Abstract
Electrochemical impedance measurements were used for the detection of single-strand DNA sequences using a peptide nucleic acid (PNA) probe layer immobilized onto Si/SiO2 chips. An epoxysilane layer is first immobilized onto the Si/SiO2 surface. The immobilization procedure consists of an epoxide/amine coupling reaction between the amino group of the PNA linker and the epoxide group of the silane. A 20-nucleotide sequence of PNA was used. Impedance measurements allow for the detection of the changes in charge distribution at the oxide/solution interface following modifications to the oxide surface. Due to these modifications, there are significant shifts in the semiconductor’s flat-band potential after immobilization and hybridization. The results obtained using this direct and rapid approach are supported by fluorescence measurements according to classical methods for the detection of nucleic acid sequences.
INTRODUCTION
Major efforts are currently being devoted to the development of devices allowing the direct (non-labelled) and rapid detection of genetic materials in liquid media. These techniques rely on the specific affinity inherent to the hybridization between single-strand DNA (ssDNA) sequences, and their complementary counterparts as a probe layer affixed to the surface of a transducer. Approaches to direct signal generation that have received much attention in recent years include surface plasmon resonance (1–9), acoustic network analysis (10–12) and quartz crystal microbalance (QCM) (13–17). In addition, the feasibility of using electrochemical techniques for the detection of biomolecules (18–24), especially those based on impedance and/or field effect measurements (25–30), is now also well established. Initial reports by our group in this area describe an electrochemical approach to detecting the hybridization of complementary strands using Si/SiO2 substrates as a transducer, and simple homooligonucleotide sequences as probes (31–34). This particular approach involves measuring variations in electrical impedance when the functionalized Si/SiO2 substrates are used as working electrodes in a standard three-electrode electrochemical cell. The Si/SiO2/ssDNA/electrolyte structure allows direct detection of the hybridization process as one monitors the variation in charge distribution within the semiconductor, caused by the changes occurring within the ssDNA probe layer.
One of the main challenges to achieving highly reproducible and more sensitive silicon-based sensor devices is the optimization of the probe layer immobilization procedures, to achieve probe layer uniformity, and the nature of the probe layer itself. In this respect much attention is now being focused on the use of synthetic peptide nucleic acid (PNA) sequences as probes (35,36). PNA is a DNA mimic in which the sugar phosphate backbone has been replaced by a pseudo peptide-like backbone, composed of N2-amithoethylglycine units linked by amide bonds. PNA is charge neutral and has the proper inter-base spacing, resulting in stronger affinity for the complementary DNA sequence. PNA has a significantly higher sensitivity and specificity making it superior to DNA as the probe in biosensors (37–40). The present paper investigates the immobilization of PNA as probe onto a Si/SiO2 electrode, and the detection of its hybridization with complementary DNA using the electrochemical impedance approach.
PNA immobilization
Various methods have been reported for the immobilization of PNA onto a transducer. Burgener and co-workers have examined the hybridization characteristics of PNA, as covalently coupled onto a sensor surface (41). This method consists of a coupling between the maleimide group attached to the N-terminal of the PNA and the sulfhydryl groups of an activated dextran matrix on the sensor surface. Another approach was used by Wang and co-workers in which the PNA, derivatized with a thiolate anchor group, was immobilized as a self-assembled monolayer onto a gold-plated QCM (42).
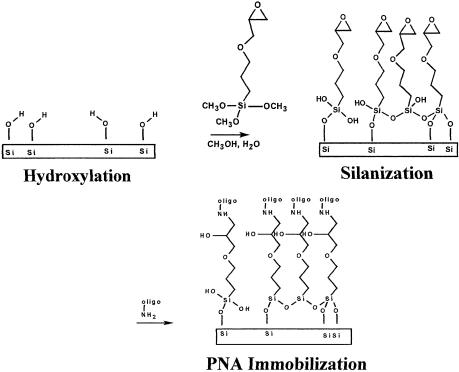
In our approach, the PNA is covalently linked to the surface of Si/SiO2 chips that have been functionalized with a silane, 3-glycidoxypropyltrimethoxysilane (GPTS) (43–45). GPTS consists of hydrolyzable methoxy groups that allow for the covalent binding of the silane to the oxide surface through siloxane bonds (referred to as silanization in Fig. 1), following the formation of silanol goups by acid treatment of the oxide (referred to as hydroxylation in Fig. 1). In addition, GPTS has an epoxide functional group that enables the binding of the PNA. Using a procedure derived from work by Lamture and co-workers (46), the covalent binding in this case is achieved through an epoxide-amine coupling reaction between the epoxide group of the GPTS and the aminolinker of the PNA oligomer (referred to as PNA immobilization in Fig. 1).
Figure 1.
Probe layer immobilization procedure.
MATERIALS AND METHODS
Silicon chips
The Si/SiO2 substrates (TRONICS Microsystems, France) are 1 × 1 cm2 with a 300 µm thick silicon layer. The Si (100) was phosphorus doped (n-type) to a density of 1015 cm–3. There is a silicon dioxide layer of 150 Å on the front. The back of the chip has a gold/chromium ohmic contact of 2000/500 Å.
Reagents
3-glycidoxypropyltrimethoxysilane (GPTS, 98%), N-N diisopropylethylamine (DPEA, 99.5%, under nitrogen), ethyl alcohol (spectrophotometric grade) were purchased from Sigma-Aldrich and used without further purification. Propane-2-ol was purchased from Analar. Hydrochloric acid and NaCl were reagent grade and purchased from Fisher. All solutions were prepared in distilled-deionized water. A Milli-RX 20 Millipore device provided distilled-deionized (d-d) water. The GPTS and N-N diisopropylethylamine were stored at 4°C.
The DNA oligonucleotides (20 base-long oligomer) were purchased from Biocorp (Montreal). The PNA oligonucleotide [20 base-long oligomer bearing an ‘O’ linker (amino linker) at the 5′ end] was purchased from PE Biosystems. The DNA oligomer is perfectly matched to the PNA oligomer that has the following sequence, 5′-AGTCCATTGCAGTCCATTGC-3′. All nucleic acids were stored at –20°C. The fluorescein-labelled DNA oligonucleotides were purchased from Biocorp (Montreal). The fluorescein-labelled PNA oligonucleotides were purchased from PE Biosystems (bearing the fluorescein label at the 3′ end).
GPTS/amine immobilization method
The glassware was thoroughly washed with d-d water before use. The Si/SiO2 substrates were cleaned by immersion in acetone for 5 min, boiling d-d water for 10 min, thorough rinsing with d-d water, and drying under nitrogen. The substrates were immersed in concentrated HCl solution. Following acid treatment, the chips were, once again, placed in boiling d-d water for 10 min, rinsed thoroughly with d-d water and dried under N2.
To determine the optimal silanization time (the best GPTS coverage), a preliminary experiment was performed using fluorescein-labelled bovine serum albumin protein (BSA, from Sigma). Chips were prepared by varying the silanization times (30, 60, 90, 120, 360 and 480 min). The chips were placed in propane-2-ol for 2 min, rinsed with d-d water for 30 s and dried under nitrogen. Each chip was coated with 10 µl of a BSA aqueous solution and the immobilization was carried out at room temperature in water-saturated atmosphere for 2 h. The washing procedure described above was performed. Fluorescence measurements were taken immediately after silanization.
Prior to silanization, the silane mixture was stirred for 5 min and the chips were immersed in a thin film of ethanol for 5 min to remove any adsorbed particles. The silane mixture consisted of a 1% GPTS solution [mixture of ethanol, water, DPEA and GPTS (23.5:1.25:0.25:0.25 v/v/v/v)]. The chips were left to react with the silane mixture under medium agitation for ∼4 h in a sealed polystyrene container. Following silanization, the substrates were immersed in propane-2-ol for 5 min (to remove excess GPTS or any contaminants), rinsed with d-d water for 30 s, and dried under N2. The substrates were stored in closed welled plates under ambient atmosphere.
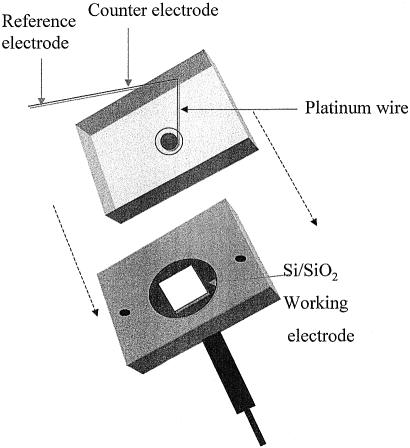
Immobilization of the probe layer was performed by placing 100 µl of a 0.02 µg/µl PNA (in 0.1 M NaCl) aqueous solution onto the modified Si/SiO2 chip forming the working electrode of a specially designed three-electrode cell (Fig. 2). Impedance measurements were taken while the reaction took place. The substrates were thoroughly rinsed with d-d water to remove any non-specifically adsorbed PNA from the surface. In order to confirm the presence of the immobilized PNA and the hybridized DNA, the oligonucleotides were labelled with a fluorescent probe, fluorescein.
Figure 2.
Schematic representation of the specially designed cell. Reference electrode (RE); counter electrode (CE); working electrode (WE).
Hybridization procedures
The hybridization was performed using fluorescein-labelled DNA. The concentration of DNA is equal to the concentration of PNA used for the immobilization. The hybridization consisted of placing 100 µl of a 0.02 µg/µl DNA in 0.1 M NaCl aqueous solution in the specially designed cell and leaving the chips to react while taking impedance measurements. The substrates were thoroughly rinsed with d-d water to remove any unhybridized DNA from the surface.
Fluorescence measurements
The chips with the immobilized fluorescein-labelled PNA strands and the fluorescein-labelled DNA strands were prepared using the procedure described above. The immobilization and hybridization steps were done in the dark. Following immobilization and hybridization, the chips were mounted on a standard microscope slide. The fluorescence measurements were taken using a Genepix 4000B fluorescence microarray scanner. The image of the chip showed the circular region where immobilization of labelled PNA had been done. The Genepix scanner aligns user-constructed blocks with features on an array. The blocks were constructed with features to our specifications. The pixel size was set at 20 µm. The diameter of the feature indicator was set to the diameter of the spot. The features were aligned on the spots. The photomultiplier tube (PMT) voltage, focus and feature diameter were set each time a new slide was scanned. The PMT voltage and focus were adjusted to ∼1000 V and 200 µm, respectively. A median intensity was taken at a wavelength of 532 nm.
Impedance measurements
A DC potential of –0.5 to 2 V was applied to the chips using a three-electrode potentiostatic set-up. An AC voltage of 100 kHz frequency and 10 mV amplitude was superimposed onto the DC potential. Impedance measurements were taken using a computer-controlled Voltalab electrochemical workstation (model PGZ 301 by Radiometer, Copenhagen), and the accompanying Voltamaster computer program was used to calculate the out-of-phase impedance (Zi) and generate the plots. All measurements were done at room temperature and conducted in the dark to avoid photogeneration of charge carriers in the semiconducting electrode.
RESULTS AND DISCUSSION
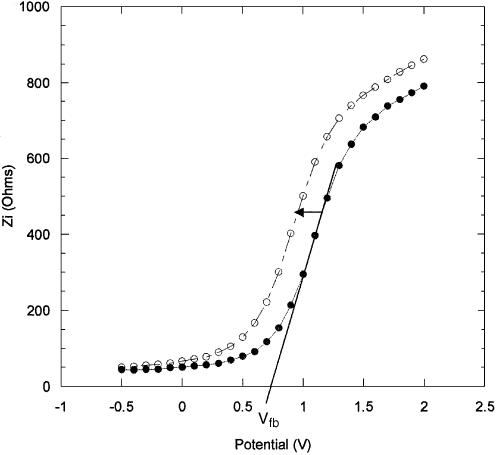
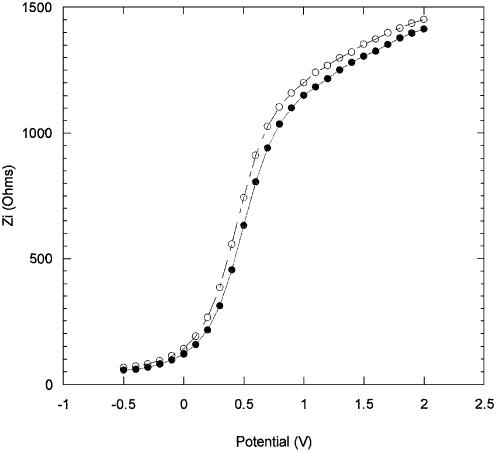
In the impedance technique the capacitance of the semiconductor/oxide/electrolyte structure is measured as a function of the DC potential applied to the working electrode. Modification of the oxide surface through immobilization and hybridization causes a change in the charge distribution within the space-charge layer of the semiconductor, resulting in a change of flat-band potential (Vfb). The change in Vfb is reflected by a displacement of the out-of-phase impedance curves along the DC potential axis, toward more negative or more positive potentials depending on the effect the surface transformations have on the aforementioned charge distribution. The values of Vfb are directly obtained by extrapolating the steeply rising portion of the Zi versus applied DC potential curves (illustrated in Fig. 5).
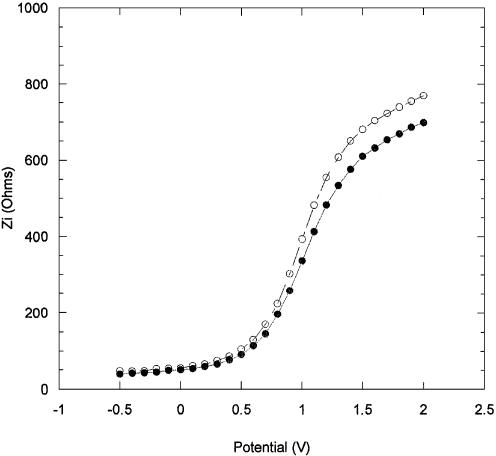
Figure 5.
Imaginary impedance as a function of the DC applied potential, (closed circle) after 12 min, and (open circle) after 60 min, immobilization of PNA.
Silanization
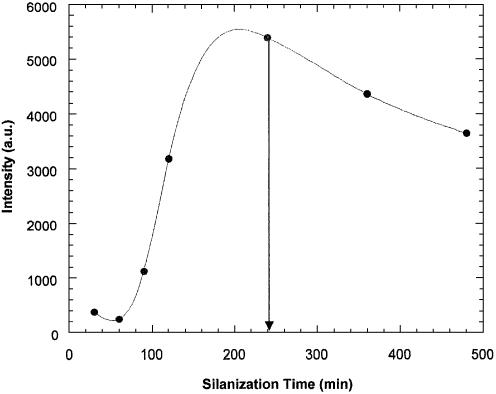
To determine the optimal silanization time, individual chips were submitted to silanization times of 30, 60, 90, 120, 360 and 480 min. Fluorescein-labelled BSA was immobilized for ∼2 h on each chip. Figure 3 shows the observed fluorescence intensities and the optimal silanization time was found to be 4 h, which corresponds to the highest median intensity of 5.4 × 103 a.u.
Figure 3.
Fluorescence intensity as a function of the silanization time.
Immobilization of the PNA probe layer
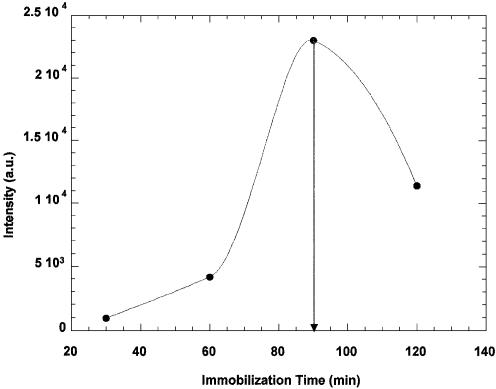
Figure 4 shows the variation of fluorescence intensity versus immobilization time for individual chips covered with fluorescently-labelled BSA. The silanization time used for these chips was 4 h. The immobilization time yielding maximum coverage was determined to be ∼1.5 h. Under these conditions, the median fluorescence intensity was of 2.3 × 104 a.u.
Figure 4.
Fluorescence intensity as a function of the immobilization time using BSA.
Figure 5 shows the shift in Zi values due to the immobilization of the PNA probe layer on GPTS modified Si/SiO2 chips. Fluorescein-labelled PNA was used in these measurements to further confirm the presence of the probe layer following the immobilization. The actual time used to immobilize the PNA probe layers was 1 h, to ensure that the probe layer was not too dense, in order not to inhibit the subsequent hybridization process. The impedance measurements were taken during the immobilization to monitor the change over time. Five successive impedance curve measurements were taken, each measurement lasting 12 min. Curve 1 in Figure 5 shows the initial measurement taken at the start of the immobilization. Curve 2 is the final measurement taken at the end of the immobilization. The curves taken in between kept their general shape and progressively shifted to more negative potentials (not shown for clarity). The overall shift in Vfb was found to be –202 mV. A blank run was performed to assess the shift due to the electrolyte alone (without PNA) and, as Figure 6 indicates, the effect of the electrolyte is minimal, causing a change in Vfb of only –8 mV. Therefore, the overall shift in Vfb attributed to immobilized PNA is of –196 mV.
Figure 6.
Imaginary impedance as a function of the DC applied potential, (closed circle) after 12 min, and (open circle) after 60 min, in contact with electrolyte (no PNA).
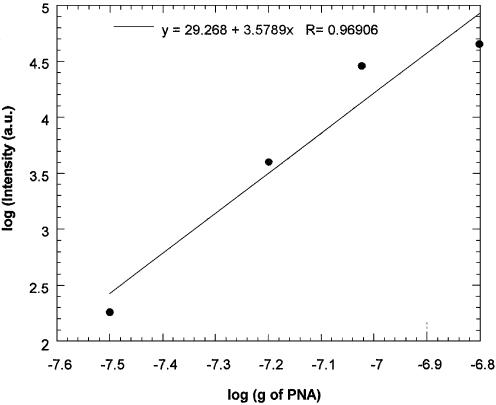
Fluorescence measurements were taken after the immobilization (following the impedance measurements). The median intensity of the PNA probe layer was determined to be 2.3 × 104 a.u.. It is interesting to note that this intensity coincides exactly with the value obtained for the fluorescent BSA experiment (Fig. 4), at maximum coverage. From the standard curve shown in Figure 7, which was generated by depositing known amounts of fluorescent PNA on Si/SiO2 chips (allowing the solvent to evaporate, but not removing any of the PNA by rinsing in this case, to preserve the known amount), the median intensity is found to correspond to a surface density of the order of 1012 strands of PNA oligomer per cm2.
Figure 7.
Standard curve of fluorescence intensity for the immobilization of PNA.
Hybridization with complementary DNA
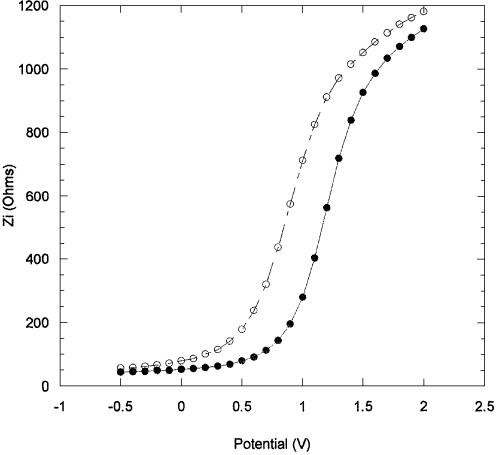
Figure 8 shows the impedance results obtained for PNA/DNA hybridization, using fluorescent-labelled DNA, performed at room temperature for 1 h. Unlabelled PNA was used as the probe layer and the chips were vigorously washed with distilled water to remove any unbound PNA prior to hybridization. The impedance measurements were taken using 0.1 M NaCl as electrolyte. As in the case of the PNA immobilization, impedance measurements were taken as the hybridization took place. Figure 9 shows the result of a blank run performed to determine if any Vfb shift was caused by the electrolyte. The effect of the electrolyte was negligible and therefore, the change in Vfb due to hybridization of DNA was found to be –375 mV. Following hybridization, the chips were washed with distilled water (room temperature) to remove any unhybridized labelled DNA. Fluorescence measurements taken immediately after the washing gave an intensity of 1.1 × 104 a.u., confirming the presence of the complementary DNA, and indicating that ∼50% of the surface immobilized PNA have been hybridized.
Figure 8.
Imaginary impedance as a function of the DC applied potential, (closed circle) after 12 min, and (open circle) after 60 min, hybridization with complementary DNA.
Figure 9.
Imaginary impedance as a function of the DC applied potential, (closed circle) after 12 min, and (open circle) after 60 min, in contact with electrolyte (no DNA).
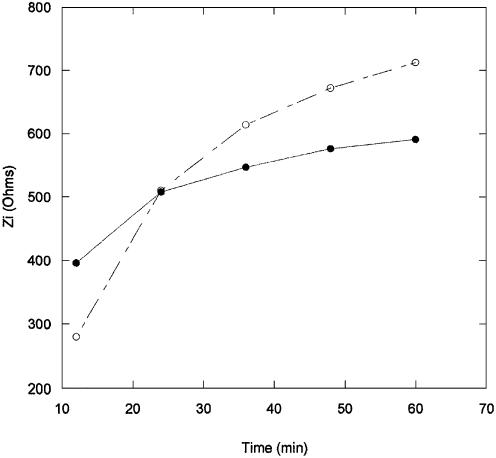
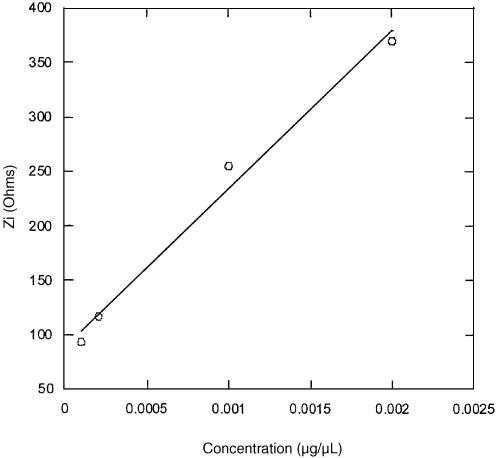
Figure 10 serves to compare the electrochemical impedance responses for the immobilization and hybridization steps, observed during the series of five measurements taken over 1 h (taken from experiments shown in Figs 5 and 8). The data are presented in the form of Zi values taken at fixed applied DC potentials of 1.1 V for the immobilization, and 1.0 V for the hybridization, as a function of time. The overall variations in Zi were found to be of ∼200 Ω and 450 Ω for the immobilization and hybridization steps, respectively. It is important to note that although the fluorescence measurements indicate a surface density of hybridized DNA which is only half that of the immobilized probe PNA, the variation in Zi (and Vfb) for hybridization is about double the value observed for immobilization. This is in accordance with the fact that the impedance-based approach to detection is sensitive to charge variations at the semiconductor/electrolyte interface, and is therefore expected to be more sensitive to the hybridization event involving charged DNA, as opposed to the immobilization of PNA. These results also show that hybridized DNA surface densities of the order of 1011 to 1012, which yield Zi variations of ∼400 Ω, can be readily measured. The sensors sensitivity to concentration variations in that range was also tested, and the results are shown in Figure 11. Individual chips were each exposed to a 100 µl amount of DNA solution at a concentration of 0.0001, 0.0002, 0.001 and 0.002 µg/µl, and left to hybridize for 1 h. The results show a good linear relationship for Zi versus DNA concentrations in this range.
Figure 10.
Imaginary impedance variations at 1.1 V for immobilization (closed circle), and 1.0 V for hybridization (open circle), as a function of time.
Figure 11.
Imaginary impedance as a function of DNA concentrations varying from 0.0001 to 0.002 µg/µl.
CONCLUSION
Reproducible immobilization of ss-PNA probe layers was achieved through an epoxide/amine coupling reaction between the PNA amino linker and the epoxide group of GPTS, followed by hybridization with the complementary DNA sequences. Electrochemical impedance measurements are seen to provide a rapid, direct and sensitive approach for the detection of the immobilization and hybridization events, as corroborated by standard fluorescence measurements. These events are indicated by significant and reproducible variations in the semiconductor flat-band potential and out-of-phase impedances.
Acknowledgments
ACKNOWLEDGEMENT
This research was made possible through support from the Canadian Institutes of Health Research.
REFERENCES
- 1.Wood S.J. (1993) DNA–DNA hybridization in real-time using biacore. Microchem. J., 47, 330–337. [Google Scholar]
- 2.Persson B., Stenhag,K., Nilsson,P., Larsson,A., Uhlen,M. and Nygren,P.A. (1997) Analysis of oligonucleotide probe affinities using surface plasmon resonance: a means for mutational scanning. Anal. Biochem., 246, 34–44. [DOI] [PubMed] [Google Scholar]
- 3.Jordan C.E., Frutos,A.G., Thiel,A.J. and Corn,R.M. (1997) Surface plasmon resonance imaging measurements of DNA hybridization adsorption and streptavidin/DNA multilayer formation at chemically modified gold surfaces. Anal. Chem., 69, 4939–4947. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson P., Persson,B., Larsson,A., Uhlen,M. and Nygren,P.A. (1997) Detection of mutations in PCR products from clinical samples by surface plasmon resonance. J. Mol. Recognit., 10, 7–17. [DOI] [PubMed] [Google Scholar]
- 5.Peterlinz K.A. and Georgiadis,R.M. (1997) Observation of hybridization and dehybridization of thiol-testhered DNA using two-color surface plasmon resonance spectroscopy. J. Am. Chem. Soc., 119, 3401–3402. [Google Scholar]
- 6.Jensen K.K., Orum,H., Nielsen,P.E. and Norden,B. (1997) Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry, 36, 5072–5077. [DOI] [PubMed] [Google Scholar]
- 7.Kai E., Sawata,S., Ikebukuro,K., Iida,T., Honda,T. and Karube,I. (1999) Detection of PCR products in solution using surface plasmon resonance. Anal. Chem., 71, 796–800. [DOI] [PubMed] [Google Scholar]
- 8.He L., Musick,M.D., Nicewarner,S.R., Salinas,F.G., Benkovic,S.J., Natan,M.J. and Keating,C.D. (2000) Colloidal au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J. Am. Chem. Soc., 122, 9071–9077. [Google Scholar]
- 9.Nelson B.P., Grimsrud,T.E., Liles,M.R., Goodman,R.M. and Corn,R.M. (2001) Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal. Chem., 73, 1–7. [DOI] [PubMed] [Google Scholar]
- 10.Su H., Kallury,K.M. R., Thompson,M. and Roach,A. (1994) Interfacial nucleic acid hybridization studied by random primer 32P labelling and liquid-phase acoustic network analysis. Anal. Chem., 66, 769–777. [Google Scholar]
- 11.Su H. and Thompson. M. (1995) Kinetics of interfacial nucleic acid hybridization studied by acoustic network analysis. Biosens. Bioelectron., 10, 329–340. [Google Scholar]
- 12.Cavic B.A. and Thompson,M. (2000) Adsorptions of plasma proteins and their elutabilities from a polysiloxane surface studied by an on-line acoustic wave sensor. Anal. Chem., 72, 1523–1531. [DOI] [PubMed] [Google Scholar]
- 13.Fawcett N., Evans,J., Chien,L. and Flowers,N. (1988) Nucleic acid hybridization detected by piezoelectric resonance. Anal. Lett., 21, 1099–1114. [Google Scholar]
- 14.Caruso F., Rodda,E., Furlong,D., Nikura,K. and Okahata,Y. (1997) Quartz crystal microbalance study of DNA immobilization and hybridization for nucleic acid sensor development. Anal. Chem., 69, 2043–2049. [DOI] [PubMed] [Google Scholar]
- 15.Okahata Y., Matsunobo,Y., Ijiro,K., Mukae,M., Murakami,A. and Makino,K. (1992) Hybridization of nucleic acids immobilized on a quartz crystal microbalance. J. Am. Chem. Soc., 114, 8299–8300. [Google Scholar]
- 16.Towery R.B., Fawcett,N.C., Zhang,P. and Evans,J.A. (2001) Genomic DNA hybridizes with the same rate constant on the QCM biosensor as in homogeneous solution. Biosens. Bioelectron., 16, 1–8. [DOI] [PubMed] [Google Scholar]
- 17.Patolsky F., Weizmann,Y. and Willner,I. (2002) Redox-active nucleic-acid replica for the amplified bioelectrocatalytic detection of viral DNA. J. Am. Chem. Soc., 124, 770–772. [DOI] [PubMed] [Google Scholar]
- 18.Millan K.M., Saraullo,A. and Mikkelsen,S.R. (1994) Voltammetric DNA biosensor for cystic fibrosis based on a modified carbon paste electrode. Anal. Chem., 66, 2943–2948. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Cai,X., Rivas,G. and Shiraishi,H. (1996) Stipping potentiometric transduction of DNA hybridization processes. Anal. Chim. Acta, 326, 141–147. [Google Scholar]
- 20.Wang J., Palecek,E., Nielsen,P.E., Rivas,G., Cai,X., Shiraishi,H., Dontha,N., Luo,D. and Farias,P.A.M. (1996) Peptide nucleic acid probes for sequence-specific DNA biosensors. J. Am. Chem. Soc., 118, 7667–7670. [Google Scholar]
- 21.Mikkelsen S.R. (1996) Electrochemical biosensors for DNA sequence detection. Electroanalysis, 8, 15–19. [Google Scholar]
- 22.Wang J., Cai,X., Rivas,G., Shiraishi,H. and Dontha,N. (1997) Nucleic-acid immobilization, recognition and detection at chronopotentiometric DNA chips. Biosens. Bioelectron., 12, 587–599. [DOI] [PubMed] [Google Scholar]
- 23.Marraza G., Chianella,I. and Mascini,M. (1999) Disposable DNA electrochemical sensor for hybridization detection. Biosens. Bioelectron., 14, 43–51. [DOI] [PubMed] [Google Scholar]
- 24.Chiti G., Marraza,G. and Mascini,M. (2001) Electrochemical DNA biosensor for environmental monitoring. Anal. Chim. Acta, 427, 155–164. [Google Scholar]
- 25.Berney H., Alderman,J., Lane,W.A. and Collins,J.K. (1997) A differential capacitive biosensor using polyethylene glycol to overlay the biolayer. Sens. Actuators B, 44, 578–584. [Google Scholar]
- 26.Patolsky F., Zayats,M., Katz,E. and Willner,I. (1999) Precipitation of an insoluble product on enzyme monolayer electrodes for biosensor applications: characterization by faradaic impedance spectroscopy, cyclic voltammetry and microgravimetric quartz crystal microbalance analyses. Anal. Chem., 71, 3171–3180. [DOI] [PubMed] [Google Scholar]
- 27.Kharitonov A.B., Shipway,A.N. and Willner,I. (1999) An Au nanoparticle/bispipyridinium cyclophane-functionalized ion-sensitive field-effect transistor for the sensing of adrenaline. Anal. Chem., 71, 5441–5443. [DOI] [PubMed] [Google Scholar]
- 28.Berggren C., Stalhandske,P., Brundell,J. and Johansson,G. (1999) A feasibility study of a capacitive biosensor for direct detection of DNA hybridization. Electroanalysis, 11, 156–159. [Google Scholar]
- 29.Berney H., West,J., Haefele,E., Alderman,J., Lane,W. and Collins,J.K. (2000) A DNA diagnostic biosensor: development, characterisation and performance. Sens. Actuators B, 68, 100–108. [Google Scholar]
- 30.Alfonta L., Katz,E. and Willner,I. (2000) Sensing of acetylcholine by a tricomponent-enzyme layered electrode using faradaic impedance spectroscopy, cyclic voltammetry and microgravimetric quartz crystal microbalance transduction methods. Anal. Chem., 72, 927–935. [DOI] [PubMed] [Google Scholar]
- 31.Souteyrand E., Cloarec,J.P., Martin,J.R., Wilson,C., Lawrence,I., Mikkelsen,S. and Lawrence,M.F. (1997) Direct detection of the hybridization of synthetic homo-oligomer DNA sequences by field effect. J. Phys. Chem. B, 101, 2980–2985. [Google Scholar]
- 32.Martin J.R., Souteyrand,E., Lawrence,M.F. and Mikkelsen,S.R. (2000) Method for analysing biological substances in a conductive liquid medium. US Patent 6,150,106.
- 33.Cloarec J.P., Deligianis,N., Martin,J.R., Lawrence,I., Souteyrand,E., Polychronakos,C. and Lawrence,M.F. (2002) Immobilization of homooligonucleotide probe layers onto Si/SiO2 substrates: characterization by electrochemical impedance measurements and radiolabelling. Biosens. Bioelectron., 17, 405–412. [DOI] [PubMed] [Google Scholar]
- 34.Marquette C., Lawrence,I., Polychronakos,C. and Lawrence,M.F. (2002) Impedance based DNA chip for direct Tm measurement. Talanta, 56, 763–768. [DOI] [PubMed] [Google Scholar]
- 35.Egholm M., Buchardt,O., Christensen,L., Behrens,C., Freler,S.M., Driver,D.A., Berg,R.H., Kim,S.K., Norden,B. and Nielsen,P.E. (1993) PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature, 365, 566–568. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen P.E., Egholm,M. and Buchardt,O. (1994) Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjugate Chem., 5, 3–7. [DOI] [PubMed] [Google Scholar]
- 37.Weiler J., Gausepohl,H., Hauser,N., Jensen,O.N. and Hoheisel,J.D. (1997) Hybridisation based DNA screening on peptide nucleic acid (PNA) oligomer arrays. Nucleic Acids Res., 25, 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross P.L., Lee,K. and Belgrader,P. (1997) Discrimination of single-nucleotide polymorphisms in human DNA using peptide nucleic acid probes detected by MALDI-TOF mass spectrometry. Anal. Chem., 69, 4197–4202. [DOI] [PubMed] [Google Scholar]
- 39.Corey D.R. (1997) Peptide nucleic acids: expanding the scope of nucleic acid recognition. TIBTECH, 15, 224–229. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen P.E. and Haaima,G. (1997) Peptide nucleic acid (PNA). A DNA mimic with a pseudopeptide backbone. Chem. Soc. Rev., 73–78. [Google Scholar]
- 41.Burgener M., Sanger,M. and Candrian,U. (2000) Synthesis of a stable and specific surface plasmon resonance biosensor surface employing covalently immobilized peptide nucleic acids. Bioconjugate Chem., 11, 749–754. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Nielsen,P.E., Jiang,M., Cai,X., Fernandes,J.R., Grant,D.H., Ozsoz,M., Beglieter,A. and Mowat,M. (1997) Mismatch-sensitive hybridization detection by peptide nucleic acids immobilized on a quartz crystal microbalance. Anal. Chem., 69, 5200–5202. [DOI] [PubMed] [Google Scholar]
- 43.Kallury K.M.R., Krull,U.J. and Thompson,M. (1988) X-ray photoelecton spectroscopy of silica surfaces treated with polyfunctional silanes. Anal. Chem., 60, 169–172. [Google Scholar]
- 44.Maskos U. and Southern,E.M. (1992) Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesised in situ. Nucleic Acids Res., 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray D.E., Case-Green,S.C., Fell,T.S., Dobson,P.J. and Southern,E.M. (1997) Ellipsometric and interferometric characterization of DNA probes immobilized on a combinatorial array. Langmuir, 13, 2833–2842. [Google Scholar]
- 46.Lamture J.B., Beattie,K.L., Burke,B.E., Eggers,M.D., Ehrlich,D.J., Fowler,R., Hollis,M.A., Kosicki,B.B., Reich,R.K. and Smith,S.R. (1994) Direct detection of nucleic acid hybridization on the surface of a charge coupled device. Nucleic Acids Res., 22, 2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]