prerequisite is the integration of the available data from many interrelated sources and experiments in order to draw relevant conclusions.
Here we describe a new automatic strategy for obtaining and administrating biologically relevant data from high-throughput investigation of cDNAs in order to systematically identify and characterize novel genes, as well as comprehensively describe the function of the encoded proteins. For this purpose, full-length cDNAs (FL-cDNAs) were chosen as targets because they contain the complete and non-interrupted protein coding regions.
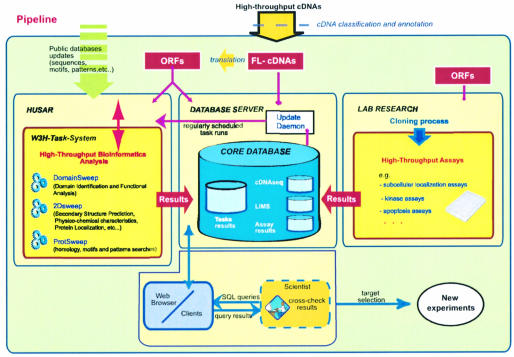
This strategy (Fig. 1) performs an automated structural and functional bioinformatic analysis of each of the investigated proteins. This includes similarity searches against protein sequence databases and specialized motif collections, the prediction of secondary structural elements, attributing each sequence to known super-families, protein localization prediction, physicochemical properties and functional domain assignment. Additionally, experimental results generated from high throughput assays are accounted for, and both bioinformatic and experimental results are integrated into a relational database system, which allows researchers to cross-check different protein features in silico.
Figure 1.
Pipeline description. A new integrated and automated strategy for obtaining and administrating data from high-throughput investigations of cDNAs. Input for the described pipeline are FL-cDNAs extracted from large-scale generated cDNAs. Verified ORFs are cloned into expression vectors to be used in high-throughput experimental assays. Additionally, all identified ORFs undergo an exhaustive bioinformatics analysis by running automated tasks (DomainSweep, 2DSweep and ProtSweep). These tasks are part of the W3H task framework, which allows the integration of heterogeneous applications to create tailor-made analysis flows. Both computational and experimental results are integrated into a relational database (‘Core Database’). The core database consists of several single databases, which allow the researcher to cross-check different protein features in silico by simply executing appropriate SQL queries via web browsers or clients.
MATERIALS AND METHODS
Core database
The core database consists of several individual databases for storing sequences, annotation, data concerning laboratory work (LIMS database), assay results (experimental-assay-database) and the data produced by tasks analyses (tasks-analysis database).
Tables in the analysis database are created depending on the structure of a single representative Extensible Markup Language (XML) file, a typical result file containing all elements that could appear in the results, via table-based mapping. An XML Complex Type containing one or more elements and/or attributes corresponds to a table and an XML Simple Type (having only one value) corresponds to a column. The hierarchical structure of the XML result files is mapped to a set of related tables where one element with one or more child elements corresponds to one entry (= row) in the first table with one or more related entries in a second table.
The tables and table relations are created in a semi-automatic fashion. First, appropriate SQL statements (table creation/alteration) are derived from the representative XML file via Extensible Style sheet Language Transformations (XSLT). In a second step, these statements are edited (to define the right data types for the columns) and then executed on the database server.
Server
A Windows 2000 Server (Microsoft) is used to host the relational-database system (MS SQL Server 2000) and a web server (MS IIS 5) for access from outside of the department. Scripts controlling the data flow of the analysis are written in VBScript and run on the server. These scripts perform database queries, invoke remote shells to start tasks on the W3H-Tasks system (1) and retrieve results, which is done by remote copy (rcp).
Data insertion and transformation
XML files are transformed by XSLT (http://www.w3.org/TR/xslt) using the MSXML ActiveX-control (Microsoft), before the data is inserted into the database. During the transformation, unique identifiers (as new elements) are introduced at the beginning of each element node. Using these identifiers, the relationship between data in related tables is established (primary/foreign key relationship). Data from the transformed XML files is loaded into the core database using the SQLXMLBulkLoad object (Microsoft) and an annotated XDR schema, which defines the element to columns and tables mapping. The schema is created automatically via XSLT using the representative XML file used for database creation. Data from statistical analysis using the software R is inserted into the database via ODBC.
Database access
Access to the database is achieved using database tools (integrated into the SQL-Server package), designed client applications (MS Access or VB applications) or through the web server using standard browsers.
Implementation of complex data and program flows for protein analysis
We have developed three new complex data and program flows for the analysis of different protein aspects: protein domain architecture (DomainSweep), homology searches (ProtSweep), physicochemical characteristics and secondary structure prediction analysis (2Dsweep).
Implementation
The protein analysis tasks have been implemented under the W3H task framework (1) that allows the integration of heterogeneous applications and methods to create tailor-made analysis flows. Some examples of already existing tasks in other biological contexts are EST-Annotator (2) for the assembly and annotation of ESTs and cDNA2Genome for the annotation of cDNAs (3).
Every single task (2Dsweep, ProtSweep, DomainSweep) has a ‘task configuration file’ containing the specification of parameters, algorithms, methods and applications to be used. This file also contains the description of the application dependencies, workflow and data flow information. By means of these descriptions and rules the W3H task framework can schedule the necessary execution of applications in parallel or sequentially as specified in the task configuration file (1).
The final output of a task is an XML file, which contains all relevant information obtained during the execution of the configuration file. This XML file can be used for further analysis (i.e. direct integration in user’s databases, additional pipeline analysis) or can be transformed by means of post-processing mechanisms into an HTML page using XSLT. At DKFZ the W3H task framework is currently used within the Heidelberg Unix Sequence Analysis Resources (HUSAR) environment (4), which allows the use and combination of all available databases and bioinformatic tools within it.
Task use in high-throughput analysis
A task analyses only one sequence at a time. For high-throughput analysis of sequences, it is necessary to initialize as many tasks as there are sequences to be analysed. On this level parallelization of sequences is managed by scripts running on the database server. These scripts invoke remote shells, as many as needed, transferring task name, protein identifier and further parameters to start the tasks on the W3H-Tasks system.
Domain architecture analysis tasks: DomainSweep. This task identifies domain architecture within a protein sequence and therefore aids in finding correct functional assignments for uncharacterized protein sequences. It employs different database search methods to scan several protein/domain family databases. Among these models, in increasing complexity, are: ProDom automatically generated protein family consensus sequences (5), PROSITE regular-expression patterns (6), BLOCKS ungapped position specific scoring matrices of sequence segments (7), PRINTS sequence motifs (8), gapped position specific scoring matrices, Prosite profile and Hidden Markov Models (HMM) such as PFAM (9). Each database model covers a slightly different, but overlapping set of protein families/domains. Each model has its own diagnostic strengths and weaknesses, and for each of these protein/domain family databases used we have established different thresholds to select trusted hits. For example in the case of the database PFAM we compare the input sequence against the HMM profile of each PFAM protein family. In principle, it is possible to decide the significance of a match upon its expectation value (E-value). However, there are a few complications such as that there are no analytical results available for accurately determining E-values for gapped alignments, especially profile HMM alignments. Therefore, we use the trusted cut-off values (TC), which exist for each PFAM family as a threshold. The TC value is the lowest score for sequences included in the family. We consider a PFAM domain hit very significant if its score is better than the corresponding TC value and at the same time has a significant E-value (<1.0).
For each of the protein/domain databases used we have established different thresholds and rules. Hits obtained from searching with HMMscan against PRINTS with an E-value <1.0 are pre-selected as trusted hits. DomainSweep uses BlocksSearcher to compare a protein query sequence with the BLOCKS database. Protein families within BLOCKS are characterized with more than one conserved segment/block. Hits with a combined E-value <0.01 (for all the identified blocks in the hit) are selected as trusted ones. We run ProdomBlast, a specialized BlastP2 search, against the ProDom database. ProdomBlast reports exactly one hit for each ProDom family detected, those hits with an E-value <0.01 are selected. In the case of PROSITE, Pfscan hits with a score greater than the cut-off n_score (normalized score, level = 0, stored in each PROSITE profile) are selected.
Afterwards, DomainSweep takes all ‘trusted positive hits’ of all individual database searches for further data interpretation in order to identify ‘significant’ hits. (i) If two or more hits belong to the same INTERPRO family. The task compares all true positive hits of the different protein family databases, grouping together those hits that are members of the same INTERPRO family/domain. (ii) If hits show the same order described in PRINTS or BLOCKS. Both databases characterize a protein family through a group of highly conserved motifs/segments in a well-defined order. The task compares the order of the identified true positive hits with the order described in the corresponding PRINTS or BLOCKS entry. Only hits in the correct order are accepted. (iii) All other trusted positive hits that do not fulfil these criteria are listed as ‘putative’.
Protein homology analysis task: ProtSweep. It performs several homology searches against protein and DNA databases sequences. It scans for homologous sequences against SWISSPROT (10), TrEMBL (10) and updates of both databases using BlastP (11). It also performs TBlastN analysis, comparing the peptide query sequence against a database named HONEST. This database includes all EMBL sequences and GenBank sequences not included in EMBL with the exception of Expressed Sequence Tags (EST), Sequence Tagged Sequences (STS), High Throughput Genome Sequences (HTG) and Genomic Survey Sequences (GSS) sequences, which usually do not contain annotation and thus cannot be used for sequence identification. The tasks select the sequences from the Blast outputs depending on a predefined cut-off value. By default, this cut-off score corresponds to an expectation value (E-value) of 0.01.
Protein location, chemical–physical properties and secondary structure prediction task: 2DSweep. This task analyses the presence of secretory signal peptides (SPScan), the amino-acid composition of the protein, its molecular weight, its isoelectric point, the distribution of protease cleavage sites and the possible sub-cellular localization of the protein [Psort (12)]. It executes two different secondary structure prediction methods using a different heuristic: Discrimination of protein Secondary Structure Class (DSC) (13) and PsiPred (14), a two-stage neural network based on position-specific scoring matrices generated by PsiBlast. Additionally, 2DSweep shows several other common measures of secondary structures such as sequence flexibility according to the Karplus–Schulz method (15), surface probability (16), antigenicity index (17), moments of hydrophobicity for each residue using two different methods, Kyte–Doolittle (18) and Hopp–Woods (19), and the presence of glycosylation points. It also shows the prediction of possible helix–turn–helix motifs [HTHScan (20)] or coiled-coil segments [CoilScan (21)] to identify sequence-specific DNA-binding structures. To complete the secondary structure prediction the task searches for transmembrane helices and intervening loop regions [TmHMM (22)].
Additional information about the tasks can be checked in the open server when clicking the help (‘?’) symbol. It is possible to access the help pages with the task descriptions from here. The three tasks used here are available as individual applications in the open server to academic users at http://genius.embnet.dkfz-heidelberg.de/menu/biounit/open-husar/. It is also possible to download results as XML files.
Public databases
All public databases used by the tasks have been implemented under the Sequence Retrieval System (SRS) (23). They are automatically updated whenever new releases become available.
Experimental assays
Experiments for determining the subcellular localization of proteins are carried out as described by Simpson et al. (24). Data entry is carried out as described by Bannasch et al. (25).
Assays on cell progression are run using an automated microscope. Upon transfection of mammalian cells with the appropriate expression vectors, the fluorescent signals of CFP/YFP fusion proteins and dyes (e.g. indicating DNA replication) are recorded. Results are stored in XML files, which are uploaded to the database server where the data is subsequently inserted into the core database; statistical analyses are carried out using the statistical package R.
In proteomics applications, the proteins are expressed in Escherichia coli, purified and spotted on glass slides. These arrays are incubated with a variety of protein kinases to identify potential substrates. To this end, individual kinases are incubated with radioactive ATP with the arrays to allow for transfer of the labelled γ-phosphoryl group from ATP to the protein. Specificity of kinase reaction is tested with kinase-specific inhibitors, which should abolish phosphorylation of the potential substrates. Incorporation of radioactivity is monitored with the help of a phosphorimager; data analysis is done using GenePix (Axon Instruments) and the statistical package R.
RESULTS AND DISCUSSION
Due to the large amount of data obtained from the annotation and validation of FL-cDNAs produced by the German cDNA Consortium (26), we were forced to find an integrated and automated strategy for conducting high throughput protein functional analyses. For this purpose, we have combined a wide variety of information sources and results from experimental assays. This strategy has allowed us to implement a pipeline (Fig. 1) for the identification and characterization of proteins.
Input data
The input for this pipeline are ORFs stemming from the annotation and validation of FL-cDNAs produced mainly by the German cDNA Consortium (26). Most of these high-throughput-generated cDNAs (HT-cDNAs) are derived from full-length clones. However, some of them present frameshifts or are produced from incompletely processed transcripts. Thus, a manual step was previously established to extract FL-cDNAs from the HT-cDNAs by an annotation and validation process (to be described elsewhere). Afterwards the FL-cDNA sequences, together with data concerning the ORFs and the deduced amino-acid sequences, are stored on the Heidelberg Unix Sequence Analysis Resources (HUSAR) system as well as in a central, relational database, which integrates all arising experimentally and theoretically determined data (Core Database).
Experimental assays
After annotation, ORFs are selected and cloned into expression vectors to be used in several high-throughput assays such as the determination of their subcellular localization (24), the examination of effects on cell progression (e.g. cell cycle control, cell proliferation or apoptosis) or assays on protein arrays after protein expression, e.g. in kinase assays (for details see Materials and Methods).
Bioinformatic analysis
Protein sequences derived from the annotated ORFs are also computationally analysed using the tasks ProtSweep, DomainSweep and 2DSweep. The tasks perform an exhaustive structural and functional analysis employing a wide variety of methods. Our strategy for assigning relevant functional roles is based on the joint use of both global (homology similarity) and local (domain and motif) sequence similarities (27). The ProtSweep approach for functional characterization of unknown proteins is based on sequence similarity to annotated proteins in existing databases. DomainSweep identifies domain architecture within the protein sequence. This task employs different search methods to scan a number of protein/domain family databases that follow several different classification schemes: hierarchical families of proteins, families of protein domains, sequence motifs or conserved regions and structural classes (for details see Materials and Methods). Each of these databases provides added-value information about protein structure, domain architecture, activity and metabolic role. The combination of these two methods presents several advantages when used as a basic approach to large-scale analysis: it improves the identification of proteins that are difficult to characterize based only on pairwise alignments and it provides an effective means of retrieving relevant biological information from vast amounts of data.
It is clear that any automatic sequence analysis implies a reasonable compromise between sensitivity and selectivity, and that no ideal recognition threshold exists that would allow for perfect separation of true and false similarities. The parameters used in each task are optimized for sensitivity in order to increase the quality of the results. For that reason our thresholds tend to be very conservative and stringent and thus the possibility of extending false positives is very limited in the domain functional assignment part. We have decided to use the functional description from INTERPRO (28) in the case of significant hits because of their high degree of confidence. Additionally, 2DSweep provides secondary structure predictions, possible protein sub-cellular localization, protease cleavage sites and peptide statistics (for details see Materials and Methods).
All relevant information obtained by the tasks’ evaluation of the single searches and individual applications is merged in a final XML file. Hence, the process of querying multiple sources and merging heterogeneous results usually applied in the investigation of unknown proteins is rendered unnecessary.
Data flow management
Following ORF annotation, the ORFs are cloned into expression vectors to be used in the described high-throughput assays. Data management concerning the cloning process is provided by client programs (MS Access or VB applications) thus acting as a laboratory information management system (25). Assay results are stored in the core database either automatically or manually using client programs (VB applications, Web Interface), depending on the assay’s automation degree.
Additionally, every ORF entering the cloning pipeline is automatically bioinformatic analysed. Scripts running on the database server regularly check the core database for the presence of new available (= annotated) ORFs. Then the described protein tasks are initialized by invoking remote shells from the server (see Materials and Methods). Tasks XML output files are transferred to the database server and their content is then inserted automatically into the core database previous transformation via XSLT.
Bioinformatic results from previously analysed ORFs are updated in regular intervals using scripts like the one mentioned above. For this purpose, the oldest entries in the analysis database belonging to one ORF are deleted and replaced by new data after the re-run of the tasks. Update intervals depend on how often the public databases are updated (e.g. BlastP2-similarity searches are run on a weekly basis).
Extensibility of the strategy
This integrated strategy allows the extensibility of the analysis pipeline at different levels. The implementation of the protein analysis tasks under the W3H tasks system provides great flexibility and extensibility. As new and improved algorithms and methodologies are developed, they can be incorporated into the protein analysis configuration file without having to redesign the entire task. It is also possible to incorporate specific sets of databases as they become available and to implement additional configuration parameters.
Due to the specific XML format of the result files, it is possible to establish a clear correlation between the XML hierarchy and existing (or to be created) database’s relations. Therefore, using one representative result file, it is possible to quickly create new tables whenever new or modified tasks and/or experimental assays are used in the pipeline.
Core database and data accessibility
The relational core database integrates and stores data from experimental results obtained from the high-throughput assays and from the bioinformatic analysis referenced by a common ORF (= protein) identifier. The database can be queried with the client tools supplied by the database system. These applications offer a high degree of flexibility but lack visualization features (e.g. results are shown in one table only). A more comprehensive user interface can be provided by custom-made client applications (developed in a high-level programming language) as stand-alone programs or applications on a web server. They wrap the appropriate SQL statements and process the returned results.
Selected data stored in the core database is publicly available via the web interface of LIFEdb (http://www.dkfz.de/LIFEdb).
The data integration in a relational database represents a major advantage since the high-level structured query language (SQL) can be used to access all data regardless of their origin. The user can pose arbitrarily complex queries to cross-check experimental results with in silico protein features by simply executing appropriate SQL statements.
The simplest use of the database is to query for any features of a protein, either predicted or experimental, and to show them corroborated. For example, the protein encoded by the clone DKFZp564B1023 has no significant similarity to proteins with an assigned function. However, the experimental assays show a nuclear localization and a motif search against the BLOCKS database gave the result ‘ATP-dependent helicase, DEAD-box’, thus indicating that this protein is potentially involved in RNA processing. Such a use can help to describe proteins annotated as ‘hypothetical’ in detail.
Other queries can be made to check for compliance between predicted results and experimental protein features in order to confirm results for further annotations. For instance, the protein encoded by the clone DKFZp761E2110 is predicted to be localized in the cytoplasm or in the nucleus but with no significantly different probabilities. However, at the moment that the experimental results are integrated into the core database the nuclear localization is confirmed.
Furthermore, the most advantageous use of the database is the selection of protein subsets with defined features by combining bioinformatic and experimental information in one query. For instance, it is possible to select subsets of potential DNA-interacting proteins by querying entries with proved nuclear localization, a basic charge and the keyword ‘DNA’ (e.g. DKFZp564C0464 is a hit in this query, which encodes a ‘DNA methyltransferase 1 associated protein 1’). Obtained subsets can be used in subsequent protein analysis or undergo further experimental assays.
Future prospects
Currently we are not using the full potential of the tasks in the pipeline because not all information provided by them is integrated in the core database. Besides the integration of missing features, ongoing work focuses on the analysis of conflicting results provided by the different methods evaluated. We are currently developing checks through the application of filtering strategies and algorithms that take into account the relationships between domain structure and homology searches (ProtSweep and DomainSweep). This filtering system takes into account the different quality of annotation of the different protein databases, both inside the individual tasks and between tasks, with the idea to assign confidence levels. Future work will also involve the extension of the assay automation, the development of further assays and their integration into the analysis pipeline.
Conclusion
Up to now several annotation web pipelines have been described, such as PEDANT (29) or web protein analysis sources such as PANAL (30), which overlap to a certain extent with the functional protein domain analysis and secondary structure predictions performed by the tasks. However, there are no descriptions in the literature about integrating large-scale experimental and computational protein analysis results and information from public databases into a relational database. Thus, this strategy constitutes a new automated approach to obtaining and administrating high-throughput functional analysis of biologically relevant data. It constitutes an example of use and integration of many different resources, dealing with issues like data access, data formats or job management, to reduce the gap between large scale experimental and bioinformatic data production and its interpretation.
The automation of this process makes it possible to reduce the user actions to solely querying the results at the end of the data pipeline in order to select relevant proteins with a desired set of features.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dorit Arlt, Ulrike Korf and Simone Schleeger for providing information about the genomics and proteomics assays, Peter Ernst and Agnes-Hotz-Wagenblatt for many useful discussions during the preparation of this manuscript and Christopher Previti for proofreading. This work has been supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the German Human Genome Project (DGHP grant 01KW0012) and the National Genome Research Network (NGFN grant 01GR0101).
REFERENCES
- 1.Ernst P., Glatting,K.H. and Suhai,S. (2003) A task framework for the web interface W2H. Bioinformatics, 19, 278–282. [DOI] [PubMed] [Google Scholar]
- 2.Hotz-Wagenblatt A., Hankeln,T., Ernst,P., Glatting,K.H., Schmidt,E.R. and Suhai,S. (2003) ESTAnnotator: A tool for high throughput EST annotation. Nucleic Acids Res., 31, 3716–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DelVal C., Glatting,K.H. and Suhai,S. (2003) cDNA2Genome: a tool for mapping and annotating cDNAs. BMC Bioinformatics, 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senger M., Flores,T., Glatting,K., Ernst,P., Hotz-Wagenblatt,A. and Suhai,S. (1998) W2H: WWW interface to the GCG sequence analysis package. Bioinformatics, 14, 452–457. [DOI] [PubMed] [Google Scholar]
- 5.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sigrist C.J., Cerutti,L., Hulo,N., Gattiker,A., Falquet,L., Pagni,M., Bairoch,A. and Bucher,P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinform., 3, 265–274. [DOI] [PubMed] [Google Scholar]
- 7.Henikoff S., Henikoff,J.G. and Pietrokovski,S. (1999) Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics, 15, 471–479. [DOI] [PubMed] [Google Scholar]
- 8.Attwood T.K., Bradley,P., Flower,D.R., Gaulton,A., Maudling,N., Mitchell,A.L., Moulton,G., Nordle,A., Paine,K., Taylor,P. et al. (2003) PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res., 31, 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 13.King R.D. and Sternberg,M.J. (1996) Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci., 5, 2298–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGuffin L.J., Bryson,K. and Jones,D.T. (2000) The PSIPRED protein structure prediction server. Bioinformatics, 16, 404–405. [DOI] [PubMed] [Google Scholar]
- 15.Karplus P.A. and Schulz,G.E. (1987) Refined structure of glutathione reductase at 1.54 Å resolution. J. Mol. Biol., 195, 701–729. [DOI] [PubMed] [Google Scholar]
- 16.Emini E.A., Hughes,J.V., Perlow,D.S. and Boger,J. (1985) Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol., 55, 836–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jameson B.A. and Wolf,H. (1988) The antigenic index: a novel algorithm for predicting antigenic determinants. Comput. Appl. Biosci., 4, 181–186. [DOI] [PubMed] [Google Scholar]
- 18.Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 19.Hopp T.P. and Woods,K.R. (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl Acad. Sci. USA, 78, 3824–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy W.N., Bailey,T.L. and Elkan,C.P. (1996) ParaMEME: a parallel implementation and a web interface for a DNA and protein motif discovery tool. Comput. Appl. Biosci., 12, 303–310. [DOI] [PubMed] [Google Scholar]
- 21.Lupas A. (1996) Prediction and analysis of coiled-coil structures. Methods Enzymol., 266, 513–525. [DOI] [PubMed] [Google Scholar]
- 22.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intel. Syst. Mol. Biol., 6, 175–182. [PubMed] [Google Scholar]
- 23.Etzold T., Ulyanov,A. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. Methods Enzymol., 266, 114–128. [DOI] [PubMed] [Google Scholar]
- 24.Simpson J.C., Wellenreuther,R., Poustka,A., Pepperkok,R. and Wiemann,S. (2000) Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep., 1, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannasch D., Mehrle,A., Glatting,K., Pepperkok,R., Poustka,A. and Wiemann,S. (2004) LIFEdb: a database for functional genomics experiments integrating information from external sources and serving as a sample tracking system. Nucleic Acids Res., 32, D505–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiemann S., Weil,B., Wellenreuther,R., Gassenhuber,J., Glassl,S., Ansorge,W., Bocher,M., Blocker,H., Bauersachs,S., Blum,H. et al. (2001) Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res., 11, 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C.H., Huang,H., Yeh,L.S. and Barker,W.C. (2003) Protein family classification and functional annotation. Comput. Biol. Chem., 27, 37–47. [DOI] [PubMed] [Google Scholar]
- 28.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P., Bucher,P. et al. (2002) InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief. Bioinform., 3, 225–235. [DOI] [PubMed] [Google Scholar]
- 29.Frishman D., Albermann,K., Hani,J., Heumann,K., Metanomski,A., Zollner,A. and Mewes,H.W. (2001) Functional and structural genomics using PEDANT. Bioinformatics, 17, 44–57. [DOI] [PubMed] [Google Scholar]
- 30.Silverstein K.A., Kilian,A., Freeman,J.L., Johnson,J.E., Awad,I.A. and Retzel,E.F. (2000) PANAL: an integrated resource for Protein sequence ANALysis. Bioinformatics, 16, 1157–1158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The relational core database integrates and stores data from experimental results obtained from the high-throughput assays and from the bioinformatic analysis referenced by a common ORF (= protein) identifier. The database can be queried with the client tools supplied by the database system. These applications offer a high degree of flexibility but lack visualization features (e.g. results are shown in one table only). A more comprehensive user interface can be provided by custom-made client applications (developed in a high-level programming language) as stand-alone programs or applications on a web server. They wrap the appropriate SQL statements and process the returned results.
Selected data stored in the core database is publicly available via the web interface of LIFEdb (http://www.dkfz.de/LIFEdb).
The data integration in a relational database represents a major advantage since the high-level structured query language (SQL) can be used to access all data regardless of their origin. The user can pose arbitrarily complex queries to cross-check experimental results with in silico protein features by simply executing appropriate SQL statements.
The simplest use of the database is to query for any features of a protein, either predicted or experimental, and to show them corroborated. For example, the protein encoded by the clone DKFZp564B1023 has no significant similarity to proteins with an assigned function. However, the experimental assays show a nuclear localization and a motif search against the BLOCKS database gave the result ‘ATP-dependent helicase, DEAD-box’, thus indicating that this protein is potentially involved in RNA processing. Such a use can help to describe proteins annotated as ‘hypothetical’ in detail.
Other queries can be made to check for compliance between predicted results and experimental protein features in order to confirm results for further annotations. For instance, the protein encoded by the clone DKFZp761E2110 is predicted to be localized in the cytoplasm or in the nucleus but with no significantly different probabilities. However, at the moment that the experimental results are integrated into the core database the nuclear localization is confirmed.
Furthermore, the most advantageous use of the database is the selection of protein subsets with defined features by combining bioinformatic and experimental information in one query. For instance, it is possible to select subsets of potential DNA-interacting proteins by querying entries with proved nuclear localization, a basic charge and the keyword ‘DNA’ (e.g. DKFZp564C0464 is a hit in this query, which encodes a ‘DNA methyltransferase 1 associated protein 1’). Obtained subsets can be used in subsequent protein analysis or undergo further experimental assays.