Abstract
A procedure for precise assembly of linear DNA constructs as long as 20 kb is proposed. The method, which we call long multiple fusion, has been used to assemble up to four fragments simultaneously (for a 10.8 kb final product), offering an additional improvement on the combination of long PCR and overlap extension PCR. The method is based on Pfu polymerase mix, which has a proofreading activity. We successfully assembled (and confirmed by sequencing) seven different linear constructs ranging from 3 to 20 kb, including two 20 kb products (from fragments of 11, 1.7 and 7.5 kb), two 10.8 kb constructs, and two constructs of 6.1 and 6.2 kb, respectively. Accuracy of the PCR fusion is greater than or equal to one error per 6.6 kb, which is consistent with the expected error rate of the PCR mix. The method is expected to facilitate various kinds of complex genetic engineering projects that require precise in-frame assembly of multiple fragments, such as somatic cell knockout in human cells or creation of whole genomes of viruses for vaccine research.
INTRODUCTION
Many recombinant DNA projects require splicing of two or more long fragments of DNA, which sometimes cannot be done using conventional restriction enzyme cloning and adapters, or which would require introduction of unwanted sequences into the junctions. If an error rate of one per 5–20 kb is acceptable, overlap extension PCR (1,2) can be employed to fuse necessary fragments precisely and quickly. Existing protocols of overlap extension PCR, however, are limited to regular (short) PCR, i.e. a limit of ∼3–4 kb (3,4). Here we describe a modification of the method allowing for creation of recombinant products as long as 20 kb, at the same time capable of fusing up to four fragments simultaneously. The new method, which we call long multiple fusion, offers far greater speed and flexibility than traditional restriction enzyme cloning. Long multiple fusion can facilitate construction of linear recombinant DNAs of varying complexity. For example, it could potentially allow a vaccine researcher to create custom-made viral genomes in a few steps. In addition, it has the potential to accelerate the effort to knock out genes in human somatic cells (5). This field has been quite slow compared with the advances made in mouse gene knockout technology. Only approximately a dozen genes have been disrupted in human somatic cells compared to several thousand genes knocked out in mouse embryonic stem cells. This is largely because somatic cell knockouts require more sophisticated targeting constructs (6).
MATERIALS AND METHODS
All primers are shown in a 5′ to 3′ direction and some chimeric primers extend over two or more lines.
Short triple fusion (3 kb)
Described here is the original experiment that served to establish the long multiple fusion approach. It demonstrates that the multiple fusion can be performed not only with long PCR, but also with short (regular) PCR (limitations are discussed later). Fragments 1 and 3 were amplified from Borrelia burgdorferi genomic DNA using primers I and II, and V and VI respectively. Kan resistance gene (middle fragment) was amplified from plasmid pBLS500 using primers III and IV. Primer sequences: I, CCTAGCTAGCG GAGAAGACCGCATCTCCTG; II, TTTACTGGATGAA TTGTTTTAGTACCTCTAGATAGCGATTTGCCATCAT CAGAGC; III, ATATCTAGAAAATTCTATCATAATTGT GGTTTCAA; IV, CTATCTAGAGGTACTAAAACAATT CATCCAGTAAA; V, TTGAAACCACAATTATGATAG AATTTTCTAGATATAATTGGGCAAAGTATAATGAA TGG; VI, CGGGATCCTTTACATGCTCAGTCATAGGA CCTG. All three PCR products are ∼1 kb in size; they were gel-purified with extraction from agarose using Qiaex II Gel purification kit (Qiagen).
Triple reaction step A. 38.2 µl water, 10 µl ThermoPol reaction buffer, 5 µl 10 mM dNTP mix (Sigma), 20 µl (100 ng) fragment 1, 20 µl (100 ng) fragment 3, 4 µl (20 ng) fragment 2 (Kan), 2 µl 100 mM MgSO4 solution, 0.8 µl Deep Vent DNA Polymerase (New England BioLabs). Cycling parameters: initial denaturation 94°C 2 min, subsequent steps 94°C 15 s, annealing at 56°C 20 s, extension 72°C 1 min, 10 cycles total, hold at 4°C.
Triple reaction step B. 72 µl water, 10 µl ThermoPol reaction buffer, 5 µl 10 mM dNTP mix (Sigma), 3 µl of unpurified PCR product from step A, 2 µl 100 mM MgSO4 solution 2 µl primer I, 2 µl primer VI (both 2 µM), 0.8 µl Deep Vent DNA Polymerase. Cycling parameters: initial denaturation 94°C 2 min, subsequent steps 94°C 15 s, annealing at 62°C 20 s, extension 72°C 2 min 30 s, 35 cycles total, final additional extension 72°C 3 min, hold at 4°C. All reactions were run with heated lid. As one can see, primers used in this short triple fusion were not nested. This is undesirable in general and will not work for long multiple fusion. The resulting 3 kb PCR product was analyzed using restriction digestion with PstI and EcoRII and electrophoresis in 1% agarose.
Long triple fusion (20 kb)
The sources of sequences for manipulations described below are p3XFLAG-CMV-9 vector (Sigma) and GM3 synthase BAC clone obtained from Incyte Genomics Inc. (screening service of CIT human BAC library release 2). BAC DNA was purified using KB-500 BAC purification kit (Incyte Genomics) and kept at 4°C. All PCR products were kept at 4°C and never frozen. The sequence of ∼50 kb human GM3 synthase (SIAT9, sialyltransferase 9) gene is available in GenBank (see Supplementary Material, ‘Whole gene’).
Some primers (chimeric ones) contained long 5′ additional sequences, which resulted in introduction of sequence homologous to the end of a prospective adjacent fragment (Figs 1 and 2). These overlaps and the use of nested primers at later stages resulted in a product (20 kb) that is somewhat shorter than the sum of lengths of the parts. Primers were designed such that the target-specific sequence (3′ part) in a chimeric primer had a melting temperature within 68–70°C, and the 5′ non-complementary portion of the primer was 40–70 nt long. Correspondingly, the annealing temperature used in PCR is 65 to 68°C in order to minimize potential secondary structures and mispriming. No special software was used for the design of the chimeric primers and their primary structure was dictated solely by the nature of the prospective recombinant joints. In contrast, all regular and nested primers were designed using Primer 3 web-based software, found at http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi, for increased specificity and these primers also had melting temperatures within the 68–70°C range. Three DNA fragments of 11.2, 1.7 and 7.5 kb were joined using this approach to create a 20 kb fusion product.
Figure 1.
Outline of long triple fusion (sizes of fragments are not to scale).
Figure 2.
Mechanism of triple fusion at step A (primers are added at step B). Individual strands of DNA fragments and their 5′ and 3′ ends are shown. The flow chart shows theoretical stages of formation of double and triple fusion products. Each stage roughly corresponds to a PCR cycle (denaturation, annealing and extension of primed 3′ ends). Overlapping ends (of identical sequence) of fragments are shown with identical shading. Note that complementary ends have different fill patterns in the schematic. Triple fusion products are then amplified using nested primers at step B (not shown).
The 11.2 kb first fragment was amplified using Herculase Hotstart DNA Polymerase kit (Stratagene): 33 µl water, 5 µl Herculase buffer, 2.5 µl dNTPs 10 mM (Sigma), 5 µl GM3 synthase gene BAC clone DNA (50 ng/µl in 5 mM Tris–HCl pH 8.5), 1 µl left primer ‘21a’ (50 µM in 5 mM Tris–HCl pH 8.5), 1 µl right primer ‘21b’ (50 µM in 5 mM Tris–HCl pH 8.5), 1 µl Herculase hot-start polymerase, 1.5 µl DMSO, 50 µl of mineral oil on top after the ingredients are carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 65°C 30 s, extension 68°C 11 min 30 s plus automatic extension 5 s per cycle, 27 cycles total, final additional extension 68°C 13 min, hold at 4°C. Primer sequences: 21a, TCTGAGAGTAACTGCCCTCTTGAC ATC; 21b, CATCTTGCTTTGAGCTCGGGTG.
Second fragment, 1.7 kb Neo-polyA cassette [using HF 2 Advantage PCR kit (Clontech)]: 37 µl water, 5 µl HF buffer, 5 µl dNTPs, 0.3 µl p3XFLAG-CMV-9 vector (1 µg/µl), 1 µl left primer ‘2a’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl right primer ‘2b’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl HF polymerase. Cycling parameters: initial denaturation 94°C 30 s, subsequent steps 94°C 15 s, annealing 65°C 40 s, extension 68°C 1 min 50 s, 26 cycles total, final additional extension 68°C 3 min, hold at 4°C. Primer sequences: 2a, GTGATTGCTCGAGGCCTTCCCTGCAATGGTACACCCGAGCTCAAAGCAAGATGATTGAACAAGATGGATTGCACGCAGGTTC; 2b, ATGCATTTTTTTCATGTCACA TTCTTCAGTAGTATAATTTAACTTGAGGATATAAAGGATCCACACTCCAGGGAATTGATCCAGACATGATAAGATACA.
Third, 7.5 kb, fragment [using EXL polymerase PCR kit (Stratagene), which can be used interchangeably with Herculase DNA polymerase throughout the whole Materials and Methods section]: 34.5 µl water, 5 µl EXL buffer, 2.5 µl dNTPs 10 mM, 2.5 µl GM3 synthase gene BAC clone DNA (100 ng/µl in water), 1 µl left primer ‘3a’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl right primer ‘3b’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl EXL polymerase, 1.5 µl DMSO, 1 µl stabilizing solution, 50 µl of mineral oil on top after the ingredients are carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 60°C 30 s, extension 68°C 7 min 30 s plus automatic extension 4 s per cycle, 26 cycles total, final additional extension 68°C 9 min, hold at 4°C. Primer sequences: 3a, TGGAGTGTGGATCCTTTATATCC; 3b, AGACCTTCTTCTGCCCATATACATC.
PCRs were individually purified using Qiaquick PCR purification kit (Qiagen), with the only modification being that final elution was performed using 30 µl of 5 mM Tris–HCl pH 8.5. The purified products were then analyzed by electrophoresis in 1% precast agarose gels (Sigma), using 5 µl of each eluate per lane and quantified using 1–10 kb DNA markers (Promega) as a standard of molecular weight. The fragments were then mixed in equimolar amounts (such that total DNA amount was no less than 0.7 µg/50 µl PCR) and spliced using long triple fusion. If the total volume of the mixture of the three DNA fragments (0.45, 0.08 and 0.3 µg of 11, 1.7 and 7.5 kb fragments, respectively) exceeded one-fifth of the next PCR, they were drop-dialyzed on nitrocellulose filters [25 mm diameter, White VCWP, 0.1 µ (Millipore)] for 1 h to remove the Qiaquick elution buffer (7) or precipitated with ethanol to increase DNA concentration. In step A, the equimolar mixture of the three fragments was cycled 13 times without primers, and in step B an aliquot from step A was run with nested primers to obtain the final 20 kb product.
Long triple fusion step A. 19.5 µl water, 5 µl Herculase buffer, 2.5 µl 10 mM dNTP mix (Sigma), 3 µl 11 kb fragment (∼0.45 µg), 1.5 µl 1.7 kb fragment (∼80 ng), 15 µl 7.5 kb fragment (∼0.3 µg), no primers, 1 µl Herculase hot-start polymerase, 2.5 µl DMSO, 50 µl of mineral oil on top after the ingredients are carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 62°C 1 min, extension 68°C 21 min, 13 cycles total, final extension 21 min, hold at 4°C. The resulting PCR was purified using Qiaquick PCR purification kit and DNA eluted with 30 µl of 5 mM Tris–HCl pH 8.5. About one-third of this DNA (10 µl) was used in the next step.
Long triple fusion step B. 28.2 µl water, 5 µl Herculase buffer, 2.5 µl 10 mM dNTP mix (Sigma), 10 µl of purified DNA from step A, 0.4 µl left primer ‘21c’ (50 µM in 5 mM Tris–HCl pH 8.5), 0.4 µl right primer ‘1d’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl Herculase hot-start polymerase, 2.5 µl DMSO, 50 µl of mineral oil on top after the ingredients are carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, combined annealing and extension 68°C 20 min 40 s plus 10 s per cycle, total 31 cycles, final extension 30 min, hold at 4°C. Primer sequences: 21c, AAAGCAGGCAATTGAAT GACAGTAATGATG; 1d, GTGTAGCATTCAAGGCCTTT TGCTATCTGG. The product of the long triple fusion was analyzed using pulse-field electrophoresis [Chef Mapper (Bio-Rad, CA)].
10.8 kb long quadruple fusion
Following is a protocol for assembling a 10.8 kb recombinant product from four fragments. The sources of DNA and principles for primer design were the same as described above.
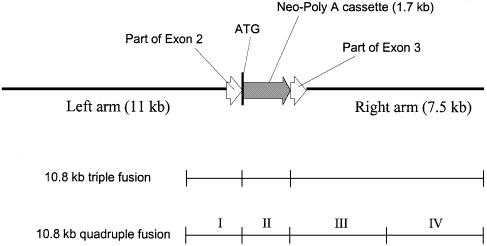
Initial fragments to be assembled, 1.9, 1.7, 3.6 and 3.9 kb (the first, second, third and fourth fragments, respectively), were amplified using conventional means: high-fidelity PCR kit, HF 2 Advantage, which is applicable for amplicons <4 kb. The principle of the long quadruple fusion is shown in Figure 3. The four fragments were amplified individually with chimeric or regular primers. The first, third and fourth PCR products (see Supplementary Material, folder 4) were amplified from human GM3 synthase gene BAC clone (Fig. 7, lower panel), and the second (1.7 kb PCR product) contained the neomycin gene and polyadenylation sequence (Neo-polyA cassette) amplified from p3XFLAG-CMV-9 vector. All PCRs were run on MJ Research Gradient Cycler PTC-225 with heated lid. All primers were synthesized by Integrated DNA Technologies, Inc.
Figure 3.
Outline of long quadruple fusion (not to scale).
Figure 7.
The structure of the neomycin-based 20 kb recombinant product. The locations of the corresponding fragments used for 10.8 kb triple fusion (Fig. 6A, lane 2) and 10.8 quadruple fusion (see Figs 3 and 9) are shown.
Initial fragments. 1.9 kb left arm (using HF 2 Advantage PCR kit): 34.5 µl water, 5 µl HF buffer, 5 µl kit dNTPs, 2.5 µl GM3 synthase gene BAC clone DNA (100 ng/µl in water), 1 µl left primer ‘1a’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl right primer ‘1b’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl HF polymerase. Cycling parameters: initial denaturation 94°C 30 s, subsequent steps 94°C 15 s, annealing 65°C 40 s, extension 68°C 2 min, 26 cycles total, final additional extension 68°C 2 min, hold at 4°C. Primer sequences: 1a, AATGAATGAGATATAACTTCTTCCATCCAA; 1b, CGTGCAATCCATCTTGTTCAATCATCTTGCTTTGAGCTCGGGTGTAC.
The 1.7 kb Neo-polyA cassette was the same as the one used in 20 kb triple fusion described in the previous section.
Third, 3.6 kb, fragment (using HF 2 Advantage PCR kit): 34.5 µl water, 5 µl HF buffer, 5 µl kit dNTPs, 2.5 µl GM3 synthase gene BAC clone DNA (100 ng/µl in water), 1 µl left primer ‘3a’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl right primer ‘QR’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl HF polymerase. Cycling parameters: initial denaturation 94°C 30 s, subsequent steps 94°C 15 s, annealing 65°C 40 s, extension 68°C 3 min 50 s, 27 cycles total, final additional extension 68°C 4 min, hold at 4°C. Primer sequences: 3a, TGGAGTGTGGATCCT TTATATCC; QR, TGGAGAGATCAGTAGGAACCAA GTC.
Fourth, 3.9 kb, fragment (using HF 2 Advantage PCR kit): 34.5 µl water, 5 µl HF buffer, 5 µl kit dNTPs, 2.5 µl GM3 synthase gene BAC clone DNA (100 ng/µl in water), 1 µl left primer ‘QL’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl right primer ‘3b’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl HF polymerase. Cycling parameters: initial denaturation 94°C 30 s, subsequent steps 94°C 15 s, annealing 65°C 40 s, extension 68°C 4 min, 27 cycles total, final additional extension 68°C 4 min, hold at 4°C. Primer sequences: QL, CCAGACTCCTTAGTCTGGCCCAGAA; 3b, AGACCTT CTTCTGCCCATATACATC.
Individual PCR products were purified and quantified as described above for the 20 kb triple fusion. After that, the four fragments carrying sequence overlaps were fused using long quadruple fusion (Fig. 3 and Fig. 7, lower panel). At each step of the long multiple fusion, DNA fragments were mixed at roughly equimolar proportions, which is essential for a high yield of a desired final product. The two left fragments (1.9 and 1.7 kb) were cycled without primers 12 times (step 1A). The same PCR protocol was used with two right fragments (3.6 and 3.9 kb, step 1B). Aliquots from these two reactions were then combined and another PCR was run for 20 cycles without primers (step 2AB). To select for the desired quadruple fusion product (Fig. 3), an aliquot of this reaction was cycled with nested primers (at the ends of the prospective 10.8 kb fusion) for 30 cycles (step 3).
Step 1A (using Herculase Hotstart DNA polymerase kit). 31.5 µl water, 5 µl Herculase buffer, 2.5 µl 10 mM dNTP mix, 5 µl 1.9 kb fragment (∼150 ng), 4 µl 1.7 kb fragment (∼150 ng), no primers, 0.5 µl Herculase polymerase, 1.5 µl DMSO, 50 µl of mineral oil on top after the ingredients were carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 55°C 1 min, extension 68°C 3 min 40 s, 12 cycles total, hold at 4°C.
Step 1B (using Herculase Hotstart DNA polymerase kit). 30.5 µl water, 5 µl Herculase buffer, 2.5 µl 10 mM dNTP mix, 4.5 µl 3.6 kb fragment (∼300 ng), 5.5 µl 3.9 kb fragment (∼300 ng), no primers, 0.5 µl Herculase polymerase, 1.5 µl DMSO, 50 µl of mineral oil on top after the ingredients were carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 55°C 1 min, extension 68°C 7 min 40 s, 12 cycles total, hold at 4°C.
Step 2AB (using Herculase Hotstart DNA polymerase kit). 39 µl water, 5 µl Herculase buffer, 2.5 µl 10 mM dNTP mix, 25 µl unpurified product of step 1A, 25 µl unpurified product of step 1B, no primers, 1 µl Herculase polymerase, 3.5 µl DMSO, 50 µl of mineral oil on top after the ingredients were carefully mixed by pipetting with a wide-nozzled tip and split into two 50 µl reactions. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 55°C 1 min, extension 68°C 11 min 30 s plus 5 s per cycle, 20 cycles total, hold at 4°C. The product was cleaned up using Qiaquick PCR purification kit, one column per 50 µl of PCR mixture, with the only modification being that the final elution was performed using 30 µl of 5 mM Tris–HCl pH 8.5.
Step 3 (using Herculase Hotstart DNA polymerase kit). 27 µl water, 5 µl Herculase buffer, 2.5 µl 10 mM dNTP mix, 10 µl eluate from step 2AB, 1 µl left primer ‘1c’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl right primer ‘1d’ (50 µM in 5 mM Tris–HCl pH 8), 1 µl Herculase polymerase, 2.5 µl DMSO, 50 µl of mineral oil on top after the ingredients were carefully mixed by pipetting with a wide-nozzled tip. Cycling parameters: initial denaturation 92°C 1 min, subsequent steps 92°C 10 s, annealing 55°C 1 min, extension 68°C 11 min 30 s plus 5 s per cycle, 30 cycles total, hold at 4°C. Primer sequences: 1c, AGCTCCTTCTCTTGTTGTTCCACCATC; 1d, GTGTAG CATTCAAGGCCTTTTGCTATCTGG. 5 µl of the PCR was analyzed on 0.5% agarose gel.
Other long triple fusions
We also assembled additional triple fusion products based on the principles outlined for the 20 kb fusion above: a 10.8 kb triple fusion product based on neomycin resistance cassette (Fig. 5 and Fig. 7, middle panel); a 20 kb product based on hygromycin resistance cassette (Figs 6 and 8); and two other products of 6.2 and 6.1 kb (Figs 10 and 11). In all cases input of DNA fragments was not less than 700 ng total per 50 µl of PCR mixture at step A (Fig. 1), and all fragments were used in equimolar amounts, (these two conditions are critical to the success). Below are the primers and templates that were used to construct the 10.8, 20 (hygro), 6.2 and 6.1 kb fusion products. All primers are shown in the 5′ to 3′ direction and some chimeric primers extend over two lines.
Figure 5.

Electrophoretic analysis of the starting material and a 10.8 kb product of long triple fusion in 0.5% agarose gel. Lane 1, 1.9 kb left arm; lane 2, 1.7 kb Neo-polyA cassette (second fragment); lane 3, DNA 1 kb step ladder (Promega), 2–10 kb; lane 4, 7.5 kb right arm; lane 5, 10.8 kb triple fusion product. Length of the product is not the same as the sum of initial fragments because of the presence of overlaps and use of nested primers.
Figure 6.
Various long triple fusion products. (A) Pulse-field electrophoresis. Lane 1, 20 kb product of fusion of fragments 11.2, 1.7 (Neo-polyA) and 7.5 kb; lane 2, 10.8 kb product of fusion of fragments 1.8, 1.7 (Neo-polyA) and 7.5 kb; lane 3, high molecular weight markers 8.3–48.5 kb (Bio-Rad), from bottom to top: 12.2, 15.0, 17.1, 19.4, 22.6, 24.8, 29.9, 33.5, 38.4 and 48.5; lane 4, 20 kb product of fusion of fragments 11.2, 1.7 (Hygro-polyA) and 7.5 kb. (B) Electrophoresis in 1% agarose (Sigma). Lane 1, 1 kb DNA step ladder (Promega), 1–10 kb; lane 2, 20 kb Neo-polyA product.
Figure 8.
The structure of the hygromycin-based 20 kb recombinant product (see also Fig. 6A, lane 4).
Figure 10.
Structure of vectors GFP One (upper panel) and GFP Two (lower panel). Each of these constructs was synthesized from three fragments: the left arm (up to exon 3 start, ∼2.3 kb), startless GFP (680 bp) and the right arm (∼3.6 kb). Exons and introns are from human GM3 synthase gene. See also Figure 11.
Figure 11.
Assembly of GFP-based targeting vectors (vectors GFP One and GFP Two in Fig. 10). Left: amplification of components from non-isogenic genomic DNA (homologous regions) and phMGFP plasmid vector (GFP protein coding region). Lane 1, GFP fragment (680 bp) amplified with chimeric primers (85 nt each) specific for targeting vector One; lane 2, GFP fragment (680 bp) amplified with chimeric primers (85 nt each) specific for targeting vector Two; lane 3, left homologous arm (2.3 kb, the same for both vectors); lane 4, right homologous arm (3.6 kb) for vector One; lane 5, right homologous arm (3.6 kb) for vector Two; lane M, DNA molecular weight markers, 1–10 kb. Right: PCR products obtained using long triple fusion method. Lane 1, targeting vector GFP One, 6.1 kb (see Fig. 10, upper panel); lane 2, targeting vector GFP Two, 6.2 kb (see Fig. 10, lower panel); lane M, DNA molecular weight markers, 1–10 kb.
10.8 kb triple fusion. Left arm 1.9 kb (using HF 2 Advantage PCR kit): template BAC clone of human GM3 synthase, primers AATGAATGAGATATAACTTCTTCCATCCAA and CGTGCAATCCATCTTGTTCAATCATCTTGCTTTG AGCTCGGGTGTAC.
1.7 kb Neo-polyA cassette (using HF 2 Advantage PCR kit): template p3XFLAG-CMV-9 vector (Sigma), primers: GTGATTGCTCGAGGCCTTCCCTGCAATGGTACACCCGAGCTCAAAGCAAGATGATTGAACAAGATGGATTGCACGCAGGTTC and ATGCATTTTTTTCATGT CACATTCTTCAGTAGTATAATTTAACTTGAGGATATAAAGGATCCACACTCCAGGGAATTGATCCAGACATGATAAGATACA.
7.5 kb right arm (using Hot Start Herculase DNA polymerase): template BAC clone of human GM3 synthase, primers: TGGAGTGTGGATCCTTTATATCC and AGACC TTCTTCTGCCCATATACATC.
Nested primers used in the final fusion step (using Hot Start Herculase DNA polymerase): AGCTCCTTCTCTTGTTGTTCCACCATC and GTGTAGCATTCAAGGCCTTTTGCTATCTGG.
20 kb Hygro-based triple fusion. Left arm 11 kb (using Hot Start Herculase DNA polymerase): template BAC clone of human GM3 synthase, primers TCTGAGAGTAACTGCCCTCTTGACATC and CAGACGTCGCGGTGAGTTCAGGCTTTTTCATCTTGCTTTGAGCTCGGGTGTACCA.
Hygro-polyA cassette (1.7 kb) (using HF 2 Advantage PCR kit): template vector pTK-Hyg (Clontech), primers: AGTGATTGCTCGAGGCCTTCCCTGCAATGGTACACCCGAGCTCAAAGCAAGATGAAAAAGCCTGAACTCACCGCGACGTCTG and TGTCACATTCTTCAGTAGTATAAT TTAACTTGAGGATATAAAGGATCCACACTCCAGCGCCAGAAATCCGCGCGGTGGTTTTTGG.
Right arm (7.5 kb) (using Hot Start Herculase DNA polymerase): template BAC clone of human GM3 synthase, primers: TGGAGTGTGGATCCTTTATATCC and AGACC TTCTTCTGCCCATATACATC.
At step A, annealing temperature of 48°C was used.
Nested primers used in the final fusion step (using Hot Start Herculase DNA polymerase): AAAGCAGGCAATTGAATGACAGTAATGATG and GTGTAGCATTCAAGGCCTTTTGCTATCTGG.
6.1 kb GFP One vector. 2.3 kb left arm (using HF 2 Advantage PCR kit): template human genomic DNA [isolated using Trizol reagent (Invitrogen)], primers GATGCAACATATAAAGGCTCTTTGA and CATAATACCTAATTGATTTTTTTCCAGA.
680 bp startless GFP cassette (using HF 2 Advantage PCR kit): template phMGFP vector (Promega), primers: CATCTCATGATTCTCACATGACTACATCATAATACCTAATTGATTTTTTTCCAGAGGCGTGATCAAGCCCGACATGAAGATCAAG and AAATAAAAACAATAAAAATAA GTAAACAAACAAACAAAAAAAACCAATGTACCTTTTAGCCGGCCTGGCGGGGTAGTCCGCTGTG.
Right arm (3.6 kb) (using HF 2 Advantage PCR kit): template human genomic DNA (isolated using Trizol reagent), primers: GGTACATTGGTTTTTTTTGTTTGTTTG and GTA TACAGTTGAGCCAAAGTGAAATTTGT.
Nested primers used in the final fusion step (using Hot Start Herculase DNA polymerase): GAAGGCCCAGCTTGTTATTAAAAGAC and TCCAAAAGAAGAGTCGTACCCAGA.
6.2 kb GFP Two vector. 2.3 kb left arm (using HF 2 Advantage PCR kit): template human genomic DNA (isolated using Trizol reagent), primers GATGCAACATATAAAGGCTCTTTGA and CATAATACCTAATTGATTTTTTTCCAGA.
680 bp startless GFP cassette (using HF 2 Advantage PCR kit): template phMGFP vector, primers: CATCTCATGATTCTCACATGACTACATCATAATACCTAATTGATTTTTTTCCAGAGGCGTGATCAAGCCCGACATGAAGATCAAG and GCATATGCTTGGCAATGTTTTCATTTAAAC CACACCAATGGGATATCTAACCTTTTTAGCCGGCCTGGCGGGGTAGTCCGCTGTG.
Right arm (3.6 kb) (using HF 2 Advantage PCR kit): template human genomic DNA (isolated using Trizol reagent), primers: AAGGTTAGATATCCCATTGGTGTGG and GACTTGGTTCCTACTGATCTCTCCA.
Nested primers used in the final fusion step (using Hot Start Herculase DNA polymerase): AGAACTAGGAGAAAAATTCAGTGAGAGC and AGACTAAGGAGTCTGGGTATTTTCCTG.
Sequencing
Sequencing of fusion points in all of the above constructs as well as comprehensive sequencing of the 10.8 kb triple fusion product was performed by Northwoods DNA, Inc. (Becida, MN) and MCLab, Inc. (South San Francisco, CA).
RESULTS
As one can see in the outline of the long triple fusion procedure (Fig. 1), homologous sequences introduced into adjacent ends of DNA fragments to be fused essentially allow the end fragments to act as giant chimeric primers on the middle fragment. This results in appearance of the longer DNA species in the mixture (Fig. 2), which consist of two or three fragments spliced together, as revealed by agarose gel analysis (data not shown). This stage requires a relatively low annealing temperature, because (i) the concentration of complementary ends is low and (ii) they are shorter than expected and thus have lower melting temperature (discussed later). This is the essence of the part A of long triple fusion (Fig. 1). Part B allows one to selectively amplify only the triple fusion product using nested primers and, at this stage, annealing temperature should be high (∼68°C for primers with a melting temperature of 70–72°C) for best specificity of primers. Nested primers are required because long PCR frequently results in products that are somewhat shorter at the ends than expected and therefore cannot be re-amplified with the original primers (data not shown).
A modification of the triple fusion allows one to join four fragments simultaneously as shown in Figure 3. In this case, there are two primerless steps. First, two pairwise reactions are run, and then the products of the two reactions are combined and cycled to produce a quadruple fusion species. In the final step, an aliquot from the second primerless step is used to amplify the final product with nested primers. Equimolar concentrations of DNA fragments at each step of a long multiple fusion are essential to reduce the appearance of side products at the final step.
We have successfully applied this method to produce recombinant DNAs of the following lengths: 3, 6.2, 10.8 and 20 kb. The error rate of our PCR fusion method was less than one error per 6.6 kb (Table 1, discussed below). Figure 4 shows the shorter fusion, 3 kb, which served as a basis for the development of the long multiple fusion method. The correct structure of the 3 kb recombinant product was confirmed by restriction analysis (three initial 1 kb fragments are not shown in Fig. 4).
Table 1. Base changes detected in the 10.8 kb triple fusion construct.
| Substitution | Position in construct | Known SNP | GenBank ID |
|---|---|---|---|
| G→A |
1141 |
G or A |
rs6547635 |
| A→T |
4463 |
T or A |
rs7561062 |
| G→A |
5824 |
G or A |
rs4832210 |
| A→G | 6077 | G or A | rs6759239 |
The construction of this product involved a total of ∼70 cycles (26 + 13 + 31) using Pfu-based PCR kits (the enzyme has a proofreading activity). Of the 10.8 kb, ∼6.6 kb were sequenced using multiple runs of about 300–500 bp (see Supplementary Material, folders 1 and ‘Whole gene’). Discrepancies between the expected sequence and the raw sequence data were detected by aligning the data using BioEdit software (19) and then verified visually in chromatographic files. Four substitutions were found (Supplementary Material, folder 1) and are presented above. Since all of these base changes correspond to known SNPs in GM3 synthase gene (go to http://www.ncbi.nlm.nih.gov/SNP/, note that the genomic data is complementary to our direct sequence of the gene), it can be concluded that the error rate of the triple fusion method is less than or equal to one error per 6.6 kb, which is consistent with the manufacturer’s data (see Discussion) (16). All known SNPs in that region of GM3 synthase gene are listed in Supplementary Material, folder 1, file ‘10.8kbNeo expected sequence.txt’.
Figure 4.

Analysis of the product of triple fusion of three 1 kb fragments in 1% agarose gel. Lane 1, PCR product; lane 2, DNA molecular weight markers. Note that primers used in step B were not nested.
Initially, we assembled the 10.8 kb construct from three fragments simultaneously (Fig. 5) using the approach outlined in Figure 1. After this first long triple fusion was confirmed by sequencing and electrophoresis, we proceeded to optimize the procedure for longer constructs (20 kb) and a greater number of fragments (four). An optimized triple fusion protocol enabled us to assemble a 20 kb product from 11.2, 1.9 and 7.5 kb fragments using the same approach. Figure 6 (A, lane 1, and B) shows the product of long triple fusion of the above DNA fragments, and one can see that the yield and purity are worse than those of the 10.8 kb product in Figure 5. As is the case with most regular long PCR products, there are some nucleoprotein aggregates at the start, which do not enter the gel (seen in Fig. 6B). The amount of these aggregates was less significant for the 10.8 kb product (Fig. 5, lane 5), nevertheless further chromatographic or gel purification of long fusion products could be required. As estimated by agarose gel electrophoresis, a 50 µl PCR yields ∼1 µg of the 20 kb product. The structure of the first 20 kb construct is shown in Figure 7, upper panel. We also assembled a 20 kb product using a different middle fragment, hygromycin resistance cassette instead of neomycin resistance (Fig 6, lane 4, and Fig. 8). Both 20 kb constructs are so-called promoterless targeting vectors (6), which contain (i) a left arm carrying a large first intron and translation regulatory sequence at the beginning of exon 2 of GM3 synthase gene, (ii) promoterless drug resistance gene with polyadenylation signal, i.e. Neo-polyA cassette (or Hygro-polyA cassette) inserted exactly at the translation start of GM3 synthase gene and replacing parts of exons 2 and 3 and all of intron 2 and (iii) a right arm of GM3 synthase gene sequence downstream of exon 3 (Figs 7 and 8). As one can see in Figure 7, the 20 kb Neo construct includes the above 10.8 kb triple fusion product (Fig. 7, middle panel) and the 10.8 kb quadruple fusion product (Fig. 7, lower panel) described below. The correct structure of the above long constructs was confirmed by sequencing (MCLab, CA) 300–500 bp across the two joints (Figs 7 and 8) between the fragments (Supplementary Material, folders 1, 2 and 3).
Figure 9 shows initial fragments 1.9 (first), 1.7 (second), 3.6 (third) and 3.9 kb (fourth) and the 10.8 kb product of long quadruple fusion. The product is of significant yield and purity, containing very few side products (Fig. 9, lane 6). It is estimated that a 50 µl PCR yields ∼3 µg of the recombinant product. To confirm the correctness of the structure of the 10.8 kb product, it was partially sequenced (MCLab, CA) (see Supplementary Material, folder 4). The sequencing covered ∼400 bp around each of the three chimeric joints and matched the expected sequence. There was no need to fuse the third and fourth fragments (3.6 and 3.9 kb) because they are part of the 7.5 kb fragment of GM3 synthase gene sequence (Fig. 7, lower panel). Nevertheless we amplified the third and fourth fragments such that they overlap by 60 nt, theoretically (less than 60 in reality due to shortening of high-fidelity PCR products, discussed below), solely to test our quadruple fusion approach, after we successfully assembled the 10.8 kb construct from the three fragments (Fig. 7, middle panel).
Figure 9.
Electrophoretic analysis of the starting material and a 10.8 kb product of long quadruple fusion in 0.5% agarose gel. Lane 1, 1.9 kb first fragment; lane 2, 1.7 kb Neo-polyA cassette (second fragment); lane 3, 3.6 kb third fragment; lane 4, 3.9 kb fourth fragment; lane 5, DNA 1 kb step ladder (Promega), 1–10 kb; lane 6, 10.8 kb recombinant product. Length of the product is not the same as the sum of initial fragments, because of overlaps and use of nested primers.
Originally we made an attempt at assembling the above constructs (Figs 7 and 8) by means of restriction digestion and ligation; however, the complexity of precise construction of a product that is longer than 8–10 kb was overwhelming. The approach proposed in this work facilitates the task significantly. Long multiple fusion approach also takes less time: 1–2 weeks compared to several months or more with the traditional restriction/ligation approach.
We also assembled shorter products (Fig. 10) from a different region of human GM3 synthase gene and a modified green fluorescent protein (Monster GFP) sequence found in phMGFP vector. Electrophoretic analysis of the component fragments and final products is shown in Figure 11. Correct structure of these 6.1 and 6.2 kb constructs was also confirmed by sequencing across fusion points (see Supplementary Material, folders 5 and 6). Sample fusion point sequence is shown in Figure 12.
Figure 12.
Sample sequence of a fusion point in 6.2 kb construct GFP Two. Expected sequence is shown as letters beneath peaks. Color code: green, A; blue, C; black, G; red, T. Double underscored region, end of intron 2; asterisk, start of exon 3; single underscored region, beginning of startless Monster GFP open reading frame. See also Figure 10, lower panel and Supplementary Material, folder 6, file NS2-SQGFP1.ab1, starting from position 48.
To verify the error rate of our method, we sequenced ∼6.6 kb of the 10.8 kb triple fusion product (shown in Figs 5 and 7), and compared the sequence to the sequence of human GM3 synthase (see Supplementary Material, folder ‘whole gene’) found in GenBank. All mismatches between the expected sequence and the raw data were verified manually by visual inspection of alignments and chromatograms. In this 6.6 kb region of completed good-quality sequence, we found four base changes (Table 1 and Supplementary Material, folder 1). Other deletions/insertions and substitutions were eliminated as erroneous reads of chromatographic peaks, since most of the sequence was covered twice or more. As shown in Table 1, all four substitutions correspond to known single nucleotide polymorphisms (SNPs) in this region of GM3 synthase (sialyltransferase 9) gene. Therefore, the changes most likely resulted from the natural differences between the genomic DNAs, our BAC clone of the gene and the DNA used in the Human Genome Project, rather than from PCR errors. It follows that the error rate of the long triple fusion method is less than or equal to one error per 6.6 kb.
DISCUSSION
The method for splicing two short fragments of DNA, also known as overlap extension PCR, was described more than a decade ago (2). Powerful as it is, the technique is limited to products of 3–4 kb in length and to fusion of no more than two pieces of DNA at a time (8). The novelty of the method proposed in this work is two-fold: first it allows one to simultaneously splice up to four fragments, rather than two, with high yield; second, it allows one to splice long PCR products (>4 kb), which is also not achievable by overlap extension PCR. Specifically, overlap extension PCR method requires several non-trivial modifications in order to be used with long PCR or with several fragments rather than two.
First, the initial fragments have to be produced with an enzyme that has a 3′→5′ proofreading activity, e.g. Pfu polymerase rather than Taq polymerase. The problem with Taq is that it leaves single A-overhangs at 3′ ends, which disrupt priming of the overlapping regions during the overlap extension reaction (9). The majority of long PCR protocols and commercial kits are predominantly based on Taq polymerase and thus cannot be easily used for overlap extension reaction. Long PCR mixtures that are based predominantly on a proofreading enzyme came on the market only recently (Herculase polymerase, EXL polymerase, PfuUltra High-Fidelity DNA polymerase: all based on Pfu enzyme); however, they are proprietary and their contents were not disclosed. Alternatively, one could generate the needed long intermediate fragments with Taq polymerase and then flush the ends, e.g. using T4 polymerase treatment (10). This would add several additional intermediate steps into the whole procedure and we did not test this approach.
Second, purification of the intermediate PCR products should be limited to ‘bind-and-wash’ or desalting methods, such as Qiaquick. Although acceptable for short PCR under 3 kb (demonstrated in Fig. 4), gel-purification of longer fragments renders them unusable for long multiple fusion (data not shown). This is most likely due to the damage to the template that results from exposure to ethidium bromide and UV light. Long PCR in general is much more sensitive to the quality of the template (11). Repeated freezing and thawing or storage in distilled water, rather than in a buffer with pH above 7, will also render a template unusable for long PCR, but not for regular PCR [(11) and our data]. If one does have to gel-purify long PCR products, it is recommended that very low concentrations of ethidium bromide be used in agarose gels (<0.15 µg/ml) and the gel never exposed to direct UV light (as when bands are cut out), which is achieved by performing all manipulations on a glass plate, thus exposing DNA only to indirect UV. We did not test if DNA purified in this way will still work for long multiple fusion, but it does work well for homologous recombination in somatic cells, suggesting low levels of DNA damage by UV. Since gel purification is undesirable, it follows then that intermediate products, such as the ones shown in Figures 5, 6, 9 and 11, should be produced with high specificity, i.e. with few or no side products. In our experience, this can be achieved most of the time if a primer-design software is used to select highly specific primers, and their melting temperature is within the 68–70°C range (accordingly, annealing temperature during long PCR is 65–68°C). Where there is little leeway for primer design (chimeric joints), in our experience the above melting temperature range (can be achieved by varying primer length) yields primers that are sufficiently specific most of the time. Primers with melting temperature outside this range can result in the appearance of non-specific side products that can worsen the purity of the product of long multiple fusion (data not shown). This is especially true for DNA fragments amplified directly from genomic DNA rather than a BAC clone or plasmid. The use of chimeric primers with genomic DNA is highly undesirable because it will produce a significant background of non-specific products. Therefore, an optimal approach would be to amplify fragments that must come from genomic DNA using regular primers only, and use chimeric primers carrying extra-long overlaps (50–70 bases) to produce adjoining fragments if these can be amplified from plasmid DNA. In fact, we used this approach with our 20 kb (Neo) triple fusion and the 6.2 and 6.1 kb GFP vectors, where we used chimeric primers only with a middle fragment, and amplified two terminal fragments with regular primers only. As it turned out, introduction of overlapping sequence to only one end of a prospective fusion point rather than both will still work if the resulting overlap is sufficiently long (55–70 bases).
Third, regardless of the type of enzyme used in long PCR, nested primers should be used for consecutive PCR manipulations with PCR products (12). We determined that reamplification of PCR products with primers used in the first round fails for several different commercial kits (Taq-based and Pfu-based, data not shown). However, the use of nested primers in the second round (reamplification), or very long primers (such as our chimeric primers 47–99 nt long) significantly improves the success of reamplification.
Fourth, it follows from the argument above that non-complementary 5′ ‘tails’ of the chimeric primers intended for long overlap extension PCR should be long, 25 nt or more, rather than short, 10–25 nt. Hypothetically, if an additional 20 nt are introduced with the chimeric primer, and 20 nt are lost after long PCR due to the ‘shortening’ phenomenon, there may not be any overlapping sequence left in the intermediate product. As mentioned in Materials and Methods, we found that, in order for a chimeric primer to work and to be specific, the critical requirement is that the complementary 3′ portion of the primer is such that its melting temperature is ∼68–70°C, which translates into a length of 24–28 nt. The 5′ portion (non-complementary) of a chimeric primer usually has little effect on quality of this kind of primer if the annealing temperature used during PCR is high (65–68°C). High annealing temperature minimizes mispriming and formation of secondary structures (9). We have successfully used chimeric primers 46–99 nt long (see Materials and Methods). Overlap extension worked even when a chimeric primer was used on only one side of a prospective joint; as already mentioned, in those cases chimeric primers contained a 50–70mer ‘tail’, e.g. adjacent primers 2b and 3a, or 21b and 2a. It should be noted that oligonucleotides longer than 30–35 nt have to be purified from ‘erroneous’ oligonucleotides arising during chemical synthesis. This adds substantially to the cost; however, if a junction is not critical (does not contain promoters, etc), purification is optional, and it does not seem to affect the yield or specificity of an overlap extension reaction. In our case, we used purified primers in the left joint, for example 1b and 2a in the 10.8 kb fusion, and unpurified primers in the right joint, such as 2b and 23a in the 20 kb fusion (Materials and Methods and Fig. 1).
All of the above modifications to overlap extension PCR are critical for the success of either a multiple fusion or creation of long recombinant products (up to 20 kb). If the above four principles are kept in mind, overlap extension PCR can be used to achieve ‘long double fusion’ as well. We did this when we assembled the 10.8 kb product in two steps: first, we spliced the left and the middle fragment (1.9 and 1.7 kb), and then we fused the 3.5 kb product with the 7.5 kb right arm (Fig. 7, middle panel). It was confirmed by sequencing to have the expected sequence (data not shown). This stepwise approach however, requires more primers and intermediate purification steps than long multiple fusion, i.e. it takes more time and work. In addition, it introduces more errors, as one has to run more total PCR cycles. Another problem with long overlap extension PCR is that we could not apply it successfully to longer products, i.e. 20 kb. It yielded many non-specific products and little, if any, of the expected ones (data not shown).
This problem can be solved if the overlap extension reaction is split into two substeps as shown in Figures 1 and 3. First 11–13 cycles are run without primers using low annealing temperature to allow for annealing and extension of overlaps; an aliquot from this reaction is then added to a fresh PCR mix and another 30 cycles are run with primers using high annealing temperature for best specificity. Thus, the approach shown in Figure 1 can be used with two long fragments as well. It should be noted that a short triple reaction, whereby one just mixes three overlapping fragments with primers and runs overlap extension PCR, has been reported (4), but our group and others have had little success with this approach (12,13). In our experience, this approach yields very little or no expected product (we performed three different reactions of this type, data not shown); thus, in the few experiments tested, it appears to be important to leave the primers out of the early cycles.
There are reports documenting reconstruction of a short DNA fragment from multiple overlapping degradation products (14,15); it should be noted, however, that the reconstruction when overlaps are already present [some in excess of 200 bp in Golenberg et al. (15)] is quite different from construction, where the main challenge is introduction of chimeric sequence at the ends of unrelated DNA pieces and finding a protocol that would produce a good yield of the expected recombinant product, especially a long one. Theoretically, it is easy to calculate that if one mixes equimolar amounts of three overlapping fragments (Fig. 2), then after a certain number of PCR cycles (without end primers), all available 3′ ends are extended, and the longest recombinant strands (corresponding to the final fusion product) will represent one third molar fraction of the final mix (Fig. 2). Analogous results can be demonstrated for two and four overlapping fragments, where double and quadruple fusion products will comprise one half and one quarter (in molar terms), respectively, of the final products. Further cycling will not increase the fraction of the expected final recombinant product because all available 3′ ends will have been exhausted very quickly. Further, it is also easy to show that if at this point primers are added that are terminal with respect to the expected recombinant product, the latter will be amplified and the percentage of the shorter (incomplete) fusion strands will be asymptotically approaching zero.
Finally, although long multiple fusion offers greater speed and flexibility for construction of recombinant molecules than traditional restriction enzyme and ligase cloning, it is expected to introduce errors, as any other PCR. We sequenced a total of 6.6 kb in the 10.8 kb triple fusion product and did not detect PCR errors other than known SNPs, suggesting that the accuracy of the triple fusion method is greater than one error per 6.6 kb. This is somewhat better than the estimates obtained from the vendor’s data, one error per 5 kb (Stratagene) as shown below. Assuming that the error rate of Herculase polymerase is 2.8 errors per cycle per megabase according to the manufacturer’s data (16), total number of substitutions introduced during long triple fusion is expected to be approximately (26 + 13 + 31) × 2.8 = 196 errors per megabase or about one base change per 5 kb. Our better than expected result can be explained by the fact that the number of DNA molecules produced by PCR increases exponentially and only errors that occur in early cycles will be present in a substantial fraction of DNA molecules. On the other hand, errors that happen in later cycles will be present only in a tiny fraction of DNA copies, and will not be detected by sequencing.
It should be noted that there will be more expected PCR errors for the long quadruple fusion due to the additional 20-cycle step. This trade off between speed and accuracy should be considered during the planning phase of a recombinant DNA project. If very high accuracy is required, then traditional restriction enzyme cloning should be used. For shorter projects (<4 kb), accuracy is expected to be better, since high-fidelity PCR can be used, which has an error rate as low as one change per cycle per megabase (17).
Since the PCR mixes used in this work allow amplification of 40–50 kb targets (16), we also tried to assemble a construct 29 kb long from fragments of 12.6, 1.7 and 15 kb using our triple fusion approach. However, this was not successful and further modifications and/or optimization of the method (or a different method) might be needed to produce constructs longer than the 20 kb product reported in this work. We also tried to perform a quadruple fusion of a 20 kb construct; this was not successful either. Thus, the limit of long triple fusion at this stage is 20 kb, while the upper limit of quadruple fusion seems to be ∼11 kb.
In conclusion, our approach combines two methods: long PCR and overlap extension PCR, and the latter is modified further, allowing one to splice multiple fragments rather than only two fragments. This new approach gives molecular biologists much wider latitude in the design of recombinant DNA. Three examples of work that would be impossible with existing methodologies include: (i) the generation of somatic cell knockouts for a wide range of human genes; (ii) generation of multiple custom-made viral genomes, e.g. for HIV vaccine research; and (iii) certain transgene experiments. This is largely due to constraints resulting from the lack of convenient restriction sites at junctions or, in the case of long DNA constructs, the multiple occurrence of otherwise useful restriction sites, which renders them useless. Thus, existing cloning methods usually force a researcher to assemble constructs that include unwanted DNA sequences. Another problem with existing methods is that few cloning vectors allow for insertion of large DNA fragments, and those that do cannot be used for construction of complicated recombinant products due to the above problems with restriction sites. Use of methylation, partial restriction digestion, recombinase, adapters and linkers can alleviate some, but not all, of these problems, at the same time making a project more complicated and time-consuming. Long multiple fusion overcomes these difficulties, which we demonstrate in this work. In light of the recent creation of a synthetic polio virus in vitro (18), one can see that our approach would make it very easy to create certain kinds of viruses or multiple strains of a virus. We expect long multiple fusion to significantly facilitate and accelerate other complex genetic engineering projects as well.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NIH grants RO1 CA61010 (S.L.) and RO1 AI48856 (F.C.C.), and by a Children’s Research Institute Fellowship (N.A.S.).
REFERENCES
- 1.Yon J. and Fried,M. (1989) Precise gene fusion by PCR. Nucleic Acids Res., 17, 4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yolov A.A. and Shabarova,Z.A. (1990) Constructing DNA by polymerase recombination. Nucleic Acids Res., 18, 3983–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pont-Kingdon G. (1997) Creation of chimeric junctions, deletions, and insertions by PCR. Methods Mol. Biol., 67, 167–172. [DOI] [PubMed] [Google Scholar]
- 4.Kuwayama H., Obara,S., Morio,T., Katoh,M., Urushihara,H. and Tanaka,Y. (2002) PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res., 30, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedivy J.M., Vogelstein,B., Liber,H.L., Hendrickson,E.A. and Rosmarin,A. (1999) Gene targeting in human cells without isogenic DNA. Science, 283, 9a. [Google Scholar]
- 6.Sedivy J.M. and Dutriaux,A. (1999) Gene targeting and somatic cell genetics—a rebirth or a coming of age? Trends Genet., 15, 88–90. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk R. and Sergeant,A. (1980) A simple method for dialysis of small-volume samples. Anal. Biochem., 105, 403–404. [DOI] [PubMed] [Google Scholar]
- 8.Horton R.M. and Pease,L.R. (1991) In McPherson,M.J. (ed.), Directed Mutagenesis: A Practical Approach. IRL Press, Oxford, UK, pp. 217–247. [Google Scholar]
- 9.Sambrook J. and Russel,D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 10.Warrens A.N., Jones,M.D. and Lechler,R.I. (1997) Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene, 186, 29–35. [DOI] [PubMed] [Google Scholar]
- 11.RedAccuTaq LA DNA polymerase mix (1999) Sigma-Aldrich, Saint Louis, MO, Technical bulletin No. MB-690.
- 12.Slack F. (1998) The Red Book Bulletin. In Ausubel,F. et al. (eds), Current Protocols in Molecular Biology (Suppl. 42). John Wiley & Sons Inc., New York, NY. [Google Scholar]
- 13.Horton R.M. (1999) In Innis,M.A. (ed.), PCR Applications: Protocols For Functional Genomics. Academic Press, San Diego, CA, pp. 141–149. [Google Scholar]
- 14.Nasidze I. and Stoneking,M. (1999) Construction of larger-size sequencing templates from degraded DNA. Biotechniques, 27, 480–484. [DOI] [PubMed] [Google Scholar]
- 15.Golenberg E.M., Bickel,A. and Weihs,P. (1996) Effect of highly fragmented DNA on PCR. Nucleic Acids Res., 24, 5026–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borns M. and Hogrefe,H. (2000) Strategies Newsletter. Stratagene, La Jolla, CA, Vol. 13, pp. 1–3. [Google Scholar]
- 17.Advantage-HF 2 PCR Kit User Manual (1999) 24 February, Clontech, Palo Alto, CA.
- 18.Cello J., Paul,A.V. and Wimmer,E. (2002) Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science, 297, 1016–1018. [DOI] [PubMed] [Google Scholar]
- 19.Hall T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser., 41, 95–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.