Abstract
Objective
Many studies have reported the prognostic predictive value of CD166 as a cancer stem cell marker in cancers of the digestive system; however, its predictive value remains controversial. Here, we investigate the correlation between CD166 positivity in digestive system cancers and clinicopathological features using meta-analysis.
Methods
A comprehensive search in PubMed and ISI Web of Science through March of 2013 was performed. Only articles containing CD166 antigen immunohistochemical staining in cancers of the digestive system were included,including pancreatic cancer, esophageal cancer, gastric cancer and colorectal cancer. Data comparing 3- and 5-year overall survival along with other clinicopathological features were collected.
Results
Nine studies with 2553 patients who met the inclusion criteria were included for the analysis. The median rate of CD166 immunohistochemical staining expression was 56% (25.4%–76.3%). In colorectal cancer specifically, the results of a fixed-effects model indicated that CD166-positive expression was an independent marker associated with a smaller tumor burden (T category; RR = 0.93, 95%, CI: 0.88–0.98) but worse spread to nearby lymph nodes (N category; RR = 1.17, 95% CI: 1.05–1.30). The 5-year overall survival rate was showed relationship with cytoplasmic positive staining of CD166 (RR = 1.47 95% 1.21–1.79), but no significant association was found in the pool or any other stratified analysis with 3- or 5- year overall survival rate.
Conclusion
Based on the published studies, different cellular location of CD166 has distinct prognostic value and cytoplasmic positive expression is associated with worse prognosis outcome. Besides, our results also find CD166 expression indicate advanced T category and N-positive status in colorectal cancer specifically.
Introduction
Although treatments for digestive system cancers have been developing rapidly, these cancers, especially pancreatic and colorectal cancer, are still responsible for a number of deaths [1]. It has been reported that a small subpopulation of cells, called cancer stem cells (CSCs), dominate the initiation, progression, relapse and metastasis of cancer. In recent years, certain cell surface markers have been reported as CSC markers in digestive system cancers, with high expression of these markers usually an indicator of poor prognosis. Among them, CD166 has been identified experimentally as a putative stem cell marker in various cancers [2]–[5] with a high capacity for sphere and xenograft formation.
CD166, also known as activated leukocyte cell adhesion molecule (ALCAM), is a highly conserved 110-kDa multidomain transmembrane type-1 glycoprotein of the immunoglobulin super family, which was first described as a CD6 ligand on leukocytes [6]. Further studies have revealed that it is broadly expressed in different tissues and cells, including neuronal, immune and epithelial cells, as well as stem cells of hematopoietic and mesenchymal origin. CD166 plays an important role in many biological activities, including T-cell activation and proliferation, angiogenesis, hematopoiesis and axon fasciculation [7]. CD166 is also closely related to various cancers, including melanoma, prostate cancer and breast cancer. Many cancers of the digestive system have also been found to have high expression of CD166; however, the prognostic outcomes of these studies are contradictory. Two earlier studies by Weichert and Horst et al [8], [9] reported positive expression of CD166 in colorectal cancer and that CD166 was an independent prognostic marker associated with poor survival rates. However, two other recent independent studies have found conflicting outcomes [10], [11].
Because the contradiction may be caused by limited sample sizes along with other factors, here, we performed a pooled analysis of all cancers of the digestive system, including pancreatic, gastric, esophageal and colorectal cancer. Although all cancer types were derived from the digestive system, heterogeneity within the different tissues may also exist. Therefore, we analyzed pancreatic and colorectal cancer independently to improve accuracy. Here, we present a meta-analysis that aims to clarify the prognostic value of CD166 in digestive system cancers based on currently published evidence. Other clinicopathological features were also examined in this study.
Materials and Methods
Search Strategy
We performed a systematic literature search of the electronic databases PubMed and ISI Web of Science up to March of 2013. Search terms included “CD166 antigen”, “ALCAM” or “activated leukocyte cell adhesion molecule” with “cancer”, “neoplasm” or “carcinoma”. The titles and abstracts of potential references were carefully examined to exclude irrelevant studies; the remaining articles within the topic of interest were reviewed in depth for their relevance.
Selection Criteria
The studies in this meta-analysis included either randomized control studies (RCTs) or observational studies (case-control or cohort) that evaluated the relationship between CD166 expression and the risk of developing a digestive system cancer. Studies were included if they met the following criteria: (a) they focused on digestive system cancers (esophageal, gastric, pancreatic and colorectal cancer); (b) they defined a CD166-positive group by immunohistochemistry; and (c) they described a correlation between CD166, clinicopathological features and survival outcome (either disease free survival or overall survival). Articles were excluded from the analyses based on the following criteria: (a) non-English papers; (b) review articles or letters; and, (c) insufficient data to determine the RR and CI, or the full text could not be found.
Based on a critical review checklist provided by the Dutch Cochrane Centre [12] and in an effort to control the quality of this meta-analysis, we examined the quality of all the included studies. Seven key points are depicted here: (a) clear definition of study population and origin of country, (b) clear definition of the type of carcinoma, (c) clear definition of the study design, (d) clear definition of the outcome assessment, (e) clear definition of the cut-off of CD166 expression, (f) clear definition of the method of CD166 assessment and (g) sufficient time of follow-up.
Data Extraction
All data were extracted by two independent reviewers. Data tables were generated to extract all relevant data from texts, tables and figures, including: author, year, country, patient number, detection method, duration of follow-up, T category, N category, distant metastasis, positive rates of CD166 overexpression, as well as overall survival (OS) rate. For articles that only provided survival data in a Kaplan-Meier curve, the software GetData Graph Digitizer 2.24 (http://getdata-graph-digitizer.com/) was applied to digitize and extract the data.
Because the cut-off score for CD166 positivity varied among the studies, we defined the CD166 positive group with respect to the original articles. Because of the high degree of malignancy and poor outcome of pancreatic cancer patients in clinic, the OS was standardized to 3-years in pancreatic cancer, and the other cancer types were standardized to 5-years.
Statistical Analysis
The statistical analysis was performed according to the guidelines proposed by the Meta-Analysis of Observational Studies in Epidemiology group. Relative risk (RR) with 95% confidence interval (95% CI) was calculated with Review Manager 4.2. The heterogeneity among studies was measured using the Q and I2 tests. A Fixed or Random model was used depending on the heterogeneity analysis. The potential for publication bias was assessed using the Begg rank correlation method and the Egger weighted regression method (software stata11.0). P value <0.05 was considered statistically significant. All P values are two-tailed.
Results
Search Results
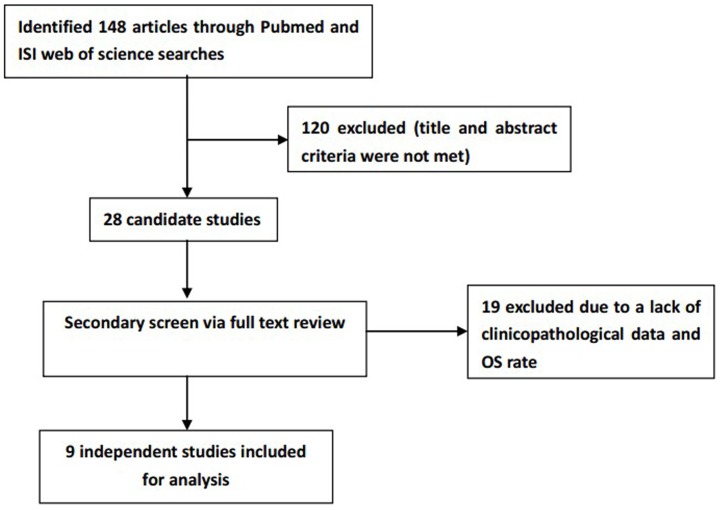
Initially, 148 articles were retrieved utilizing the search strategy above. From the title and abstract review, 120 of the articles were excluded due to non-human experiments, non-digestive system cancer-related studies, or non-original articles (e.g., review, letter). Of the remaining articles, 19 were excluded because they did not provide clinicopathological data, particularly the OS rate [2]–[4], [13]–[28]. Finally, a total of 9 studies were included in the meta-analysis with 2553 participants. All of these studies explicitly assessed the expression of CD166 and risk of cancer death by immunohistochemical staining (Figure 1).
Figure 1. Flow chart for the selection of articles to include.
Study Characteristics
All features of the 9 eligible studies are listed in Table 1. Among them, 7 were from Germany, 1 from Japan and 1 from Switzerland. Furthermore, 2 studies focused on pancreatic cancer, 4 focused on colorectal cancer, and 1 focused on each of pancreatic neuroendocrine, esophageal and gastric cancer. A total of 2553 patients with a median of 188 (from 38 to 1274) per study were included. The TNM stage and tumor grade was reported in 8 and 7 studies, respectively. Furthermore, 6 studies determined CD166 expression only on the membrane, and 3 studies stained CD166 on both the membrane and within the cytoplasm. Tissue microarrays for CD166 expression analysis were utilized in 5 studies. Three studies used whole tissue immunochemistry staining and 1 study applied both methods (Table 1). In 6 studies, none of the patients received neo-adjuvant radio- or chemotherapy prior to surgery [8]–[10], [29]–[31]. Pertaining directly to colorectal cancer, patients with tumor recurrence in Horst’s study were treated with chemotherapy, radiation therapy or surgical resection when possible. In Lugli’s study, 478 patients received post-operative therapy. In Kahlert’s study, 88 cases had an R0 resection, and 9 cases had an R1 resection. For gastric cancer, all patients had an R0 resection with at least a D1 lymph node dissection.
Table 1. Characteristics of included studies.
| NO. | Author | Year | Country | Number of Patients | Organ | Duration of Follow-up | Methods | Staining patterns | Cut off scores (pos/neg) | T category (T1–2/3–4) | N category (P/N) | Distant metastasis(M0/M1) | Grade (1–2/3) | OS (5 years) |
| 1 | Tachezy | 2012 | germany | 192 | pancretic CA | 0–193 months (median 14 months) | TMA | membrane staining | membrane staining positive(72/120) | H(105/15); L(65/7) | NA | NA | H(68/52); L(38/34) | H(29/91); L(6/66)* |
| 2 | Tachezy | 2011 | germany | 38 | pancretic neuroendocrine CA | 7–168 months (median 45 months) | TMA | membrane staining | 2+ >30% and 3+ (28/10) | NA | NA | H(4/23);L(5/5) | H(26/2); L(9/1) | H (23/51);L (3/7)* |
| 3 | Kahlert | 2009 | germany | 97 | pancretic CA | median 18.5 months | whole tissue | membrane and cytoplasmic staining | membrane and cytoplasmic staining (30/67) | H(6/24); L(12/55) | H(6/24); L(12/55) | NA | H(15/15); L(46/21) | H (0/30) L(19/48)* |
| 4 | Tachezy | 2012 | germany | 289 | esophageal CA | 0–178 months (median 16 months) | TMA | membrane staining | 1+ >30% or≥2+ (204/85) | H(87/117); L(35/50) | H(75/129); L(26/59) | H(166/38); L(72/13) | H(128/76); L(39/46) | H (43/163) L(30/55)* |
| 5 | Ishigami | 2011 | japan | 142 | gastric cancer | 0.3–104.5 months (median 18.6 months) | whole tissue | membrane and cytoplasmic staining | membrane and cytoplasmic staining (36/106) | H(28/8); L(79/27) | H(23/13); L(41/65) | NA | NA | H (20/16) L (71/35)* |
| 6 | weichert | 2004 | germany | 111 | colorectal CA | median 47 months | whole tissue | membrane and cytoplasmic staining | membrane and cytoplasmic staining (34/77) | NA | NA | NA | NA | H (20/14) L (61/16)* |
| 7 | Horst | 2009 | germany | 110 | colorectal CA | 4.8–162 months (median 94.8 months) | TMA | membrane staining | membrane staining positive (70/40) | H(22/48); L(17/28) | H(12/54); L(7/33) | NA | H(63/7); L(36/4) | H (42/28) L (33/7)* |
| 8 | Lugli | 2010 | switzerland | 1274 | colorectal CA | 0–80 months | TMA and whole tissue | membrane staining | membrane staining positive (775/499) | H(165/594); L(72/417) | H(417/329); L(228/253) | H(275/63); L(70/18) | H(658/100); L(438/49) | H (465/310) L (274/225) |
| 9 | Tachezy | 2012 | germany | 300 | colorectal CA | 1–180 months (median 39 months) | TMA | membrane staining | membrane staining positive (229/71) | H(59/170); L(14/57) | H(115/114); L(31/40) | H(168/61); L(53/18) | H(200/29); L(50/21) | H (135/94); L(26/45)* |
CA: cancer; H: high expression; L: low expression; NA:not available; TMA: tissue microarray.
data read by GetData Graph Digitizer.
Correlation of CD166 to Clinical Features
The correlation of CD166 membrane expression with overall T category, N category, distant metastasis and tumor grade is illustrated in Figures S1, S2, S3 and S4. The results suggest that CD166 correlated more with T1 and T2 category patients (pooled RR = 0.94, 95% CI: 0.89–0.99) and N-positive patients (RR = 1.20, 95% CI: 1.09–1.32). However, in colorectal cancer specifically, CD166 expression was associated with more advanced T category (RR = 0.93, 95% CI: 0.88–0.98) and N-positive status (RR = 1.17, 95%CI 1.05–1.30), and it did not show any relationship with other kinds of digestive tumors. Furthermore, we stratified the extracted data by geographic area, staining pattern, follow-up time and sample size: studies in Europe showed CD166 expression was related with T1 and T2 category patients (RR = 0.94, 95% CI: 0.89–0.99; membrane staining: RR = 0.94, 95% CI: 0.89–0.99) and N-positive status (RR = 1.17, 95% CI: 1.06–1.30; membrane staining: RR = 0.94, 95% CI: 0.89–0.99). Membranous staining of CD166 also revealed related with T1 and T2 category patients (RR = 0.94, 95% CI: 0.89–0.99), and both staining patterns were showed associated with N-positive status (membrane staining: RR = 0.94, 95% CI: 0.89–0.99; membrane & cytoplasmic staining RR = 1.51, 95% CI: 1.09–2.10). Studies with shorter (RR = 1.34, 95% CI: 1.04–1.71) or longer follow-up times (RR = 1.17, 95% CI: 1.05–1.30) both showed a positive relationship between CD166 expression and N status. The same result was also found in both studies with smaller (RR = 1.40, 95% CI: 1.02–1.92) or larger sample sizes (RR = 1.18, 95% CI: 1.06–1.30). However, there was no clear association between CD166 expression and other clinicopathological features including distant metastasis (RR = 1.10, 95% CI: 0.85–1.43) or tumor grade (RR = 0.90, 95% CI: 0.63–1.27) in either the overall or stratified analyses (Table 2).
Table 2. Results of meta-analysis on CD166 expression.
| T category(T3,4 vs. T1,2) | N category(positive vs. negative) | Distant metastasis(M1 vs. M0) | Grade(grade 3 vs. grade 1,2) | OS 3year(death vs. survive) | OS 5year(death vs. survive) | |||||||
| N1 | RR(95%CI) | N2 | RR(95%CI) | N3 | RR(95%CI) | N4 | RR(95%CI) | N5 | RR(95%CI) | N6 | RR(95%CI) | |
| Over all | 7 | 0.94(0.89–0.99) | 6 | 1.20(1.09–1.32) | 4 | 1.10(0.85–1.43) | 7 | 0.90(0.63–1.27) | 2 | 0.90(0.80–1.02) | 9 | 1.11(0.88–1.04) |
| Cancer typea | ||||||||||||
| PC | 2 | 1.04(0.81–1.34) | 1 | 1.12(0.46–2.69) | 1 | 1.70(0.90–3.23) | 3 | 1.07(0.82–1.39) | 2 | 0.90(0.80–1.02) | 3 | 1.04(0.70–1.55) |
| EC | 1 | 0.98(0.79–1.21) | 1 | 1.20(0.83–1.74) | 1 | 1.22(0.68–2.17) | 1 | 0.69(0.53–0.90) | 0 | NA | 1 | 1.22(1.03–1.45) |
| GC | 1 | 0.87(0.44–1.74) | 1 | 1.65(1.17–2.33) | 0 | NA | 0 | NA | 0 | NA | 1 | 1.35(0.85–2.12) |
| CRC | 3 | 0.93(0.88–0.98) | 3 | 1.17(1.05–1.30) | 2 | 0.98(0.71–1.36) | 3 | 0.81(0.35–1.92) | 0 | NA | 4 | 1.18(0.64–2.16) |
| Geographic area | ||||||||||||
| Europe | 6 | 0.94(0.89–0.99) | 5 | 1.17(1.06–1.30) | 4 | 1.10(0.85–1.43) | 7 | 0.90(0.63–1.27) | 2 | 0.90(0.80–1.02) | 8 | 1.09(0.85–1.39) |
| Asia | 1 | 0.87(0.44–1.74) | 1 | 1.65(1.17–2.33) | 0 | NA | 0 | NA | 0 | NA | 1 | 1.35(0.85–2.12) |
| Staining pattern | ||||||||||||
| Membrane | 5 | 0.94(0.89–0.99) | 4 | 1.17(1.06–1.30) | 4 | 1.10(0.85–1.43) | 6 | 0.81(0.56–1.15) | 1 | 0.95(0.80–1.02) | 6 | 0.96(0.74–1.24) |
| Membrane &cytoplasmic | 2 | 0.95(0.74–1.21) | 2 | 1.51(1.09–2.10) | 0 | NA | 1 | 1.60(0.96–2.64) | 1 | 0.83(0.70–0.98) | 3 | 1.47(1.21–1.79) |
| Follow time (month)b | ||||||||||||
| <37.5 | 4 | 0.99(0.84–1.16) | 3 | 1.34(1.04–1.71) | 1 | 1.22(0.68–2.17) | 3 | 0.96(0.63–1.45) | 2 | 0.90(0.80–1.02) | 4 | 1.15(0.86–1.54) |
| ≥37.5 | 3 | 0.93(0.88–0.99) | 3 | 1.17(1.05–1.30) | 3 | 1.06(0.79–1.42) | 4 | 0.81(0.37–1.75) | 0 | NA | 5 | 1.11(0.71–1.76) |
| Sample sizec | ||||||||||||
| <188 | 3 | 1.01(0.84–1.21) | 3 | 1.40(1.02–1.92) | 1 | 1.70(0.90–3.23) | 3 | 1.37(0.87–2.18) | 1 | 0.83(0.70–0.98) | 5 | 1.49(1.24–1.79) |
| ≥188 | 4 | 0.93(0.88–0.98) | 1.18(1.06–1.30) | 3 | 1.04(0.78–1.38) | 4 | 0.79(0.53–1.19) | 1 | 0.95(0.80–1.12) | 4 | 0.87(0.66–1.15) |
PC:pancretic cancer, EC: esophageal cancer, GC: gastric cancer, CRC: colorectal cancer.
median of followup time among all studies included.
median of sample size among all studies included.
CD166 Expression and 3- or 5-year Overall Survival Rate
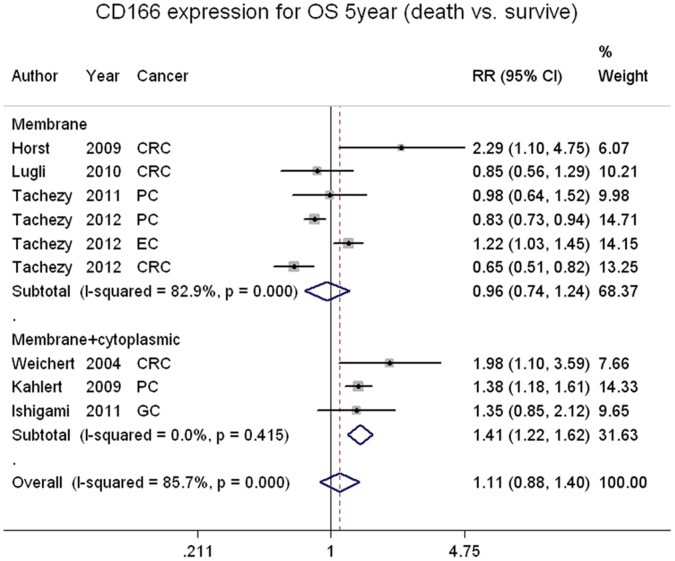
The 5-year overall survival rate was extracted from 9 studies, which was composed of 4 colorectal cancer studies, 1 gastric, 1 esophageal and 3 pancreatic cancer study and two studies of pancreatic cancer were also examined with 3-year overall survival. The pooled 5-year overall survival rates of CD166-positive and CD166-negative patients were 57.3% (767/1339) and 33.7% (523/1214), respectively. The 3-year overall survival rates were 25.3% (38/150) and 12.2% (17/139) for the same patients, respectively. The pool analysis did not show significant association between overall survival rate and CD166 status (Figures S5 and S6), however, the stratified group based on staining pattern revealed membrane and cytoplasmic staining was related with worse prognosis (RR = 1.47 95%CI 1.21–1.79), but the membrane staining alone could not show any prediction value (Table 2, Figure 2).
Figure 2. CD166 stratified on staining pattern and 5-year overall survival rate in digestive cancer patients.
Publication Bias
Heterogeneity testing and publication bias analyses were performed among the studies based on membrane and cytoplasmic staining. Results indicate that the funnel plots are almost symmetric and that the P values of Begg’s and Egger’s tests were 0.296 and 0.533, respectively (Table S1). Thus, no evidence for publication bias in the meta-analysis was found.
Discussion
Many carcinomas of the digestive system have been considered cancer stem cell-related diseases in recent years, including esophageal, gastric, pancreatic and colorectal cancer [32]–[35]. In an effort to identify these small populations of cells, a number of cell surface proteins have been identified as CSC markers. Many studies have demonstrated experimentally that CD166 can enrich for CSC-like cells in a variety of cancers [4], [5]. Moreover, Levin et al [4] have found that CD166 is expressed at low levels in differentiated intestinal cells but robustly expressed on the surface of cells within the stem cell niche at the base of a crypt, which strongly infers its relationship with stem cell properties. Although high expression of CSC markers are usually considered as a prognosis of poor outcome, several contradictions to this generalization exist in published studies on the putative CSC-marker CD166 [21].
To investigate the basis for these contradictory conclusions, the staining methods among the studies in this meta-analysis were compared. Although the same antibody was used in each of these studies, the staining methods have varied between analyzing both membranous and cytoplasmic staining, whereas others take only membranous staining in consideration. Tachezy et al [11] found that CD166 was predominantly expressed at the cell membrane and that cytoplasmic staining intensity was related to the intensity of membranous staining and did not occur in the absence of membranous staining; thus, only membranous staining was considered in their study. The other three studies in this meta-analysis that focused on membranous staining including colorectal and pancreatic cancer, all concluded that high CD166 expression is a positive marker for good prognosis [10], [14], [36]. In contrast, the three studies in our analysis that analyze both membrane and cytoplasmic staining intensity propose that CD166 high expression contributes to poor clinical outcome [8], [30], [31]. Interestingly, in oral, breast and ovarian carcinomas, decreased membranous and increased cytoplasmic expression of CD166 is also associated with worse prognosis [37]–[39].
CD166 has been demonstrated to participate in the metastatic cascade of cancer cells. CD166-mediated intercellular adhesion involves interactions between the amino terminal D1 domain of opposing receptor molecules on two cells and is strengthened by lateral oligomerization of neighboring molecules on the cell surface, which engage the membrane proximal domains D4 and D5 [40]. These data suggest that high CD166 expression could impede cancer cell release from a local lesion. Furthermore, it has been shown that CD166/ALCAM can be actively cleaved by ADAM17/TACE-mediated proteolysis [41]. In ovarian cancer, pharmacologic inhibition of ADAM proteins, or specific silencing of ADAM17/TACE, hampered shedding of CD166 expressed on the cell surface. Interestingly, CD166/ALCAM can be translocated from the cell surface to the cytoplasm via a clathrin-dependent pathway. Specifically, soluble CD166/ALCAM (sALCAM) binds to scFv I/F8 to form a chimera, which induces endocytosis of the membrane-bound CD166/ALCAM. Recombinant sALCAM chimeric molecules inhibit the adhesive function of CD166/ALCAM through a competitive binding effect, which results in increased cancer cell motility [42], [43]. Van Kempen et al [44] also found that disruption of CD166 self-interaction was associated with tumor cell motility and metastasis. These studies all suggest that CD166 shedding from the cell surface may predict tumor progression and poor prognosis.
Colon cancer is a classical model for tumor progression studies because of its natural development from crypt stem cells to adenomas to fully formed carcinomas [45]; CD166 is highly expressed on the surface of crypt cells in this disease. However, both cell surface and cytoplasmic expression of CD166 is apparent in early adenoma formation in ApcMin/+ mice, human colorectal cancer and metastatic disease. Furthermore, only a subset of CD166 positive cells co-localize with the proliferation marker Ki67 [4]. These observations suggest that specific subcellular localization of CD166 could be used as a clinical prognostic marker because the loss of CD166 cell surface expression appears to be a precursor for tumor progression.
Part of the shedding CD166 would release into the tumor environment and circulation. A few studies have gone so far as to examine the shedding of CD166/ALCAM into blood serum. Two studies in this meta-analysis of esophageal and pancreatic cancer found a significant upregulation of CD166 in the blood serum of patients, but this observation only had a prognostic value in the esophageal cancer patients, as no significant correlation was found between elevated tissue expression and serum level in the pancreatic cancer patients [29], [36]. Klasingam et al. [46] have described significantly high sALCAM in the blood serum of breast cancer patients, but no prognostic data were reported. Variation in sALCAM among studies may have multiple causes: sALCAM must pass through the barrier of tumor tissue and vascular endothelial cells to be flushed into the blood stream, and sequential sectioning has failed to establish a direct relationship between ADAM17/TACE and ALCAM [31]. These data imply that the level of sALCAM in circulation is unstable and that it is inappropriate to use it for estimating prognoses.
Although the evidence addressed above may imply that cell surface expression of CD166 would be a positive prognostic marker and that the shedding of CD166, in other words, cytoplasmic CD166 expression would predict the reverse outcome, but in our stratified analysis, only cytoplasmic staining showed close relationship with poor prognosis, and the result of membrane staining of CD166 in unclear because two of the included studies provided significantly contradictory results [9], [29]. Recently, a colorectal cancer study may have provided a succinct method to assess the prognostic value of CD166 [21]. They found that an elevated mRNA level of CD166 was associated with poor outcome, yet intact membranous CD166 protein (co-localized extracellular and intracellular domain) is associated with improved outcome. With a novel method that stained the extracellular and intracellular domains of CD166 separately, they found that the extracellular domain of CD166 underwent shedding while the intracellular epitope remained. Thus, they concluded that shedding of the extracellular domain of CD166 correlated with patient outcome rather than loss of expression, which was previously considered the prognostic value of CD166. Unfortunately, the antibody applied in previous studies could not differentiate the subcellular epitope of CD166 in immunohistochemistry, making it difficult for a pathologist to accurately judge whether the protein was located on the cell surface, greatly affecting the prognostic capacity of CD166 expression.
This meta-analysis is subject to a few limitations. First, the number of studies included is relatively small, particularly for gastric and esophageal cancer. Second, 8 of the 9 studies were from Europe, including 7 from Germany, 1 from Switzerland and only one from Asia. Distinct site differences are believed to exist and could cause publication bias. Third, the criteria for determining positive or negative expression of CD166 varied among studies. Six studies only studied membranous CD166 expression while the rest also took cytoplasmic expression under consideration. Finally, although we tried to identify the disease free survival rate, these data were almost entirely missing from these studies. Most importantly, based on our meta-analysis of previous studies and systematic review of related articles indicates that the biological function of CD166 in tumor progression is complicated and that determining its subcellular location could be the key for accurate prognostic predictions. Thus, a more standardized staining method should be employed in future studies.
Supporting Information
CD166 expression and T category.
(TIF)
CD166 expression and N category.
(TIF)
CD166 expression and tumor grade.
(TIF)
CD166 expression and distant metastasis.
(TIF)
CD166 expression and 5-year overall survival rate.
(TIF)
CD166 expression and 3-year overall survival rate.
(TIF)
Heterogeneity test and publication bias analyses among studies included.
(DOC)
PRISMA Flow Diagram.
(DOC)
PRISMA Checklist.
(DOC)
Funding Statement
This work was supported by the Zhejiang Provincial Program for the cultivation of High-level Innovative Health Talents (JH), the 151 Talent Project of Zhejiang Province (JH), the Key project of Zhejiang Traditional medicine (No. 2012ZZ010), the Key discipline of Health ministry of Zhejiang Province (11-CX11) and the Key cultivate program of national science foundation (No. 91019005). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29. [DOI] [PubMed] [Google Scholar]
- 2. Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, et al. (2007) Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 104: 10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, et al. (2008) Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 105: 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, et al.. (2010) Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 139: 2072–2082 e2075. [DOI] [PMC free article] [PubMed]
- 5. Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, et al. (2012) Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One 7: e42564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowen MA, Patel DD, Li X, Modrell B, Malacko AR, et al. (1995) Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med 181: 2213–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen A SG, Zijlstra A. (2011) ALCAM. doi:101038/mpa00412601 [DOI] [PMC free article] [PubMed]
- 8. Weichert W, Knosel T, Bellach J, Dietel M, Kristiansen G (2004) ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol 57: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horst D, Kriegl L, Engel J, Kirchner T, Jung A (2009) Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest 27: 844–850. [DOI] [PubMed] [Google Scholar]
- 10. Lugli A, Iezzi G, Hostettler I, Muraro MG, Mele V, et al. (2010) Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer 103: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tachezy M, Zander H, Gebauer F, Marx A, Kaifi JT, et al. (2012) Activated leukocyte cell adhesion molecule (CD166)–its prognostic power for colorectal cancer patients. J Surg Res 177: e15–20. [DOI] [PubMed] [Google Scholar]
- 12. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, et al. (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 13. Patel BB, Yu Y, Du J, Levi E, Phillip PA, et al. (2009) Age-related increase in colorectal cancer stem cells in macroscopically normal mucosa of patients with adenomas: a risk factor for colon cancer. Biochem Biophys Res Commun 378: 344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tachezy M, Zander H, Marx AH, Gebauer F, Rawnaq T, et al. (2011) ALCAM (CD166) expression as novel prognostic biomarker for pancreatic neuroendocrine tumor patients. J Surg Res 170: 226–232. [DOI] [PubMed] [Google Scholar]
- 15. Hong X, Michalski CW, Kong B, Zhang W, Raggi MC, et al. (2010) ALCAM is associated with chemoresistance and tumor cell adhesion in pancreatic cancer. J Surg Oncol 101: 564–569. [DOI] [PubMed] [Google Scholar]
- 16. Leavell BJ, Van Buren E, Antaki F, Axelrod BN, Rambus MA, et al. (2012) Associations between markers of colorectal cancer stem cells and adenomas among ethnic groups. Dig Dis Sci 57: 2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarkar D, Shields B, Davies ML, Muller J, Wakeman JA (2012) BRACHYURY confers cancer stem cell characteristics on colorectal cancer cells. Int J Cancer 130: 328–337. [DOI] [PubMed] [Google Scholar]
- 18. Melin C, Perraud A, Akil H, Jauberteau MO, Cardot P, et al. (2012) Cancer stem cell sorting from colorectal cancer cell lines by sedimentation field flow fractionation. Anal Chem 84: 1549–1556. [DOI] [PubMed] [Google Scholar]
- 19. Muraro MG, Mele V, Daster S, Han J, Heberer M, et al. (2012) CD133+, CD166+CD44+, and CD24+CD44+ phenotypes fail to reliably identify cell populations with cancer stem cell functional features in established human colorectal cancer cell lines. Stem Cells Transl Med 1: 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, et al. (2011) Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res 17: 6934–6943. [DOI] [PubMed] [Google Scholar]
- 21.Hansen AG, Freeman TJ, Arnold SA, Starchenko A, Jones-Paris CR, et al.. (2013) Elevated ALCAM shedding in colorectal cancer correlates with poor patient outcome. Cancer Res. [DOI] [PMC free article] [PubMed]
- 22. Borlak J, Meier T, Halter R, Spanel R, Spanel-Borowski K (2005) Epidermal growth factor-induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene 24: 1809–1819. [DOI] [PubMed] [Google Scholar]
- 23. Verma A, Shukla NK, Deo SV, Gupta SD, Ralhan R (2005) MEMD/ALCAM: a potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology 68: 462–470. [DOI] [PubMed] [Google Scholar]
- 24. Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, et al. (2011) MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene 30: 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS Jr, Crawford H, et al. (2009) Phenotypic subpopulations of metastatic colon cancer stem cells: genomic analysis. Cancer Genomics Proteomics 6: 19–29. [PubMed] [Google Scholar]
- 26. Varghese S, Burness M, Xu H, Beresnev T, Pingpank J, et al. (2007) Site-specific gene expression profiles and novel molecular prognostic factors in patients with lower gastrointestinal adenocarcinoma diffusely metastatic to liver or peritoneum. Ann Surg Oncol 14: 3460–3471. [DOI] [PubMed] [Google Scholar]
- 27.Hwang WL, Yang MH, Tsai ML, Lan HY, Su SH, et al.. (2011) SNAIL regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology 141: 279–291, 291 e271–275. [DOI] [PubMed]
- 28.Lee HJ, Eom DW, Kang GH, Han SH, Cheon GJ, et al.. (2012) Colorectal micropapillary carcinomas are associated with poor prognosis and enriched in markers of stem cells. Mod Pathol. [DOI] [PubMed]
- 29. Tachezy M, Effenberger K, Zander H, Minner S, Gebauer F, et al. (2012) ALCAM (CD166) expression and serum levels are markers for poor survival of esophageal cancer patients. Int J Cancer 131: 396–405. [DOI] [PubMed] [Google Scholar]
- 30. Ishigami S, Ueno S, Arigami T, Arima H, Uchikado Y, et al. (2011) Clinical implication of CD166 expression in gastric cancer. J Surg Oncol 103: 57–61. [DOI] [PubMed] [Google Scholar]
- 31. Kahlert C, Weber H, Mogler C, Bergmann F, Schirmacher P, et al. (2009) Increased expression of ALCAM/CD166 in pancreatic cancer is an independent prognostic marker for poor survival and early tumour relapse. Br J Cancer 101: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Todaro M, Francipane MG, Medema JP, Stassi G (2010) Colon cancer stem cells: promise of targeted therapy. Gastroenterology 138: 2151–2162. [DOI] [PubMed] [Google Scholar]
- 33. Chen T, Yang K, Yu J, Meng W, Yuan D, et al. (2012) Identification and expansion of cancer stem cells in tumor tissues and peripheral blood derived from gastric adenocarcinoma patients. Cell Res 22: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lonardo E, Hermann PC, Mueller MT, Huber S, Balic A, et al. (2011) Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell 9: 433–446. [DOI] [PubMed] [Google Scholar]
- 35. Chen Y, Li D, Wang D, Liu X, Yin N, et al. (2012) Quiescence and attenuated DNA damage response promote survival of esophageal cancer stem cells. J Cell Biochem 113: 3643–3652. [DOI] [PubMed] [Google Scholar]
- 36. Tachezy M, Zander H, Marx AH, Stahl PR, Gebauer F, et al. (2012) ALCAM (CD166) expression and serum levels in pancreatic cancer. PLoS One 7: e39018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van den Brand M, Takes RP, Blokpoel-deRuyter M, Slootweg PJ, van Kempen LC (2010) Activated leukocyte cell adhesion molecule expression predicts lymph node metastasis in oral squamous cell carcinoma. Oral Oncol 46: 393–398. [DOI] [PubMed] [Google Scholar]
- 38. Mezzanzanica D, Fabbi M, Bagnoli M, Staurengo S, Losa M, et al. (2008) Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin Cancer Res 14: 1726–1733. [DOI] [PubMed] [Google Scholar]
- 39. Burkhardt M, Mayordomo E, Winzer KJ, Fritzsche F, Gansukh T, et al. (2006) Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J Clin Pathol 59: 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Kempen LC, Nelissen JM, Degen WG, Torensma R, Weidle UH, et al. (2001) Molecular basis for the homophilic activated leukocyte cell adhesion molecule (ALCAM)-ALCAM interaction. J Biol Chem 276: 25783–25790. [DOI] [PubMed] [Google Scholar]
- 41. Bech-Serra JJ, Santiago-Josefat B, Esselens C, Saftig P, Baselga J, et al. (2006) Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Mol Cell Biol 26: 5086–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piazza T, Cha E, Bongarzone I, Canevari S, Bolognesi A, et al. (2005) Internalization and recycling of ALCAM/CD166 detected by a fully human single-chain recombinant antibody. J Cell Sci 118: 1515–1525. [DOI] [PubMed] [Google Scholar]
- 43. Rosso O, Piazza T, Bongarzone I, Rossello A, Mezzanzanica D, et al. (2007) The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res 5: 1246–1253. [DOI] [PubMed] [Google Scholar]
- 44. van Kempen LC, Meier F, Egeblad M, Kersten-Niessen MJ, Garbe C, et al. (2004) Truncation of activated leukocyte cell adhesion molecule: a gateway to melanoma metastasis. J Invest Dermatol 122: 1293–1301. [DOI] [PubMed] [Google Scholar]
- 45.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, et al.. (2012) Lineage Tracing Reveals Lgr5+ Stem Cell Activity in Mouse Intestinal Adenomas. Science. [DOI] [PubMed]
- 46. Kulasingam V, Zheng Y, Soosaipillai A, Leon AE, Gion M, et al. (2009) Activated leukocyte cell adhesion molecule: a novel biomarker for breast cancer. Int J Cancer 125: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD166 expression and T category.
(TIF)
CD166 expression and N category.
(TIF)
CD166 expression and tumor grade.
(TIF)
CD166 expression and distant metastasis.
(TIF)
CD166 expression and 5-year overall survival rate.
(TIF)
CD166 expression and 3-year overall survival rate.
(TIF)
Heterogeneity test and publication bias analyses among studies included.
(DOC)
PRISMA Flow Diagram.
(DOC)
PRISMA Checklist.
(DOC)