Abstract
The basic helix–loop–helix factor Math5 (Atoh7) is critical for the determination of retinal ganglion cell (RGC) fate in mice. Recently, genome-wide association studies have identified the ATOH7 locus as a major determinant of variation in the human optic disc area, which is directly correlated with the RGC number. These studies suggest that the level of Math5 expression may determine the ultimate number of RGCs. To test this hypothesis, we systematically compared optic nerve area and RGC axon number in C57BL/6J congenic Math5+/– and +/+ mice at young adult and neonatal ages by transmission electron microscopy. Optic disc area and RGC abundance were not significantly different in adults, but heterozygotes had thinner optic nerves and 25–30% fewer RGCs at birth than wild-type littermates (P < 0.05). Our results suggest that Math5 dosage is important for the genesis, but not the ultimate number, of RGCs. Our findings highlight the importance of ganglion cell culling as a compensatory mechanism for retinal homeostasis, and support a quantitative role for Math5 in RGC specification.
Keywords: apoptosis, Atoh7, culling, glaucoma, genome-wide association studies, Math5, neurogenesis, optic disc area, optic nerve, retinal ganglion cell
Introduction
Humans and mice show marked intraspecies variations in the total number of retinal ganglion cells (RGCs) and optic disc area, which are directly correlated measures [1–5]. In principle, this variation may arise from differences in RGC genesis or in the extent of developmental culling. In mice, multipotent retinal progenitors exit the cell cycle and differentiate as RGCs between embryonic day E11 and postnatal day P0 (birth), with peak genesis at E14 [6,7]. RGC axons coalesce to form the optic nerve and project to brain visual centers. More than 60% of new RGCs are lost by apoptosis during the period of synaptic refinement, which occurs during the first postnatal week in mice and during late gestation in primates [2,8–11]. RGC survival is believed to reflect the formation of functional synaptic connections in the brain [10].
The Math5 (Atoh7) basic helix–loop–helix transcription factor is critical for the specification of ganglion cells [12]. Math5 mutant mice lack the vast majority (>95%) of RGCs [13,14], and recessive loss-of-function mutations cause congenital retinovascular disease in humans [15–17], with optic nerve aplasia being the primary phenotype in both species. Recent genome-wide association studies (GWAS) have identified the ATOH7 locus as a major determinant of variation in the optic disc area in normal human populations [3–5] and a contributing factor in the susceptibility to glaucoma disease [18,19]. However, a substantial cohort of RGCs are Math5-independent, and only a small fraction of Math5+ cells adopt the RGC fate [20].
Given the importance of Math5 in the development of RGC, we examined the quantitative effects of Math5 gene dosage on ganglion cell abundance. We found that mice with a 50%reduction in gene dosage (Math5+/–) generate fewer RGCs than Math5+/+ mice, but postnatal compensatory mechanisms equalize the number of ganglion cells between these genotypes. Our results highlight the robustness of mammalian retinal development, show the buffering capacity of the initial RGC pool, and implicate Math5 as a limiting determinant of RGC abundance.
Materials and methods
Processing optic nerves for transmission electron microscopy
All mouse procedures were approved by the University of Michigan Committee on the Use and Care of Animals (UCUCA). Congenic Math5+/– mice (Atoh7tm1Gla [13]) were systematically maintained on a C57BL/6J background (N18 generation). Young adult (P30) Math5 heterozygotes and control littermates were anesthetized with 100 mg/kg ketamine and 5 mg/kg xylazine and perfused transcardially with 15 ml 1.25% glutaraldehyde (GA) 1% paraformaldehyde (PFA), followed by 10 ml 2.5% GA 2% PFA. For P1 optic nerves, eyelids were incised and whole heads were fixed by immersion in 2.5% GA 2% PFA for 1 h at room temperature. Eyes were then carefully dissected with optic nerves attached and postfixed in 1.25% GA 1% PFA overnight at 41°C.
Fixed eyes were washed three times in 0.1M phosphate buffer (pH 7.4), and optic nerves were transected 2–3mm from the sclera. The cut nerves were washed three times in 50mM sodium cacodylate phosphate buffer, postfixed in 1% osmium tetroxide 50mM sodium cacodylate for 1 h at 41°C, dehydrated through a graded ethanol series, infiltrated with 100% propylene oxide, and embedded in EMBed-812 resin (Electron Microscopy Sciences, Hatfield, Pennsylvania, USA). For light microscopy, semithin (0.5 µm) sections were stained with toluidine blue. For transmission electron microscopy (TEM), ultrathin (75 nm) sections were stained with 7% uranyl acetate and Reynold’s lead citrate, and imaged using a Philips CM-100 electron microscope (Andover, Massachusetts, USA).
Quantitative analysis of retinal ganglion cell axons
For each genotype and age, axons were counted in 3–4 optic nerves. Ten fields were systematically sampled from central (four) and peripheral (six) areas at ×10 500 magnification (103 µm2). The optic nerve area was measured from low-power (×380–530) TEM images by manually tracing the perimeter using ImageJ software (NIH, Bethesda, Maryland, USA). The total number of RGCs per optic nerve was extrapolated by multiplying the average RGC density from 10 fields by the optic nerve area. The results were compared between genotypes using Student’s t-test.
Results
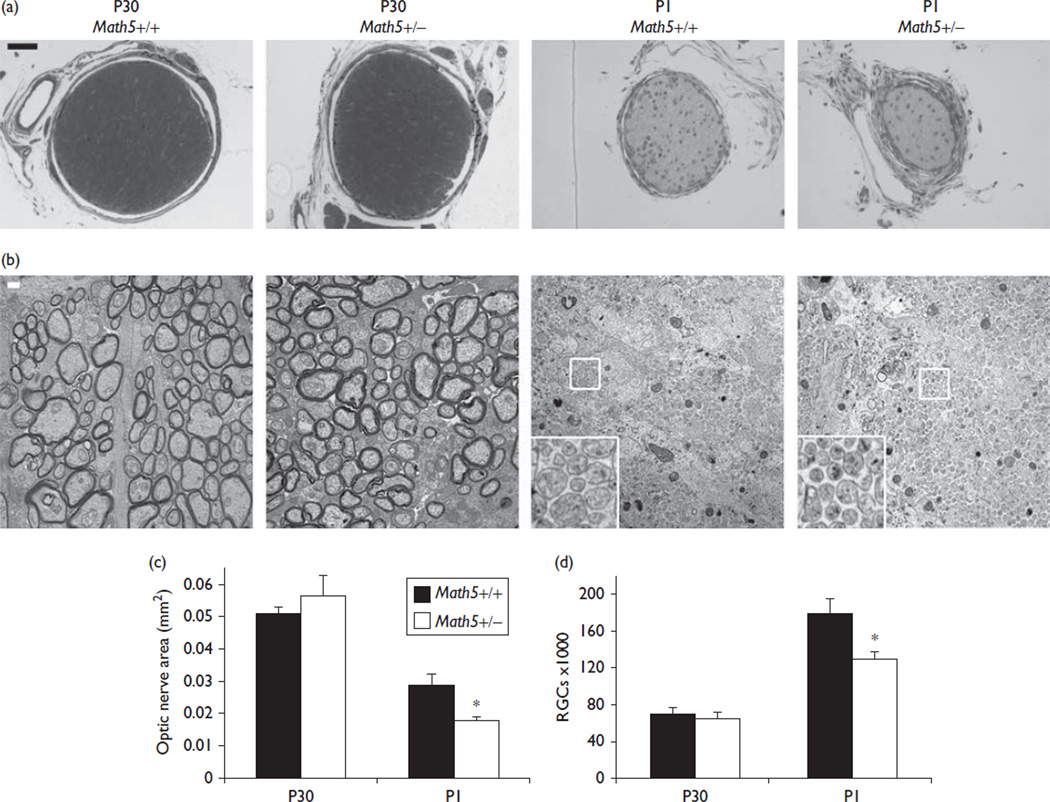
To isolate the role of Math5 dosage in determining RGC abundance, we systematically measured the optic nerve area and the total number of RGCs in neonatal (P1) and young adult (P30) mice (Fig. 1). These ages were chosen to reflect the size of the RGC population before and after the period of postnatal culling, respectively [9,11]. To identify ganglion cells definitively, we counted axon fibers in optic nerves of congenic (N18) C57BL/6J Math5 heterozygous and wild-type mice by TEM [1,2]. In adult mice, the optic nerve area and RGC number did not differ between these genotypes (Fig. 1, Table 1), and were comparable with previously reported values for C57BL/6J strains [1,2]. In contrast, neonatal Math5+/– mice showed a marked reduction in the optic nerve area (36%, 0.018 vs. 0.028mm2, P<0.03) and RGC number (20%, 129 000 vs. 178 000 per eye, P<0.04). Surprisingly, the density of RGC axons was higher in P1 Math5+/– mice than that in wild-type littermates (7.5 vs. 6.3 million axons/mm2, P<0.04), despite a marked decrease in the optic nerve area. This difference did not persist in adults, suggesting a transient deficiency of optic nerve glia.
Fig. 1.
Heterozygous Math5 effect on retinal ganglion cell (RGC) number and optic nerve size. (a) Toluidine blue-stained semithin sections of P30 (left) or P1 (right) optic nerves. (b) High-power (×10 500) transmission electron microscopy images of representative ultrathin optic nerve sections. (c, d) Quantitative analysis of (c) the optic nerve area and (d) the overall RGC abundance. The cross-sectional area and RGC number are significantly reduced in Math5+/– mice at P1, but not P30. *P<0.05. Scale bar, 50 µm in (a), 200 nm in (b).
Table 1.
Optic nerve metrics in Math5+ / – and Math5+ / + animals
| Math5 genotype | Age | n | Axons counteda | Optic nerve area (mm2) [a] | RGC density×106 (axons/mm2) [b] | Total RGCs [a×b] |
|---|---|---|---|---|---|---|
| + / + | P30 | 3 | 1440±80 | 0.051±0.002 | 1.39±0.07 | 71000±6000 |
| + / – | P30 | 3 | 1200±100 | 0.056±0.006 | 1.2±0.1 | 65000±7000 |
| + / + | P1 | 4 | 6500±400 | 0.028±0.004 | 6.3±0.4 | 178000±16000 |
| + / – | P1 | 4 | 7700±100 | 0.018±0.001 | 7.5±0.1 | 129000±8000 |
RGC, retinal ganglion cell.
Axons were counted in high-quality areas and normalized to the entire image area. Errors are reported as SEM.
Given the wide variation in RGC abundance among inbred mouse strains [1], we also compared residual RGC axons between genetically diverse Math5 – /– mice, which retain only B4% of RGCs [21]. There was no apparent difference in the density of TuJ1+ axons between C57BL/6J congenic (N18) and outcrossed (129S1×C57BL/6J) F2 Math5 – / – mice (data not shown).
Discussion
The absolute number of ganglion cells per eye, and the photoreceptor-to-RGC ratio, determine the anatomy of the optic nerve and ultimate spatial integration of visual signals in the retina. These parameters appear to be precisely controlled; yet, few genetic factors have been identified for variations in the optic disc area and RGC number. In mice, one quantitative trait locus (Nnc1) for RGC abundance was defined between C57BL/6J and DBA/2J inbred strains [22]. Recently, human GWAS identified the ATOH7 locus as the major single determinant of variation in the optic disc area in Asian and White populations [3–5]. Because no common coding variants were identified in individuals with normal vision or optic nerve disease [17,18], this effect is most likely because of differences in transcriptional regulation. Indeed, the GWAS peak is centered over rs3858145, an SNP located 20 kb upstream from the ATOH7 promoter, within a conserved transcriptional enhancer [15]. Math5 mRNA is transiently expressed in retinal progenitors during or immediately following the terminal cell cycle [20] and is reduced in Math5 heterozygotes [13]. In principle, a specific threshold level may be required during histogenesis to confer RGC competence to individual progenitor cells or the retinal population. The Math5 bHLH protein, for example, may represent the limiting component in a multimeric transcriptional complex that initiates the ganglion cell development program. In this model, decreasing Math5 expression would decrease RGC genesis. Conversely, Math5 overexpression would not be expected to alter RGC abundance, and this prediction holds true in Crx > Math5 transgenic mice, which express Math5 broadly in a large population of retinal progenitor cells [23]. Sequence variants in human ATOH7 enhancer regions may cause quantitative or qualitative differences in the spatiotemporal pattern of Math5 expression.
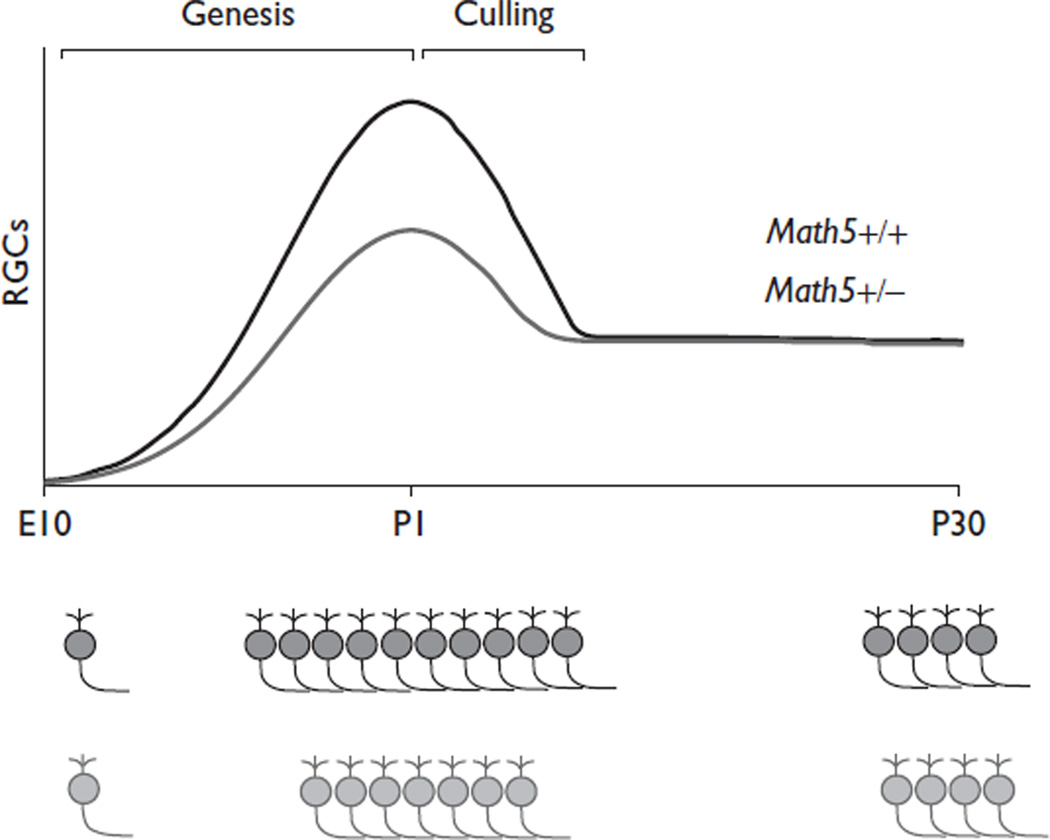
In adult mice, a 50% reduction in Math5 activity did not alter the number of RGCs or the optic nerve crosssectional area. However, it remains unclear whether humans carrying one null ATOH7 allele [15,17] have a reduced number of ganglion cells. This is an intriguing possibility because GWAS data connect ATOH7 to variations in the optic nerve area [3–5], and one patient with optic nerve hypoplasia was found to be heterozygous for an allele (A47T) with reduced function [3,17]. In open-angle glaucoma, patients experience progressive death of ganglion cells during adulthood. Visual impairment is not clinically significant until 40–50% of RGCs are lost, but field differences are detectable by perimetry with a 25–30% reduction in ganglion cells [24]. The potentially discordant effects of the ATOH7 genotype on adult RGC number between humans and mice may indicate species differences in genetic heterogeneity, which was absent in our comparison between inbred littermates, or the degree of developmental culling. The overall extent of culling is comparable in mice and humans, involving two-thirds of new RGCs [8,11], but the genetic background can influence birth and death independently [2]. Our results suggest that Math5 dosage affects RGC genesis, but other mechanisms compensate to ensure the proper number of functional ganglion cells in adults (Fig. 2). Although 25–30% fewer RGCs are generated in Math5+/– mice, fewer are lost during the culling period [9,11]; therefore, the adult optic nerve is similar to the wild type (Fig. 1). These results show an important ‘fate-buffering’ capacity in the retina and provide an instructive example of developmental compensation for a genetic defect [25].
Fig. 2.
Model of retinal ganglion cell (RGC) genesis and culling in Math5+/+ and +/– mice. In Math5 heterozygotes, fewer ganglion cells are produced than in wild-type mice during genesis stages (E11–P0). However, adult Math5+/– and +/+ mice have a similar number of RGCs. Therefore, fewer ganglion cells undergo apoptosis during the neonatal RGC culling period in Math5+/– mice.
Conclusion
Our results show that Math5 (Atoh7) dosage has a significant impact on the genesis of RGCs, but not on the ultimate number of ganglion cells. These findings highlight genetic differences in RGC development and culling, and the role of Math5 in these processes.
Acknowledgements
The authors are grateful to Jeff Harrison and the UM microscopy and image analysis laboratory for technical assistance with EM processing, and to David Turner and Roman Giger for valuable discussions and critical reading of the manuscript. This research was funded by grants from the National Institutes of Health (EY14259) and The Glaucoma Foundation to T.G. L.P. was supported by NIH T32 grants EY13934 and GM07863.
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Williams RW, Strom RC, Rice DS, Goldowitz D. Genetic and environmental control of variation in retinal ganglion cell number in mice. J Neurosci. 1996;16:7193–7205. doi: 10.1523/JNEUROSCI.16-22-07193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strom RC, Williams RW. Cell production and cell death in the generation of variation in neuron number. J Neurosci. 1998;18:9948–9953. doi: 10.1523/JNEUROSCI.18-23-09948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macgregor S, Hewitt AW, Hysi PG, Ruddle JB, Medland SE, Henders AK, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010;19:2716–2724. doi: 10.1093/hmg/ddq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramdas WD, van Koolwijk LM, Ikram MK, Jansonius NM, de Jong PT, Bergen AA, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khor CC, Ramdas WD, Vithana EN, Cornes BK, Sim X, Tay WT, et al. Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum Mol Genet. 2011;20:1864–1872. doi: 10.1093/hmg/ddr060. [DOI] [PubMed] [Google Scholar]
- 6.Drager UC. Birth dates of retinal ganglion cells giving rise to the crossed and uncrossed optic projections in the mouse. Proc R Soc Lond B Biol Sci. 1985;224:57–77. doi: 10.1098/rspb.1985.0021. [DOI] [PubMed] [Google Scholar]
- 7.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 8.Provis JM, van Driel D, Billson FA, Russell P. Human fetal optic nerve: overproduction and elimination of retinal axons during development. J Comp Neurol. 1985;238:92–100. doi: 10.1002/cne.902380108. [DOI] [PubMed] [Google Scholar]
- 9.Farah MH, Easter SS., Jr Cell birth and death in the mouse retinal ganglion cell layer. J Comp Neurol. 2005;489:120–134. doi: 10.1002/cne.20615. [DOI] [PubMed] [Google Scholar]
- 10.O’Leary DD, Fawcett JW, Cowan WM. Topographic targeting errors in the retinocollicular projection and their elimination by selective ganglion cell death. J Neurosci. 1986;6:3692–3705. doi: 10.1523/JNEUROSCI.06-12-03692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young RW. Cell death during differentiation of the retina in the mouse. J Comp Neurol. 1984;229:362–373. doi: 10.1002/cne.902290307. [DOI] [PubMed] [Google Scholar]
- 12.Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix–loop–helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 13.Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, et al. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JA, Goldman D, Glaser T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan K, Logan CV, McKibbin M, Sheridan E, Elcioglu NH, Yenice O, et al. Next generation sequencing identifies mutations in Atonal homolog 7 (ATOH7) in families with global eye developmental defects. Hum Mol Genet. 2011;21:776–783. doi: 10.1093/hmg/ddr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasov L, Masud T, Khaliq S, Mehdi SQ, Abid A, Oliver ER, et al. ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds197. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JH,Wang D, Huang C, Zheng Y, Chen H, Pang CP, et al. Interactive effects of ATOH7 and RFTN1 in association with adult-onset primary open angle glaucoma. Invest Ophthalmol Vis Sci. 2012;53:779–785. doi: 10.1167/iovs.11-8277. [DOI] [PubMed] [Google Scholar]
- 19.Fan BJ, Wang DY, Pasquale LR, Haines JL, Wiggs JL. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci. 2011;52:1788–1792. doi: 10.1167/iovs.10-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brzezinski JA, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–485. doi: 10.1016/j.neuron.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Williams RW, Strom RC, Goldowitz D. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J Neurosci. 1998;18:138–146. doi: 10.1523/JNEUROSCI.18-01-00138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasov L, Glaser T. Pushing the envelope of ganglion cell fate: context dependent function of Math5 (Atoh7) Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harwerth RS, Quigley HA. Visual field defects and retinal ganglion cell losses in patients with glaucoma. Arch Ophthalmol. 2006;124:853–859. doi: 10.1001/archopht.124.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]