Abstract
A simple and highly sensitive method for the detection of genomic DNA in tissue samples is described. It is based on amperometric detection of target DNA by forming an analyte/polymeric activator bilayer on a gold electrode. The biotinylated target DNA is hybridized to oligonucleotide capture probes immobilized on the gold electrode, forming the first layer. A subsequent binding of glucose oxidase– avidin conjugate to the target DNA and the introduction of a second layer of a redox polymer to the electrode, via layer-by-layer electrostatic self-assembly, allow for electrochemical detection of the catalytic oxidation current of glucose in a PBS solution. Less than 2.0 fg of rat genomic DNA, for both regulated and house-keeping genes, can be easily detected in 2.5 µl droplets. The proposed procedure shows very high specificity for genomic DNA in a RT–PCR mixture.
INTRODUCTION
Abnormalities in the expression of specific genes have been linked to a large and increasing number of diseases. The identification of over- or under-expressed genes provides a promising framework for an earlier diagnosis (1–5). There are ever-increasing needs for highly sensitive methods for single gene expression analysis. The most commonly used methods for the quantification of gene expression include northern blotting (1), ribonuclease protection (2) and reverse transcription–polymerase chain reaction (RT–PCR), both competitive and real-time RT–PCR (3–5). The first two methods require 10–100 µg of total RNA and can detect single mRNAs at the 106–107 copy levels. Such quantities can be easily isolated from bulk tissues. But if one has very small amounts of tissues or has a need to separate only certain types of cells for analysis, northern blotting and ribonuclease protection techniques are not feasible. RT–PCR can theoretically amplify a single nucleic acid molecule to millions and thus could be very useful for analyzing samples of very small sizes. However, the amplified nucleic acids are only visible or countable after labeling with either luminescent or radioactive labels. The sensitivity of the detection technique associated with the RT–PCR sets the limit of PCR based methods.
Alternative sequence-specific detection methods have recently been developed. For example, the development of bioelectronic DNA analysis systems has attracted substantial research efforts (6–8). Optical (9,10), electrochemical (11,12) and microgravimetric, quartz-crystal microbalance (6,13,14) transduction methods have been reported for the detection of DNA hybridization events. However, the quantification of gene expression has proven difficult owing to the limited sensitivity of the existing RNA/DNA detection techniques. Sensitive gene detection is one of the challenges in today’s medicine and diagnostics.
Recent advances in DNA bioelectronics have addressed the amplified electronic transduction of nucleic acid hybridization events. Nucleic acid–enzyme conjugates were employed as bioelectrocatalysts for the electrochemical transduction of nucleic acid recognition processes (15). Biocatalytic conjugates that associate to nucleic acid recognition pairs and stimulate the precipitation of an insoluble product on electrodes were used as an amplification system for DNA sensing (16,17). Similarly, nucleic acid functionalized liposomes (18) or nanoparticles (19) were used as particulate labels for the amplification of the DNA sensing processes. More recently, Zhang et al. reported a detection limit of 0.50 fM for a 38-base oligonucleotide using nucleic acid–enzyme conjugates as bioelectrocatalysts (20). However, of the many proposed biocatalytic schemes, only a few attempts were made in analyzing genomic DNA in real samples. More frequently, the methodology development was solely based upon very short synthetic oligonucleotides, usually 20–50 bases long. The applications of these methods in genomic nucleic acid analysis often prove to be different, and in many instances, difficult.
TP53 is one of the most frequently mutated genes in human cancers. Somatic (non-inherited) mutations in TP53 occur in about half of all human cancers. Normally, damage to cellular DNA initiates increased expression of TP53, which leads to arrest of the cell cycle. This interruption permits DNA repair to occur before the cell resumes the cell cycle and normal cell proliferation. If the DNA repair is not successful, then the cell undergoes apoptosis, or cell death. When TP53 mutates, DNA-damaged cells are not arrested and DNA repair does not take place. TP53 mutations prevent the cell from undergoing programmed cell death (apoptosis) in response to the DNA damage. Instead, the damaged cells continue to grow and divide in an unregulated way, which can lead to cancerous tumors.
In this paper, an electrochemical detection method based on the formation of a DNA/polymeric activator bilayer on a gold electrode surface for detecting genomic DNA in rat liver tissues was proposed. To have a better applicability of the method, a house-keeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and a regulated tumor protein gene 53 (TP53) in the rat liver tissues were chosen as our target analytes. A detection limit of 1.0 fM was demonstrated for both genes. Different from sandwich-type assays previously published (20,21), by incorporating multiple enzyme labels into the target cDNA chain, the enzyme label/base ratio was kept practically constant, and therefore the assay sensitivity and detection limit were very similar regardless the length of the genes, which are superior to the sandwich-type assays.
MATERIALS AND METHODS
Materials
Unless otherwise stated, reagents were obtained from Sigma-Aldrich (St Louis, MO) and used without further purification. Glucose oxidase–avidin D conjugate (Gox-A) was purchased from Vector Laboratories (San Diego, CA). The electron-conducting redox polymer used in this study was a poly(vinylpyridine-co-acrylamide) copolymer partially pyridine complexed with an Os(4,4′-dimethyl-2,2′-bipyridine)2Cl+/2+ (PVP-PAA-Os). Synthesis of the redox polymer was described elsewhere (22). All primers used for RT–PCR were custom-made by 1st base Pte Ltd (Singapore). The primer sequences were as follows: GAPDH sense, 5′-ATGGTGAAGGTCGGTGTCAA-3′; GAPDH antisense, 5′-TTACTCCTTGGAGGCCATGT-3′; TP53 sense, 5′-ATGGAGGATTCACAGTCGGA-3′ and TP53 antisense, 5′-TCAGTCTGAGTCAGGCCC-3′. All other oligonucleotides were custom-made by Alpha-DNA (Montreal, Canada). Oligonucleotide capture probes used in this work for detecting the GAPDH and TP53 cDNA are listed in Table 1.
Table 1. Oligonucleotide capture probes used in this study.
| Detection of TP53 cDNA | |
| Capture probe | 5′-HS-(CH3)6-T12ATGGAGGATTCACAGTCGGA-3′ |
| 5′-HS-(CH3)6-T12TCAGTCTGAGTCAGGCCCCA-3′ | |
| Detection of GAPDH cDNA | |
| Capture probe | 5′-HS-(CH3)6-T12TTACTCCTTGGAGGCCATGTAGG-3′ |
| 5′-HS-(CH3)6-T12ATGGTGAAGGTCGGTGTCAACGG-3′ | |
| Control experiment | |
| Capture probe | 5′-HS-(CH3)6-T12CCTCTCGCGAGTCAACAGAAACG-3′ |
mRNA extraction and synthesis of multiple biotinylated cDNA
mRNA in the rat liver tissues was extracted with the Dynabeads® mRNA DIRECT™ Kit (Dynal ASA, Oslo, Norway) according to the manufacturer-recommended protocol. Reverse transcription was performed with 10 ng of mRNA in a total volume of 20 µl containing 1× buffer for enhanced avian myeloblastosis virus reverse transcriptase (eAMV-RT) (50 mM Tris–HCl, pH 8.3, 40 mM KCl, 8.0 mM MgCl2, 1.0 mM DTT, eAMV-RT from Sigma-Aldrich), 500 µM of each dNTP, 1.0 µM of 3′ antisense-specific primer, 20 U RNase inhibitor and 20 U eAMV-RT. First strand cDNA synthesis was carried out by incubating the mixture on a DNA thermal cycler (Gene Amp PCR System 9700, Applied Biosystems, Foster City) at 56°C for 50 min. The cDNA was subsequently used as template for the PCR amplification and biotinylation.
PCR was performed with 2.0 µl of the RT-reaction mixture in a total volume of 50 µl containing 1× AccuTaq buffer from Sigma-Aldrich (5.0 mM Tris–HCl, 15 mM ammonium sulfate, pH 9.3, 2.5 mM MgCl2, 0.1% Tween 20), 0.40 µM each primer, 2.5 U JumpStart AccuTaq LA DNA polymerase and 10 mM dNTP (Roche, Basel, Switzerland). For biotinylated cDNA synthesis, different amounts of biotin-modified nucleotides (Biotin-16-dUTP from Roche, Germany or biotin-21-dUTP from Clontech, Palo Alto, CA) were added into the reaction mixture to have different degree of biotinylation of the cDNAs.
Amplification was performed using the following profile: initial denaturation and eAMV-RT inactivation at 95°C for 5 min, then 35 cycles of amplification at 95°C for 30 s, 55.5°C for 1 min, and 72°C for 2 min. A final extension step of 10 min at 72°C was added to ensure full-length synthesis. After amplification, the PCR products were electrophoresed on 1.0% agarose gel and visualized fluorescently with ethidium bromide staining.
Capture probe immobilization
The preparation and pretreatment of gold substrates were as previously described (23). Briefly, prior to capture probe adsorption, the gold substrate were exposed to oxygen plasma for 5–10 min and then immersed in absolute ethanol for 20 min to reduce the oxide layer. DNA capture probe monolayer was adsorbed by immersing the gold electrode in PBS solution of 100 µg/ml DNA capture probe for 16–24 h. After adsorption, the electrodes were copiously rinsed with PBS and soaked in PBS for 20 min, rinsed again, and blown dry with a stream of air, a procedure aimed removing any non-specifically adsorbed materials. The surface density of capture probes, assessed electrochemically by the use of cationic redox probe, according to the procedure proposed by Steel et al. (24), was found to be in the range of 1.03–1.25 × 10–11 mol/cm2. To improve the quality and stability of the monolayer, capture probes coated gold electrode was immersed into an ethanolic solution of 5.0 mg/ml 11-mercaptoundecanoic acid (MUA), according to the proposed procedure (25). Unreacted MUA molecules were removed and the electrode was thoroughly washed by immersion in stirred ethanol for 10 min and followed by thorough rinsing with ethanol and water. The electrode was ready after air dry.
Target DNA hybridization and enzyme labeling
Biotinylated GAPDH and TP53 cDNA transcribed from the mRNAs extracted from the rat liver tissues were used as detection targets. A 10 mM Tris–HCl and 1.0 mM EDTA (TE) solution containing 0.10 M NaCl was used as hybridization buffer. Hybridization was carried out in a 55°C water bath for 30 min. Target cDNA were denatured at 95°C for 5 min and cooled in an ice bath before being added onto the electrode surface. After hybridization, the electrode was exposed to a 2.5 µl aliquot of 5.0 mg/ml GOx-A for 30 min and rinsed thoroughly with PBS three times, a procedure aimed to remove any non-DNA related GOx uptake. To ensure the maximal loading of the redox polymer, the electrode was then exposed for at least 10 min to a 2.5 µl aliquot of 5.0 mg/ml PVP-PAA-Os redox polymer solution and rinsed with PBS briefly afterwards.
Electrochemical measurement
Electrochemical measurements were carried out in a Faraday cage with a low-noise CH Instruments Model 660A electrochemical workstation in conjunction with a Pentium computer. Cyclic voltammetry was conducted in both PBS and a PBS solution containing 20 mM glucose. An Ag/AgCl electrode was used as the reference electrode and a platinum wire as the counter electrode. The working area of the gold electrode was 5.8 mm2. Amperometric measurements were carried out at 0.36 V. All potentials reported in this work were referred to the Ag/AgCl reference electrode.
RESULTS AND DISCUSSION
Detection scheme
The scheme for detecting DNA through direct hybridization and formation of the DNA/polymeric activator bilayer is shown in Figure 1. Prior to the test, a mixture of the thiolated oligonucleotides, served as capture probes, and thiol molecules were immobilized onto the gold electrode surface through self-assembly. To minimize non-hybridization related uptake of the target DNA, anionic thiol molecules were used to form the blocking component of the mixed monolayer. An additional merit of the anionic thiols is its dual function of attracting oppositely charged redox polymer, maximizing and stabilizing the redox polymer layer while repelling the anionic GOx-A on the electrode. The electrode was then exposed to the target analyte solution. Following hybridization to its complementary biotinylated target DNA, an enzyme label was introduced to the system via avidin–biotin interaction. A redox polymer, acting as a mediator for the enzymatic reaction, was then brought to the electrode surface through electrostatic layer-by-layer self-assembly. The redox polymer layer electrochemically activates the enzyme labels attached to the target DNA. In the presence of substrate molecules, the current generated from enzyme catalytic oxidation of the substrate was detected amperometrically. The catalytic current correlates directly to the target analyte concentration in the sample solution.
Figure 1.
Schematic illustration of DNA assay using the DNA/redox polymer bilayer model.
Synthesis of biotinylated cDNA from mRNA during RT–PCR
Full-length cDNAs of house-keeping gene GAPDH and regulated gene TP53 were synthesized from mRNA isolated from the rat liver tissues. Multiple biotin labels were incorporated into the cDNA chain using biotin-modified nucleotide, biotin-dUTP, during primer extension. Figure 2 shows gel electrophoresis results of the amplified GAPDH and TP53 cDNA. Lanes 1 and 4 in Figure 2 are control experiments without the addition of biotin-dUTP. The bands are in good agreement with the size of the full-length rat TP53 (lane 1, 1176 bp) and GAPDH gene (lane 4, 1002 bp). As can be seen in Figure 2, gel electrophoresis results showed successful labeling and amplification of both genes. Different amounts of biotin-modified nucleotide were mixed with dNTP during the PCR amplification to examine the labeling efficiency. Lanes 2 and 3 correspond to different ratios of biotin-16-dUTP/dTTP. The higher the ratio of biotin-16-dUTP/dTTP, the higher the band appeared in the gel image. The lower mobility shift of the PCR product obtained with the higher ratio of biotin-16-dUTP/dTTP (lane 3) suggests a higher ratio of incorporated biotin to the cDNA. At fixed biotin-16-dUTP/dTTP ratios, duplicated PCR amplifications showed fairly good reproducibility. However, the amplification efficiency was reduced with increasing the ratio of biotin-modified nucleotide to normal nucleotide. These results suggested that the bulky side chains of biotin-modified nucleotides do not prevent polymerase from using them as substrates for the cDNA synthesis, but do have certain influence on the efficiency and thereby the yield of the PCR reaction. The same phenomena were also observed for the GAPDH amplification (lane 4–6). However, due to a longer aliphatic chain (21 carbons) attached to the dUTP, the amplification and labeling efficiency was much less than those with the 16-carbon side chain used for the amplification of TP53 cDNA (lane 1–3).
Figure 2.
PCR amplification of the full-length rat TP53 cDNA (lane 1–3) and full-length GAPDH cDNA (lanes 4–6) with increased ratio of biotin-dUTP/dTTP. Lane M, DNA size marker; lanes 1–3, corresponding to biotin-16-dUTP/dTTP ratio of 0/100, 35/65 and 65/35 respectively. Lanes 4–6, corresponding to biotin-21-dUTP/dTTP ratio of 0/10, 1/10 and 2/10 respectively.
Monolayer quality evaluation and electroactivating effect of the redox polymer
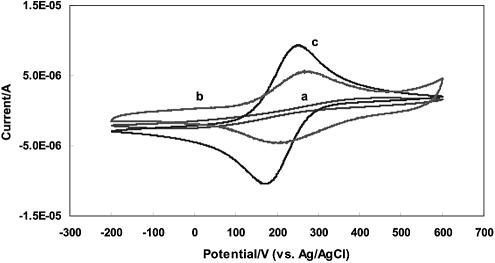
The formation of the mixed self-assembled monolayer on a gold electrode was routinely monitored by optical ellipsometric, contact angle and surface coverage measurements. All the data obtained indicate a single compact mixed molecular layer coated on the gold electrode. As expected, the obvious pathway of electron transfer between the monolayer coated electrode and the electroactive species in solution would be via electron tunneling across the insulating monolayer. The electron tunneling barrier characteristics of the capture probe monolayer and the mixed monolayer were investigated by cyclic voltammetry in a 0.50 M Na2SO4 solution containing 2.5 mM ferricyanide (Fig. 3). As shown in Figure 3a, irreversible voltammetric waves for Fe(CN)63–/4– with a very large peak-to-peak potential separation, >400 mV at 100 mV s–1, compared with the value of 59 mV for a reversible process obtained at a bare gold electrode (not shown), were observed at the mixed monolayer coated gold electrode, indicating that the monolayer impedes electron transfer between the gold electrode and the solution species (25). The redox currents were significantly reduced and completely lost their reversible character. The currents are mainly caused by an electron tunneling process across the monolayer. However, since the redox polymer is positively charged and the electrode is negatively charged, a brief soak of the electrode in the 5.0 mg/ml PVP-PAA-Os solution, resulted in the formation of a DNA/redox polymer bilayer on the electrode via the layer-by-layer electrostatic self-assembly (26). As illustrated in Figure 3b, the bilayer coated electrodes exhibited exactly as expected for a highly reversible surface immobilized redox couple with little change after exhaustive washing with water and PBS and after numerous repetitive potential cycling between –0.4 and +0.8 V, revealing a highly stable surface immobilized electrostatic bilayer on gold electrode (27). Such results ascertain that all of the osmium redox centers are allowed to reach the electrode surface and proceed to reversible heterogeneous electron transfer. The total amount of bound osmium redox centers, 1.8–8.0 × 10–10 mol/cm2, depending on the amount of nucleic acid bound to the electrode, was estimated from the area under either the oxidation or the reduction current peak corrected from the background current. Subsequent voltammetric tests in the ferricyanide solution showed a voltammogram identical to that obtained at a bare gold electrode (Fig. 3c). These changes are attributed to the decrease in the electron tunneling pathway due to the formation of the bilayer which brought the osmium redox centers to the closest possible proximity of the electrode surface, and more importantly, the much faster electron transfer rate of the osmium redox centers in the bilayer which minimizes the effect of electron tunneling across the underlying insulating monolayer and mediates electron transfer from solution species to the electrode surface. As shown later, the presence of nucleic acids and the GOx labels in the film did not appreciably alter the electrochemistry of the redox polymer. Later experiments in glucose solution also showed that the GOx labels in the bilayer retain their activities.
Figure 3.
Cyclic voltammograms of a gold electrode (a) coated with a mixed self-assembled monolayer in 2.5 mM K3Fe(CN)6 and 0.50 M Na2SO4, (b) with DNA/redox polymer bilayer in PBS, (c) with DNA/redox polymer bilayer in 2.5 mM K3Fe(CN)6 and 0.50 M Na2SO4. Scan rate, 100 mV/s. For clarity, the current scale in (b) was multiplied by 10.
Hybridization and feasibility study of target cDNA detection
In a preliminary hybridization test, the PCR amplification mixture was used as analyte without further purification. Our first detection target is the biotinylated GAPDH cDNA in the mixture. Prior to hybridization, the PCR mixture was denatured at 95°C for 5 min to separate DNA double helix. Oligonucleotides with sequence complementary to 3′ rat GAPDH gene were immobilized on the electrode surface and served as the capture probes. Upon hybridization at 55°C for 30 min, target GAPDH cDNA in the mixture was selectively bound by its complementary capture probe and immobilized on the electrode surface. Repeated rinsing with the hybridization buffer completely washed off all the non-hybridization related nucleic acids. The enzyme labels were brought to the electrode surface via biotin–avidin interaction during subsequent incubation with the GOx-A. Typical cyclic voltammograms of the electrode hybridized with the target analyte are shown in Figure 4. Figure 4A are the voltammograms of the electrode with capture probes complementary to GAPDH cDNA in the PBS buffer (curve a) and in a 20 mM glucose solution (curve b) after hybridization. Obvious catalytic current was observed in the presence of glucose due to the presence of glucose oxidase in the bilayer. While in a control experiment, non-complementary capture probes failed to capture any GAPDH cDNA from the RT–PCR mixture solution and thereby no enzyme labels were able to bind to the electrode surface. Similar voltammograms (curves a and b in Fig. 4B) were then obtained in PBS and PBS-containing glucose. No detectable catalytic current in voltammetry was noticed.
Figure 4.
Cyclic voltammograms of electrode after hybridization with the GAPDH cDNA in the PBS (curve a) and 20 mM glucose solution (curve b) with (A) capture probe complementary to the GAPDH cDNA, (B) capture probe non-complementary to the GAPDH cDNA. Scan rate, 10 mV/s.
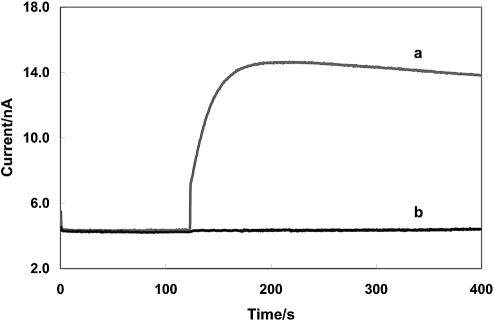
When the completed electrode was immersed in PBS, the oxidation current in amperometry increased 10.2 nA at 0.36 V (versus Ag/AgCl) upon adding 40 mM glucose to the PBS (Fig. 5a). In an identical experiment (control experiment) where non-complementary capture probes were immobilized on the electrode surface, negligible change of current was observed (Fig. 5b). The amperometric results complemented the cyclic voltammetric data obtained earlier and confirmed again that the GAPDH cDNA was successfully detected from the PCR mixture with high specificity. As expected, the amperometric signal is strongly dependent on the redox polymer loading, the oxidation current increased with increasing the amount of redox polymer up to 4.0 × 10–10 mol/cm2 and then started to level off. It was found that maximal loading of 8.0–12.0 × 10–10 mol/cm2 could easily be achieved after 5–10 min of adsorption in the 5.0 mg/ml redox polymer solution. To safeguard the amperometric sensitivity, maximal loading was always used for DNA detection. Under optimized experimental conditions, a dynamic range was found to be from 2.0 fM to 1.0 pM with a detection limit of 0.50 fM estimated from 3-fold of noise. It was found that a practically constant current (saturation current) was observed at DNA concentration of 2.9–3.3 pM.
Figure 5.
Amperometric responses of electrodes after hybridization with the GAPDH cDNA in the PCR mixture (a) with capture probe complementary to the GAPDH cDNA, (b) with capture probe non-complementary to the GAPDH cDNA. Working potential: 0.36 V, 40 mM glucose.
Analysis of rat TP53 gene
Since TP53 is one of the most important cancer markers, techniques that could offer sensitive detection and accurate quantification of this gene will help to facilitate earlier diagnosis and prognosis. Many studies on cancer have focused attention on the TP53 as a possible diagnostic and prognostic tumor marker. In this study, the aforementioned nucleic acid biosensor was applied for the detection of the full-length rat TP53 cDNA. The target TP53 cDNA was synthesized from TP53 mRNA as described in an earlier section. Multiple biotin labels were incorporated into the cDNA chain during primer extension. The amplification was confirmed by the gel electrophoretic data (Fig. 2). The total amount of TP53 cDNA after the PCR amplification was 17.2 ng/µl (22.5 pM). The PCR product was diluted to different concentrations with the TE buffer before use. TP53 cDNA solutions at 10, 50, 100, 200, 500 and 800 fM were tested. The TP53 cDNA in the PCR mixture was brought to the electrode surface by its complementary capture probes. After enzyme labels and the redox polymer were introduced, catalytic current was detected at 0.36 V, which directly corresponds to the amount of the TP53 cDNA. Similar to the GAPDH cDNA tests, for control experiment, non-complementary capture probes were immobilized on the electrode. As depicted in Figure 6, the current increased linearly with the concentration of the TP53 cDNA within this range. However, the current response no longer followed the linear relationship when the analyte concentration was further reduced. The lowest detectable concentration of this gene was found to be ∼1.0 fM. Taking the sample volume into consideration, as few as 1500 copies of DNA molecules were successfully detected using the proposed approach. To the best of our knowledge, this is the lowest reported amount of genomic DNA detected electrochemically.
Figure 6.
Amperometric responses of different TP53 concentration in 2.5 µl droplets. Working potential: 0.36 V, 40 mM glucose.
Compared with previous results based on the sandwich-type assays, the sensitivity of genomic cDNA analysis was greatly improved by adopting the multiple enzyme labeling scheme and the result was comparable to that obtained with short synthetic oligonucleotides. In the sandwich-type assay reported earlier, the ratio of enzyme label and target DNA molecule was fixed at unit. The amount of capture probe immobilized on the electrode surface and hybridization efficiency determined the amount of target DNA bound to the surface and thereby the amount of enzyme labels in spite of the size of the genes. However, in our proposed model, multiple biotin labels on a single cDNA chain greatly increased the enzyme label loading, corresponding responses from enzymatic reaction were increased, and hence the sensitivity and detection limit of the nucleic acid biosensor were substantially improved when working with long nucleic acid molecules. For example, for a 1000 bp DNA, if there is one enzyme label for every 50 base pairs, the overall signal could, theoretically, increase by 20-fold. By labeling the nucleic acid sample with multiple enzyme molecules, the enzyme label/base ratio remains more or less constant for both synthetic oligonucleotides and genomic nucleic acid samples, which, in turn, generated analytical signals of similar sensitivities. Indeed, the sensitivity obtained in this work is comparable to that observed with the short synthetic oligonucleotide (50mer) of the sandwich-type approach (21), the maximal enzyme/base ratio was estimated to be 1/25–1/30, and more importantly the detection limit is independent of DNA length in the range of 100–2000 bp.
CONCLUSIONS
Full-length cDNAs of a house-keeping gene GAPDH and a regulated gene TP53 from rat liver tissues were selectively detected at femtomolar levels using the analyte/polymeric activator bilayer setup. By labeling the nucleic acid sample with multiple enzyme molecules, the sensitivity was greatly improved and increased by 15–20-fold compared with the sandwich-type assay. The lowest detectable amount of TP53 was found to be around 1500 copies. This approach may allow direct detection of genomic genes in real samples from total RNA extracts without any PCR amplification. We are currently exploiting the feasibility of direct detection of genomic nucleic acids immediately after cells are lysed.
REFERENCES
- 1.Watson J., Gilman,M., Witkowski,J. and Zoller,M. (1992) Recombinant DNA, 2nd Edn. W.H. Freeman and Company, New York, NY. [Google Scholar]
- 2.Chan S.D.H., Dill,K., Blomdahl,J. and Wada,H.G. (1996) Nonisotopic quantification of mRNA using a novel RNase protection assay: measurement of erbB-2 MRNA in tumor cell lines. Anal. Biochem., 242, 214–220. [DOI] [PubMed] [Google Scholar]
- 3.Revillion F., Hornez,L. and Peyrat,J.P. (1997) Quantification of c-erbB-2 expression in breast cancer by competitive RT-PCR. Clin. Chem., 43, 2114–2120. [PubMed] [Google Scholar]
- 4.Cottrez F., Auriault,C., Capron,A. and Groux,H. (1994) Quantitative PCR: validation of the use of a multispecific internal control. Nucleic Acids Res., 22, 2712–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Totze G., Sachinidis,A., Vettre,H. and Ko,Y. (1996) Competitive reverse transcription/polymerase chain reaction for the quantification of p53 and mdm2 mRNA expression. Mol. Cell. Probes, 10, 427–433. [DOI] [PubMed] [Google Scholar]
- 6.Boon E.M., Ceres,D.M., Drummond,T.G., Hill,M.G. and Barton,J.K. (2000) Mutation detection by electrocatalysis at DNA-modified electrode. Nat. Biotechnol., 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen S.R. (1996) Electrochemical biosensor for DNA sequence detection. Electroanal., 8, 15–19. [Google Scholar]
- 8.Rodriguez M. and Bard,A.J. (1990) Electrochemical studies of the interaction of metal chelates with DNA. 4. Voltammetric and electrogenerated chemiluminescent studies of the interaction of tris(2,2’-bipyridine)osmium(II) with DNA. Anal. Chem., 62, 2658–2662. [DOI] [PubMed] [Google Scholar]
- 9.Jordan C.E., Frutos,A.G., Thiel,A.J. and Corn,R.M. (1997) Surface plasmon resonance imaging measurements of DNA hybridization adsorption and streptavidin/DNA multilayer formation at chemically modified gold surfaces. Anal. Chem., 69, 4939–4947. [DOI] [PubMed] [Google Scholar]
- 10.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley S.O., Barton,J.K., Jackson,N.M. and Hill,M.G. (1997) Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjugate Chem., 8, 31–37. [DOI] [PubMed] [Google Scholar]
- 12.Kelly S.O., Boon,E.M., Barton,J.K., Jackson,N.M. and Hill,M.G. (1999) Single-based mismatch detection based on charge transduction through DNA. Nucleic Acids Res., 27, 4830–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardea A., Dagan,A., Ben-Dov,I., Amit,B. and Willner,I. (1998) Amplified microgravimetric quartz crystal microbalance analysis of oligonucleotide complexes: a route to Tay-Sachs biosensor device. Chem. Commun., 839–840. [Google Scholar]
- 14.Wang J. (2000) Survey and summary from DNA biosensors to gene chip. Nucleic Acids Res., 28, 3011–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruana D.J. and Heller,A. (1999) Enzyme-amplified amperometric detection of hybridization and of a single base pair mutation in an 18-base oligonucleotide on a 7 μm diameter microelectrode. J. Am. Chem. Soc., 121, 769–774. [Google Scholar]
- 16.Patolaky F., Lichtenstein,A. and Willner,I. (2001) Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol., 19, 253–257. [DOI] [PubMed] [Google Scholar]
- 17.Patolsky F., Lichtenstein,A., Kotler,M. and Willner,I. (2001) Electronic transduction of polymerase or reverse transcriptase induced replication processes on surfaces: highly sensitive and specific detection of viral genomes. Angew. Chem. Int. Edit., 40, 2261–2265. [DOI] [PubMed] [Google Scholar]
- 18.Patolsky F., Lichtenstein,A. and Willner,I. (2000) Amplified microgravimetric quartz-crystal-microbalance assay of DNA using oligonucleotide-functionalized liposomes or biotinylated liposomes. J. Am. Chem. Soc., 122, 418–419. [Google Scholar]
- 19.Patolsky F., Ranjit,K.T., Lichtenstein,A. and Willner,I. (2000) Dendritic amplification of DNA analysis by oligonucleotide-functionalized Au-nanoparticles. Chem. Commun., 12, 1025–1026. [Google Scholar]
- 20.Zhang Y., Kim,H.H. and Heller,A. (2003) Enzyme-amplified amperometric detection of 3000 copies of DNA in a 10-µl droplet at 0.5 fM concentration. Anal. Chem., EST 2.4. [DOI] [PubMed] [Google Scholar]
- 21.Xie H., Zhang,C. and Gao,Z. (2004), Amperometric detection of nucleic acid at femtomolar levels with a nucleic acid/electrochemical activator bilayer on gold electrode, Anal. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 22.Gao Z., Binyamin,G., Kim,H.H., Barton,S.C., Zhang,Y. and Heller,A. (2002) Electrodeposition of redox polymers and codeposition of enzymes by coordinative crosslinking. Angew. Chem. Int. Edit., 41, 810–813. [DOI] [PubMed] [Google Scholar]
- 23.Gao Z., Siow,K.S. and Chan,H. (1995) Self-assembled conducting polymer monolayers of poly(3-octylthiophene) on gold electrode. Synth. Met., 75, 5–10. [Google Scholar]
- 24.Steel A.B., Herne,T.T. and Tarlov,M.J. (1998) Electrochemical quantitation of DNA immobilized on gold. Anal. Chem., 70, 4670–4677. [DOI] [PubMed] [Google Scholar]
- 25.Yang D., Zi,M., Chen,B. and Gao,Z. (1999) Separation of pin-hole and electron-tunneling processes at self-assembled monolayer on gold electrode. J. Electroanal. Chem., 470, 114–119. [Google Scholar]
- 26.Decher G. (1997) Fuzzy nanoassemblies: toward layered polymeric multicomposites. Science, 277, 1231–1237. [Google Scholar]
- 27.Bard A.J. and Faulkner,L.R. (2001) Electrochemical Methods. John Wiley & Sons, New York, NY, p. 590. [Google Scholar]