Abstract
The 22q13.3 deletion causes a neurodevelopmental syndrome, also known as Phelan-McDermid syndrome (MIM #606232), characterized by developmental delay and severe delay or absence of expressive speech. Two patients with hemizygous chromosome 22q13.3 telomeric deletion were referred to us when brain-imaging studies revealed cerebellar vermis hypoplasia (CBVH). To determine whether developmental abnormalities of the cerebellum are a consistent feature of the 22q13.3 deletion syndrome, we examined brain-imaging studies for 10 unrelated subjects with 22q13 terminal deletion. In 7 cases where the availability of DNA and array technology allowed, we mapped deletion boundaries using comparative intensity analysis with single nucleotide polymorphism (SNP) microarrays. Approximate deletion boundaries for 3 additional cases were derived from clinical or published molecular data. We also examined brain-imaging studies for a patient with an intragenic SHANK3 mutation. We report the first brain-imaging data showing that some patients with 22q13 deletions have severe posterior CBVH, and one individual with a SHANK3 mutation has a normal cerebellum. This genotype-phenotype study suggests that the 22q13 deletion phenotype includes abnormal posterior fossa structures that are unlikely to be attributed to SHANK3 disruption. Other genes in the region, including PLXNB2 and MAPK8IP2, display brain expression patterns and mouse mutant phenotypes critical for proper cerebellar development. Future studies of these genes may elucidate their relationship to 22q13.3 deletion phenotypes.
Keywords: cerebellum, chromosome, deletion, SHANK3
INTRODUCTION
Numerous patients with terminal deletions of chromosome 22q13 have been reported [Bonaglia et al., 2011; Dhar et al., 2010; Marshall et al., 2008; Phelan and Rogers 1993; Sarasua et al., 2011; Sebat et al., 2007; Szatmari et al., 2007; Wilson et al., 2003]. The hemizygous terminal chromosome 22q13 deletion syndrome is characterized by neonatal hypotonia, global developmental delay, normal to accelerated growth, absent to severely delayed speech, autistic behavior and minor dysmorphic features [Havens et al., 2004; Phelan and Rogers 1993]. Deletion sizes range from 130 Kb to >9 Mb, with the smallest reported deletion harboring the SHANK3 gene, which encodes a scaffolding protein that localizes to the postsynaptic density of excitatory synapses [Baron et al., 2006; Bonaglia et al., 2011; Wilson et al., 2003]. SHANK3 is strongly expressed in the cerebral cortex and cerebellum and has been proposed as the major cause for both the neurological features of the 22q13 deletion syndrome and for a monogenic form of autism [Bonaglia et al., 2001; Durand et al., 2007; Moessner et al., 2007].
Most 22q13 deletion patients have intellectual disability and severe delay or absence of expressive speech, while mixed evidence for correlations between deletion size and observed clinical features have been found [Phelan and Rogers 1993; Sarasua et al., 2011; Wilson et al., 2003]. Here, we report that cerebellar and posterior fossa malformations are underappreciated features of the 22q13 deletion syndrome in some cases. We discuss the potential role for additional genes, including PLXNB2 and MAPK8IP2, within the 22q13 terminal deletion region involved in hindbrain development.
MATERIALS AND METHODS
Subjects
Protocols were approved by Institutional Review Boards at all participating universities, and written informed consent was obtained from all subjects. Three patients (LR04-276, LR07-054 and LR07-144) were referred to us due to abnormal brain imaging. The remaining 7 patients were ascertained due to 22q13 deletion, most identified clinically by fluorescent in situ hybridization (FISH). Brain imaging results were reviewed independently and blind to deletion size by A.J.B and W.B.D. Diagnosis of cerebellar vermis hypoplasia (CBVH) was based on qualitative reduced size of the vermis recognized when the top of the vermis was located below the mid-tectum or the bottom above the level of the obex/nucleus gracilis, and enlarged size of the cistern magna [“mega cisterna magna” (MCM)] recognized when it appeared enlarged below and extended behind the cerebellum.
Microarray-based deletion breakpoint analysis
Genomic DNA was isolated from peripheral blood lymphocytes or saliva using standard methods. Genome-wide SNP genotyping was performed for 6 probands (4 with both parents) using either the Illumina HumanHap550 BeadChip (Illumina, Inc., San Diego, CA) according to the manufacturer’s instructions at the Cincinnati Children’s Hospital Medical Center or the Illumina Human610-quad BeadChip by the Center for Applied Genomics at the Children’s Hospital of Philadelphia. Illumina signal intensity data was initially analyzed using cnvPartition 1.2.1 (Illumina, Inc., San Diego, CA). Signal intensity data for one additional patient previously genotyped using the Affymetrix 500K Array [Moessner et al., 2007] was exported using dchip (http://biosun1.harvard.edu/complab/dchip/) and analyzed together with the other 6 probands. Copy number gains and losses were determined by Nexus 4.0 (BioDiscovery, Inc., El Segundo, CA) using genotyping signal intensity data and thresholds of 0.2 and -0.17, respectively (Suppl. Fig. S1–2). Two patients were analyzed for genomic copy number changes, one using the SignatureChip® (Signature Genomic Laboratories, LLC, Spokane, WA) BAC array (data not shown) and one using an Agilent oligonucleotide array (Agilent Technologies, Santa Clara, CA), as previously described [Klopocki et al., 2011] (Suppl. Fig. S3). Approximate breakpoints were derived from published molecular data for an additional patient [Delahaye et al., 2009]. In total, we obtained 22q13 breakpoint data for 10 probands.
RESULTS
We obtained cross-sectional brain-imaging studies for 10 patients with deletions of 22q13 (Table 1), ascertained because of cerebellar malformation or 22q13 deletion. In general, the brain imaging studies (Figure 1) show abnormalities in all patients. Corpus callosum thinning was observed in 9/10, abnormally thin white matter in 7/10, and enlarged ventricles in 8/10 subjects. We found definite CBVH in 3/10 (including 2 with definite MCM), subtle CBVH in 3/10, and subtle MCM in 3/10 subjects. We found MCM in one patient and normal brain imaging in another patient with reported del 22q13 but no molecular confirmation of deletion size, so these subjects were not included for further analysis (data not shown).
Table 1.
Summary of clinical features and chromosome 22q13 terminal deletion characterization.
| Brain imaging | Deletion mapping | |||||||
|---|---|---|---|---|---|---|---|---|
| Age1 | CC thin2 |
WM thin3 |
VMEG4 | CBVH5 | PF large6 |
Microarray7 | Deletion chr22q138 | |
| LR08-0449 | 6wk | ++ | + | R>L | + | − | Agilent 244K | 40771519–4969143210 |
| LR08-022 | 5.5y | + | ++ | + | − | − | Illumina Hap550 | 44616760–49691432 |
| LR07-054 | 9y | + | + | R>L | ++ | ++ | Illumina 610Q | 46202165–49691432 |
| LR07-14411 | 18y | − | + | − | ++ | ++ | Affymetrix 500K | 46260450–49691432 |
| LR04-276 | 7.5y | + | + | L>R | ++ | ++ | Illumina 610Q | 46491552–49691432 |
| LR08-046 | 2.5y | + | + | L>R | − | − | Illumina 610Q | 46894487–49691432 |
| LR08-043 | 3.1y | + | − | − | + | − | Signature BAC | 47457855–49691432 |
| LR09-26012 | NA | + | − | + | + | + | FISH, qPCR | 47921342–49518504 |
| LR08-020 | 10y | + | − | + | − | + | Illumina Hap550 | 48576196–49691432 |
| LR08-02113 | 5y | + | + | + | − | + | Illumina Hap550 | 48777410–49691432 |
| LR09-29014 | 14y | − | − | − | − | − | Normal | SHANK3, c.962A>G |
| Normal | 11.5y | − | − | − | − | − | NA | NA |
| Case B15 | 13y | + | + | + | + | + | FISH | 44279749–4969143216 |
NA indicates not applicable.
Additional notes:
Age [in weeks (wk) or years (y)] at MRI;
Hypogenesis (thinning) of corpus callosum (CC);
abnormally thin white matter (WM);
enlarged ventricles;
cerebellar vermis hypoplasia (CBVH);
enlarged posterior fossa (PF) consistent with “mega-cisterna magna”;
microarray platform used;
chromosome 22 deletion in NCBI Build 36/hg18;
46,XX,der(22);t(7;22)(7q36.2;22q13.2)mat, therefore also has dup(7)(q36.2qter);
presumed telomeric because deletion appears to extend past telomeric probe (ch22:49565816–49565875);
also reported in Moessner et al., 2007; Marshall et al., 2008 (NA0039);
also reported in Delahaye et al., 2009;
fragile X premutation carrier with 70 CGG repeats;
46,XX,der(22);t(14;22)(q32.33;q13.31)pat; also reported in Moessner et al., 2007 (SK0007);
reported in Tabolacci et al., 2005;
proximal deletion boundary reported between FISH probes RP1-102D24 and CTA-1109B5.
Figure 1. Brain images of patients with 22q13 terminal deletion or SHANK3 mutation.
T1-weighted midsagittal magnetic resonance images in one control subject, 10 subjects with 22q13 deletion, and one subject with an intragenic mutation of SHANK3 (LR09-90). The upper and lower limits of the vermis are marked by horizontal dashed white lines in each image.
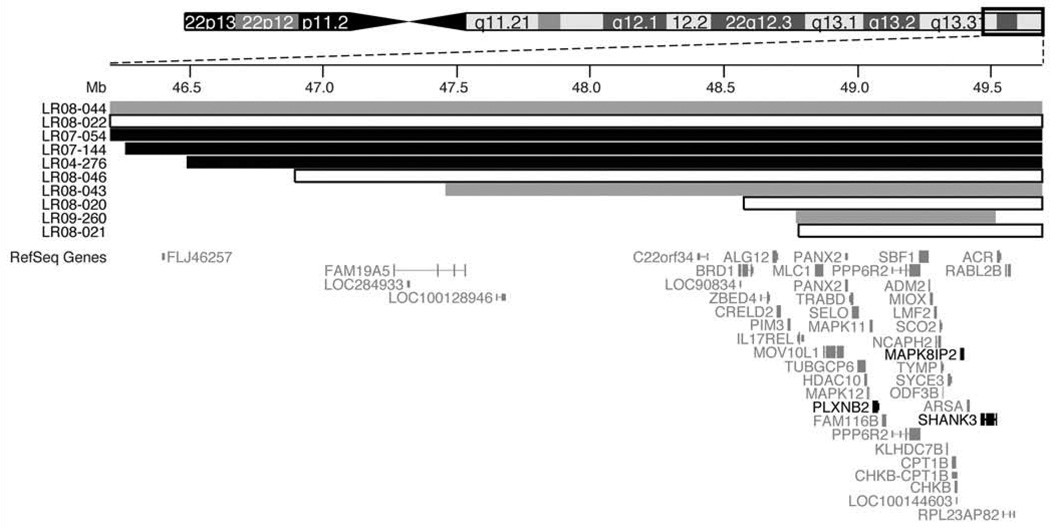
In 7 patients where the availability of DNA and array technology allowed, we mapped the deletion boundaries by using comparative intensity analysis with SNP microarrays (Supplementary Fig. S1–2). Approximate breakpoints for 3 patients were determined from molecular karyotyping (LR08-043, LR08-44) or published report (LR09-60). Deletions ranged in size from ~900 kb to >7 Mb with an average deletion size of 3 Mb (Figure 2). The 3 individuals with the most severe CBVH/MCM phenotypes have intermediate deletions. Surprisingly, the 2 individuals with the largest deletions have normal posterior fossa size and either normal vermis or mild CBVH, while the two individuals with the smallest deletions have normal vermis size and mildly enlarged posterior fossa. These data suggest influence of modifying factors from the undeleted chromosome or elsewhere in the genome.
Figure 2. Physical map of the 22q13 deletion locus.
Schematic of 22q13.31-qter (UCSC Genome Browser Mar 2006, chr22:46,000,000–49,691,432) drawn to scale shows deletions associated with normal and abnormal posterior fossa brain imaging. Deletions associated with CBVH+MCM (black) or CBVH (grey) are displayed. The deletion in two probands (LR08-44, LR08-22) extends beyond field of view. RefSeq genes are shown. PLXNB2 or MAPK8IP2 produce developmental cerebellar phenotypes with homozygous loss in mouse. Mutation or deletion of SHANK3 has been associated with autism.
DISCUSSION
We report brain-imaging studies in 10 patients with deletion 22q13 that show CBVH, enlarged posterior fossa or both in 8/10 patients without features of cerebellar atrophy, and confirm prior reports of thin corpus callosum and ventriculomegaly. While we have only single imaging studies on these patients, comparison of scans from children aged 6 weeks to 18 years (Table 1) showed similar features. In particular, the thin corpus callosum and white matter and enlarged ventricles were not more severe in older patients. While few brain-imaging studies have been reported for individuals with 22q13 deletions, posterior fossa or cerebellar abnormalities were previously noted for seven patients, thin corpus callosum in another seven, and ventriculomegaly in at least three [Bonaglia et al., 2011; Doheny et al., 1997; Lindquist et al., 2005; Philippe et al., 2008; Tabolacci et al., 2005]. However, figures of brain imaging studies were rarely provided. We reviewed the scan available in Fig. 2 of Tabolacci et al., 2005, and find changes very similar to those observed in our cohort, including mild CBVH and mildly enlarged posterior fossa (Table 1).
SHANK3 is strongly expressed in the cerebral cortex and cerebellum and has been proposed as the major cause for both the neurological features of the 22q13 deletion syndrome and for a monogenic form of autism [Bonaglia et al., 2001; Durand et al., 2007; Moessner et al., 2007]. Cerebellar dysfunction alone may explain many autism symptoms by affecting cognitive and motor behavior, as well as connectivity between the cerebellum and other brain systems [Allen 2006; Allen and Courchesne 2003; Schmahmann 2004; Tavano et al., 2007]. Reports of two neurodevelopmental cerebellar phenotypes in the autistic brain, CBVH and decreased Purkinje cell (PC) number [Bauman and Kemper 2005; Bloss and Courchesne 2007; DiCicco-Bloom et al., 2006], further suggest that genes regulating cerebellar development may also confer autism susceptibility in these patients.
Several Shank3 mouse mutants have recently been developed [Bangash et al., 2011; Bozdagi et al., 2010; Peca et al., 2011; Wang et al., 2011]. Though no gross cerebellar malformations were reported in these mice, specific investigation of cerebellar development has yet to be performed. We examined one Shank3 mouse mutant [Bozdagi et al., 2010] using integrated brain-imaging data and head CT scans (data not shown). No obvious cerebellar or posterior fossa phenotypes were detected in adult Shank3 heterozygous or homozygous mutant mice, suggesting Shank3 disruption does not impact cerebellar development in mice.
Two other genes in the 22q13 deletion region, PLXNB2 and MAPK8IP2, are strong candidates for cerebellar phenotypes. Plxnb2 and Mapk8ip2 are expressed in the mouse hindbrain and abnormal phenotypes of the developing cerebellum are observed in homozygous mutants for each gene (the heterozygous phenotypes were not described). Plxnb2 is expressed in embryonic mesenchyme and the external granule cell (GC) layer of the postnatal cerebellum [Perala et al., 2005], and is a target of Atoh1, a transcription factor critical for GC development [Klisch et al., 2011]. Plxnb2 knockout mice have a perinatal lethal phenotype that includes reduced cerebellar fissure formation and aberrant granule cell proliferation and differentiation [Friedel et al., 2007]. Mapk8ip2 encodes the islet brain-2 protein, a scaffolding protein broadly expressed in mouse brain with enrichment at postsynaptic densities, including within the cerebellum [Giza et al., 2010]. Mapk8ip2 null mice have deficits in PC dendritic aborization and synaptic transmission, but apparent normal cerebellar foliation and PC localization [Giza et al., 2010; Kennedy et al. 2007]. These mice also display behavioral phenotypes proposed to be relevant for autism including deficits in motor, learning, and social interaction paradigms [Giza et al., 2010]. In humans, PLXNB2 and MAPK8IP2 are highly expressed in cerebellar vermis [Jones et al., 2009] (http://human.brain-map.org/); their role in human cerebellar phenotypes requires further investigation.
While our series is relatively small, the high rate of cerebellar and/or posterior fossa abnormalities – seen in 8/10 subjects – suggests that developmental abnormalities of the posterior fossa and cerebellum are a common feature of the deletion 22q13 syndrome. We also confirm the high rate of thin corpus callosum and ventriculmegaly. Based on our analysis of the gene content, we propose that MCM-CBVH observed in the Phelan-McDermid syndrome, a contiguous gene deletion syndrome which includes SHANK3, are likely due to contributions from two or more genes in the region, possibly including PLXNB2 and MAPK8IP2. Our data in a single patient with an intragenic SHANK3 mutation suggests that SHANK3 disruption is not sufficient to produce CBVH, though examination of additional patients is warranted.
Supplementary Material
Acknowledgements
We are grateful for generous support and family participation from the 22q13 Deletion Foundation. We thank Christian Marshall for providing Affymetrix 500k data, Takeshi Sakurai and Joe Buxbaum for Shank3 mouse brains, Brian Roman and Kate Waimey for mouse brainimaging analysis. K.A.A. was supported by an Autism Speaks fellowship.
Footnotes
Conflict of Interest.
The authors declare no conflict of interest.
REFERENCES
- Allen G. Cerebellar contributions to autism spectrum disorders. Clin Neurosci Res. 2006;6(3–4):195–207. [Google Scholar]
- Allen G, Courchesne E. Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: an fMRI study of autism. Am J Psychiatry. 2003;160(2):262–273. doi: 10.1176/appi.ajp.160.2.262. [DOI] [PubMed] [Google Scholar]
- Bangash MA, Park JM, Melnikova T, Wang D, Jeon SK, Lee D, Syeda S, Kim J, Kouser M, Schwartz J, Cui Y, Zhao X, Speed HE, Kee SE, Tu JC, Hu JH, Petralia RS, Linden DJ, Powell CM, Savonenko A, Xiao B, Worley PF. Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism. Cell. 2011;145(5):758–772. doi: 10.1016/j.cell.2011.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311(5760):531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005;23(2–3):183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Bloss CS, Courchesne E. MRI neuroanatomy in young girls with autism: a preliminary study. J Am Acad Child Adolesc Psychiatry. 2007;46(4):515–523. doi: 10.1097/chi.0b013e318030e28b. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, Bonati MT, Giglio S, Guerrini R, Osimani S, Marelli S, Zucca C, Grasso R, Borgatti R, Mani E, Motta C, Molteni M, Romano C, Greco D, Reitano S, Baroncini A, Lapi E, Cecconi A, Arrigo G, Patricelli MG, Pantaleoni C, D'Arrigo S, Riva D, Sciacca F, Dalla Bernardina B, Zoccante L, Darra F, Termine C, Maserati E, Bigoni S, Priolo E, Bottani A, Gimelli S, Bena F, Brusco A, di Gregorio E, Bagnasco I, Giussani U, Nitsch L, Politi P, Martinez-Frias ML, Martinez-Fernandez ML, Martinez Guardia N, Bremer A, Anderlid BM, Zuffardi O. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid Syndrome. PLoS Genet. 2011;7(7):e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69(2):261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1(1):15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye A, Toutain A, Aboura A, Dupont C, Tabet AC, Benzacken B, Elion J, Verloes A, Pipiras E, Drunat S. Chromosome 22q13.3 deletion syndrome with a de novo interstitial 22q13.3 cryptic deletion disrupting SHANK3. Eur J Med Genet. 2009;52(5):328–332. doi: 10.1016/j.ejmg.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Dhar SU, del Gaudio D, German JR, Peters SU, Ou Z, Bader PI, Berg JS, Blazo M, Brown CW, Graham BH, Grebe TA, Lalani S, Irons M, Sparagana S, Williams M, Phillips JA, 3rd, Beaudet AL, Stankiewicz P, Patel A, Cheung SW, Sahoo T. 22q13.3 deletion syndrome: clinical and molecular analysis using array CGH. Am J Med Genet A. 2010;152A(3):573–581. doi: 10.1002/ajmg.a.33253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCicco-Bloom E, Lord C, Zwaigenbaum L, Courchesne E, Dager SR, Schmitz C, Schultz RT, Crawley J, Young LJ. The developmental neurobiology of autism spectrum disorder. J Neurosci. 2006;26(26):6897–6906. doi: 10.1523/JNEUROSCI.1712-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, McDermid HE, Harum K, Thomas GH, Raymond GV. Cryptic terminal rearrangement of chromosome 22q13.32 detected by FISH in two unrelated patients. J Med Genet. 1997;34(8):640–644. doi: 10.1136/jmg.34.8.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel RH, Kerjan G, Rayburn H, Schuller U, Sotelo C, Tessier-Lavigne M, Chedotal A. Plexin-B2 controls the development of cerebellar granule cells. J Neurosci. 2007;27(14):3921–3932. doi: 10.1523/JNEUROSCI.4710-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giza J, Urbanski MJ, Prestori F, Bandyopadhyay B, Yam A, Friedrich V, Kelley K, D'Angelo E, Goldfarb M. Behavioral and cerebellar transmission deficits in mice lacking the autism-linked gene islet brain-2. J Neurosci. 2010;30(44):14805–14816. doi: 10.1523/JNEUROSCI.1161-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens JM, Visootsak J, Phelan MC, Graham JM., Jr 22q13 deletion syndrome: an update and review for the primary pediatrician. Clin Pediatr (Phila) 2004;43(1):43–53. doi: 10.1177/000992280404300106. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10(11):821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- Kennedy NJ, Martin G, Ehrhardt AG, Cavanagh-Kyros J, Kuan CY, Rakic P, Flavell RA, Treistman SN, Davis RJ. Requirement of JIP scaffold proteins for NMDA-mediated signal transduction. Genes Dev. 2007;21(18):2336–2346. doi: 10.1101/gad.1563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108(8):3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Lohan S, Brancati F, Koll R, Brehm A, Seemann P, Dathe K, Stricker S, Hecht J, Bosse K, Betz RC, Garaci FG, Dallapiccola B, Jain M, Muenke M, Ng VC, Chan W, Chan D, Mundlos S. Copy-number variations involving the IHH locus are associated with syndactyly and craniosynostosis. Am J Hum Genet. 2011;88(1):70–75. doi: 10.1016/j.ajhg.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist SG, Kirchhoff M, Lundsteen C, Pedersen W, Erichsen G, Kristensen K, Lillquist K, Smedegaard HH, Skov L, Tommerup N, Brondum-Nielsen K. Further delineation of the 22q13 deletion syndrome. Clin Dysmorphol. 2005;14(2):55–60. [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81(6):1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472(7344):437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perala NM, Immonen T, Sariola H. The expression of plexins during mouse embryogenesis. Gene Expr Patterns. 2005;5(3):355–362. doi: 10.1016/j.modgep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Phelan K, Rogers C. Phelan-McDermid Syndrome. In: Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviewsTM. Seattle (WA): University of Washington, Seattle; 2005. May 11, [Updated 2011 Aug 25]. [Internet].; 1993-. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1198/ [Google Scholar]
- Philippe A, Boddaert N, Vaivre-Douret L, Robel L, Danon-Boileau L, Malan V, de Blois MC, Heron D, Colleaux L, Golse B, Zilbovicius M, Munnich A. Neurobehavioral profile and brain imaging study of the 22q13.3 deletion syndrome in childhood. Pediatrics. 2008;122(2):e376–e382. doi: 10.1542/peds.2007-2584. [DOI] [PubMed] [Google Scholar]
- Sarasua SM, Dwivedi A, Boccuto L, Rollins JD, Chen CF, Rogers RC, Phelan K, DuPont BR, Collins JS. Association between deletion size and important phenotypes expands the genomic region of interest in Phelan-McDermid syndrome (22q13 deletion syndrome) J Med Genet. 2011;48(11):761–766. doi: 10.1136/jmedgenet-2011-100225. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD. Disorders of the cerebellum: ataxia, dysmetria of thought, and the cerebellar cognitive affective syndrome. J Neuropsychiatry Clin Neurosci. 2004;16(3):367–378. doi: 10.1176/jnp.16.3.367. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Roge B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bolte S, Feineis-Matthews S, Herbrecht E, Schmotzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabolacci E, Zollino M, Lecce R, Sangiorgi E, Gurrieri F, Leuzzi V, Opitz JM, Neri G. Two brothers with 22q13 deletion syndrome and features suggestive of the Clark-Baraitser syndrome. Clin Dysmorphol. 2005;14(3):127–132. [PubMed] [Google Scholar]
- Tavano A, Grasso R, Gagliardi C, Triulzi F, Bresolin N, Fabbro F, Borgatti R. Disorders of cognitive and affective development in cerebellar malformations. Brain. 2007;130(Pt 10):2646–2660. doi: 10.1093/brain/awm201. [DOI] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20(15):3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40(8):575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.