Abstract
γ-Tubulin is a ubiquitous and highly conserved component of centrosomes in eukaryotic cells. Genetic and biochemical studies have demonstrated that γ-tubulin functions as part of a complex to nucleate microtubule polymerization from centrosomes. We show that, as in other organisms, Caenorhabditis elegans γ-tubulin is concentrated in centrosomes. To study centrosome dynamics in embryos, we generated transgenic worms that express GFP::γ-tubulin or GFP::β-tubulin in the maternal germ line and early embryos. Multiphoton microscopy of embryos produced by these worms revealed the time course of daughter centrosome appearance and growth and the differential behavior of centrosomes destined for germ line and somatic blastomeres. To study the role of γ-tubulin in nucleation and organization of spindle microtubules, we used RNA interference (RNAi) to deplete C. elegans embryos of γ-tubulin. γ-Tubulin (RNAi) embryos failed in chromosome segregation, but surprisingly, they contained extensive microtubule arrays. Moderately affected embryos contained bipolar spindles with dense and long astral microtubule arrays but with poorly organized kinetochore and interpolar microtubules. Severely affected embryos contained collapsed spindles with numerous long astral microtubules. Our results suggest that γ-tubulin is not absolutely required for microtubule nucleation in C. elegans but is required for the normal organization and function of kinetochore and interpolar microtubules.
INTRODUCTION
Centrosomes, the complex and dynamic organelles that serve as microtubule-organizing centers in animal cells, have intrigued biologists for more than a century (Wilson, 1925). They generally contain a pair of centrioles surrounded by a meshwork of pericentriolar material (Kellogg et al., 1994). Orderly duplication of the centriole pair and centrosome before mitosis ensures that the mitotic spindle will have 2 poles and that each daughter cell will inherit a centrosome. In mitotic cells, centrosomes appear to seed the growth of most spindle microtubules, with the less dynamic “minus ends” embedded in the centrosome and the more dynamic “plus ends” distal (see Kellogg et al., 1994). The aster-shaped arrays of microtubules emanating from the 2 centrosomes interact with each other and with chromosomes to organize a bipolar spindle. A normal spindle performs 2 essential roles, segregating chromosomes and controlling the position and contraction of the cleavage furrow. Because of the key role of centrosomes in mitosis and cell division, analysis of their composition and function is an area of intense investigation.
The best-characterized component of the centrosome is γ-tubulin, which is organized into stable, lock washer–shaped, open-ring complexes (Gunawardane et al., 2000; Oakley, 2000). The diameter of these γ-tubulin ring complexes (γ-TuRCs) is ∼25 nm, which coincides with the diameter of most microtubules (Moritz et al., 1995a,b). A currently favored model is that centrosome-associated γ-TuRCs serve as templates to nucleate the polymerization of α/β-tubulin dimers into microtubules. The hypothesis that γ-tubulin is required for microtubule nucleation in cells is supported by several lines of evidence. Mutants null for the γ-tubulin gene of Aspergillus fail to assemble spindles (Oakley et al., 1990). Antibody inhibition or depletion of γ-tubulin prevents microtubule nucleation from centrosomes in cultured mammalian cells and in Xenopus egg extracts (Joshi et al., 1992; Felix et al., 1994). Isolated centrosomes depleted of γ-TuRCs lose their ability to nucleate microtubules, and addition of γ-TuRCs restores microtubule nucleation potential (Schnackenberg et al., 1998). Finally, purified γ-TuRCs can nucleate microtubules in vitro (Zheng et al., 1995; Wiese and Zheng, 2000). In contrast, other lines of evidence suggest that γ-tubulin is not absolutely required to seed microtubule growth in cells. Null γ-tubulin mutations in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Drosophila impair but do not completely block the assembly of mitotic spindles (Horio et al., 1991; Sobel and Snyder, 1995; Sunkel et al., 1995; Wilson and Borisy, 1998).
We report here analysis of γ-tubulin localization and tests of γ-tubulin function in Caenorhabditis elegans early embryos. For this investigation and to advance the study of microtubule-dependent processes in general, we developed methods to express and observe γ-tubulin, β-tubulin, and histone H2B fused to green fluorescent protein (GFP) in living embryos. The large size (∼30 × 50 μm) and transparency of C. elegans embryos make them an excellent system for observing the formation and function of microtubules during a stereotyped developmental sequence. In addition, the advent of RNA-mediated interference (RNAi; Guo and Kemphues, 1995; Fire et al., 1998), combined with the completed C. elegans genome sequence (Consortium, 1998) and a collection of sequence-tagged cDNA clones (Y. Kohara, personal communication), makes function–disruption studies in the early embryo a powerful analytical approach. Our results suggest that centrosomes do not require γ-tubulin for microtubule nucleation and growth. However, it appears that centrosomes do require γ-tubulin to generate microtubules capable of participating in bipolar spindle assembly and function.
MATERIALS AND METHODS
Worm Strains
C. elegans strains were maintained as described by Brenner (1974). C. elegans N2 variety Bristol was used for RNAi analysis and was the wild-type parent of all GFP strains generated. Strain DG800, unc-32(e189) tnDf2 III/eT1 (III;IV), carries a deficiency that removes the gene encoding γ-tubulin (Furuta et al., 2000).
RNAi
The γ-tubulin cDNA clones yk496c1 and yk80h7 were obtained from Yuji Kohara (National Institute of Genetics, Mishima, Japan). Phagemid DNA was prepared as described in the Stratagene (La Jolla, CA) ExAssist protocol. Sense and antisense strands of RNA were synthesized with the use of the MEGAscript in vitro transcription kit (Ambion, Austin, TX), annealed, and injected at 0.5–1 mg/ml into young adult hermaphrodites. yk80h7 also was subcloned to generate nonoverlapping fragments for making double-stranded RNA (dsRNA). yk80h7 was cut with XhoI and BspHI to release the 5′ fragment and with BspHI and NotI to release the 3′ fragment. Each was inserted into the Bluescript SK vector (Stratagene). Identical phenotypes were observed with the use of dsRNA made from either of these 2 nonoverlapping portions of the C. elegans γ-tubulin gene, confirming that our RNAi approach was gene specific. For all dsRNAs injected, moderately affected embryos (with stunted bipolar spindles) began to appear 18–22 h after injection of the parent worm. We presume that those embryos were partially depleted for γ-tubulin. A few hours after the appearance of moderately affected embryos, severely affected embryos (with totally collapsed spindles) were observed. The defects observed likely represent the consequences of a severe (perhaps complete) loss of γ-tubulin. This interpretation was supported by comparing defects caused by RNAi of γ-tubulin in wild-type hermaphrodites with defects caused by RNAi in hermaphrodites that were heterozygous for a γ-tubulin deficiency. The severity of phenotypes was indistinguishable. All embryos produced during the moderate and severe RNAi windows died. All RNAi embryos described in RESULTS were obtained from injected mothers 18–28 h after injection.
Antibodies and Immunofluorescence Microscopy
To generate antisera against γ-tubulin, 2 peptides (amino acids 34–52 and 391–410; see Figure 2) were synthesized and conjugated to keyhole limpet hemocyanin (Research Genetics, Huntsville, AL). They were used together to immunize rabbits (Cocalico Biologicals). Antibodies were affinity purified by passing serum over a column of γ-tubulin peptides coupled to epoxy-activated agarose (Pierce, Rockford, IL). Antibodies were eluted with 0.2 M glycine and 150 mM NaCl, pH 2.0, dialyzed in PBS, and concentrated. For immunofluorescence staining of embryos, gravid adult hermaphrodites were cut, fixed, and stained as described previously (Strome and Wood, 1983). The primary antibodies used were affinity-purified anti-γ-tubulin at 1:10, mouse 4A1 anti-α-tubulin at 1:45 (a gift from M. Fuller, Stanford University School of Medicine, Stanford, CA; Piperno and Fuller, 1985), mouse PA3 anti-nucleosome at 1:100 (a gift from M. Monestier, Temple University School of Medicine, Philadelphia, PA; Monestier et al., 1994), and rabbit anti-acetylated histone H4 at 1:500 (a gift from D. Allis, University of Virginia, Charlottesville, VA; Lin et al., 1989). The secondary antibodies used were flourescein-conjugated goat anti-mouse immunoglobulin G at 1:150 and Texas Red–conjugated goat anti-rabbit immunoglobulin G at 1:250 (Jackson ImmunoResearch, West Grove, PA). Images were collected on a Bio-Rad (Hercules, CA) MRC600 scanning confocal microscope and manipulated to generate figures in NIH Image (version 1.62f, developed by Wayne Rasband, National Institutes of Health, and available on the Internet at http://rsb.info.nih.gov/nih-image/) and Photoshop (Adobe Systems, Palo Alto, CA).
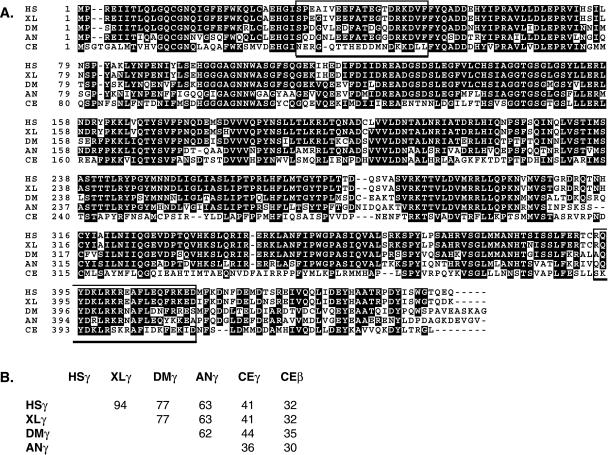
Figure 2.
Alignment of γ-tubulin sequences. (A) Sequences are from Homo sapiens (HS; GenBank accession number AF225971), Xenopus laevis (XL; M63446), Drosophila melanogaster (DM; AJ010552), Aspergillus nidulans (AN; X15479), and C. elegans (CE, CAA80164.1). Amino acids on black backgrounds are shared by three or more of the γ-tubulins shown. The open boxes enclose the C. elegans peptide sequences used to generate antibodies. (B) Grid showing the percent amino acid identities between pairs of γ-tubulins.
Generation of Transgenic Worms Expressing GFP::Tubulins and GFP::Histone
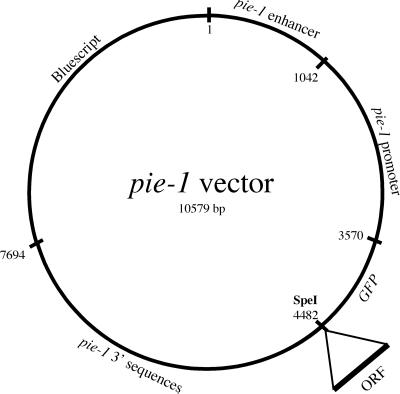
Sequences from the pie-1 gene were used to construct a vector designed to express GFP fusion proteins in the adult germ line and in early embryos. A PCR-based strategy, with the use of sequence information from Y49E10 (Consortium, 1998), was used to clone a 7.7-kb genomic fragment containing the pie-1 gene (Reese et al., 2000). BamHI sites introduced at both ATG and TAA during PCR amplification were used to replace the pie-1 open reading frame with GFP from pPD103.87, which contains the S65C mutation and 3 synthetic introns (A. Fire, S. Xu, J. Ahnn, and G. Seydoux, personal communication). The GFP is followed by a unique SpeI site, into which can be cloned open reading frames of interest (Figure 1). Addition of the third intron of pie-1 upstream of the pie-1 promoter was found to increase expression levels (our unpublished data). Into this vector, cDNA sequences encoding γ- or β-tubulin or histone were inserted. pJH4.67 contains γ-tubulin (tbg-1, F58A4.8); pJH4.66 contains β-tubulin (tbb-2, C36E8.5); and pJH4.52 contains histone H2B (F54E12.4) sequences.
Figure 1.
pie-1::GFP expression vector. Noncoding sequences from pie-1, a maternally expressed gene, were used to construct a vector that will express GFP fusion proteins in the adult germ line and in early embryos. A unique SpeI site was used to fuse open reading frames of interest to the carboxyl terminus of GFP.
To generate transgenic worms expressing GFP fusion proteins, wild-type adult hermaphrodites were injected with a mixture of DNAs that can recombine to form “complex” extrachromosomal arrays (Kelly et al., 1997): 75 μg/ml genomic DNA (linearized with PvuII), 0.2 μg/ml pRF4 rol-6(su1006) DNA (linearized with SmaI), and 0.2–0.4 μg/ml GFP construct (linearized with SacII). Roller progeny (F1 generation) were picked and screened for production of Roller F2 worms, which were examined for expression of GFP fusion proteins. Typical numbers were 35 worms injected; 50 Roller F1s produced; 15 Roller F1s produced Roller F2s; and 1–6 of those F2s displayed germ-line expression of GFP fusion proteins. During initial experiments, in which worms were propagated at 16–20°C, germ-line expression was generally restricted to the F2 generation. In later experiments, we observed that propagation of worms at 25°C resulted in 2 significant improvements: GFP signals were generally brighter, and germ-line GFP expression was heritable over many generations. In one experiment, F4 generation adult hermaphrodites that no longer expressed detectable GFP::histone at 16°C were shifted to 25°C. A significant proportion of the F5 worms examined (7 of 22) showed renewed and robust germ-line expression of GFP::histone. Control worms maintained at 16°C did not display any reactivation of GFP::histone expression. After observing the dramatic effect of temperature on brightness and heritability of GFP expression, we routinely maintained GFP worms at 25°C. By picking worms with a bright GFP signal for propagation at each generation, we have been able to maintain GFP-expressing lines for >100 generations. In general, GFP hermaphrodites were healthy and fertile, indicating that the levels of GFP-tagged tubulins and histone produced by arrays did not have adverse dominant effects on worms. Occasionally GFP::β-tubulin worms were shorter than usual and had stunted tails; we passaged and worked with normal-appearing siblings.
The bombardment method of Praitis et al. (2001) was used to generate a strain containing pie-1::GFP::β-tubulin (from pJH4.66) integrated into the genome. Ten bombardments were performed, producing 4 stably transformed lines, only 1 of which expressed GFP. Expression of GFP in this strain (WH204) is stable and does not require high-temperature growth of worms. This strain was used to generate Movie 4.
Imaging Live Embryos
Embryos were mounted in M9 buffer (Brenner, 1974) on 2–3.5% agarose pads and covered with a coverslip. Observation by Nomarski optics was done on a Zeiss (Thornwood, NY) Axioplan microscope. Images were captured with the use of NIH Image (version 1.62f) and a Hamamatsu C2400-00 camera and video controller with an Argus-10 image processor (Hamamatsu City, Japan). Positions of pronuclei and their migration rates were determined with the use of a tracking program for NIH Image written by A. Pilling (unpublished data). Observation of GFP fluorescence was done on a multiphoton fluorescence excitation microscope built by J. White and D. Wokosin (University of Wisconsin), with the use of a 60× oil immersion objective and 900-nm excitation (from a Ti-sapphire laser) in the direct detection mode (Wokosin et al., 1996). Optical sections of ∼0.5 μm were viewed at intervals of 2.5–4.3 s and collected as a time series with the use of Bio-Rad MRC1024 software. Stacks of images were manipulated in NIH Image (version 1.62f) and assembled into figures with the use of Adobe Photoshop. Imaging of GFP proteins in embryos did not impair development, indicating that the multiphoton illumination was not deleterious under the conditions used.
RESULTS
Identification of the C. elegans γ-Tubulin Gene
Database searches revealed the existence of only 1 recognizable γ-tubulin gene (F58A4.8) in the C. elegans genome (Consortium, 1998). The next most similar sequence in the C. elegans genome is β-tubulin (30–35% identity). Figure 2 shows an alignment of the amino acid sequence predicted from F58A4.8 with sequences of γ-tubulins from human, frog, fruit fly, and fungus. The C. elegans protein is divergent relative to the other γ-tubulins. Despite the divergence, its behavior parallels that of γ-tubulins from other species, as detailed below and also recently reported by Bobinnec et al. (2000).
γ-Tubulin Is Concentrated in Centrosomes in Early Embryos
To study the distribution of γ-tubulin in fixed embryos, we used peptides generated from 2 different regions of the predicted C. elegans protein to generate antisera (see Figure 2, boxes). After affinity purification, the antibodies stained centrosomes brightly (Figure 3). In newly fertilized embryos, γ-tubulin antibodies did not stain the maternal meiotic spindle, which is barrel shaped and lacks centrioles and asters (Albertson and Thomson, 1993). Centrosome staining was seen first as 2 small dots near the sperm pronucleus at the posterior pole (Figure 3A). This is consistent with recruitment of maternal γ-tubulin by sperm-supplied centrioles. The small dots of γ-tubulin grew into bright spheres as the male and female pronuclei migrated to meet in the posterior hemisphere of the embryo. The pronuclei then moved to the center of the embryo and rotated 90 deg to align the centrosomes on the anterior–posterior axis (Figure 3, B and C). During anaphase, the spheres appeared hollow, presenting ring-like profiles (Figure 3D). During telophase, in agreement with previous work (Strome, 1986; Keating and White, 1998; Bobinnec et al., 2000), the profile of the anterior pole was fairly round, but the posterior pole appeared flattened (i.e., elongated transverse to the anterior–posterior axis; see Figure 4A). In later-stage embryos, γ-tubulin antibodies stained tiny centrosomes in interphase cells and larger centrosomes in mitotic cells (Figure 3E). Anti-γ-tubulin immunofluorescence signal from centrosomes was reduced below detection in embryos from hermaphrodites injected with dsRNA made from the γ-tubulin gene (Figure 3F), confirming the specificity of the antibodies.
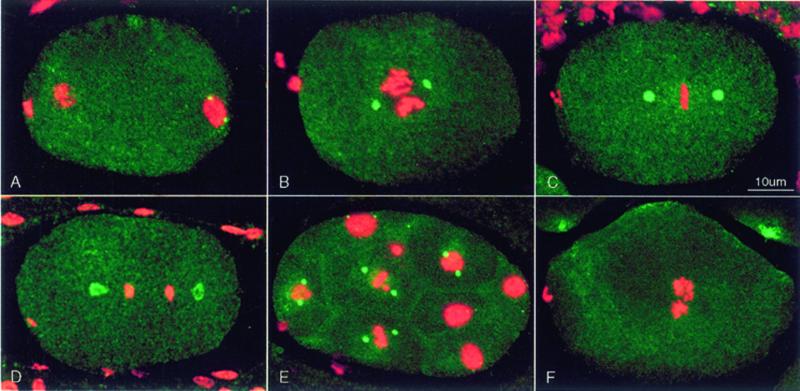
Figure 3.
Distribution of γ-tubulin seen by immunofluorescence. Fixed embryos were stained with affinity-purified rabbit anti-γ-tubulin (green) and monoclonal PA3 anti-nucleosome antibody (red), which reveals chromatin. Each image is a projection of a Z series from a scanning confocal fluorescence microscope. Embryos in this and all subsequent figures are oriented with anterior to the left. (A) Early pronuclear migration. γ-Tubulin staining is concentrated in 2 small centrosomes associated with the male pronucleus at the posterior end. Centrosomes are not seen associated with the female pronucleus or polar body at the anterior end. (B) The pronuclei have met, and a prometaphase spindle has started rotation to align on the anterior–posterior axis. Centrosomes have recruited more γ-tubulin and appear larger. (C) In metaphase, the chromosomes are tightly packed on the spindle equator, and centrosomes have enlarged. (D) In anaphase, the centrosomal γ-tubulin has taken the form of hollow spheroids. The posterior centrosome is beginning to flatten. (E) Multicellular embryo showing pairs of bright γ-tubulin–containing centrosomes in mitotic cells. Interphase cells contain tiny γ-tubulin–containing centrosomes (e.g., top surface of the uppermost nucleus). (F) Detectable anti-γ-tubulin staining of centrosomes was eliminated by RNAi using dsRNA corresponding to the C. elegans γ-tubulin sequence.
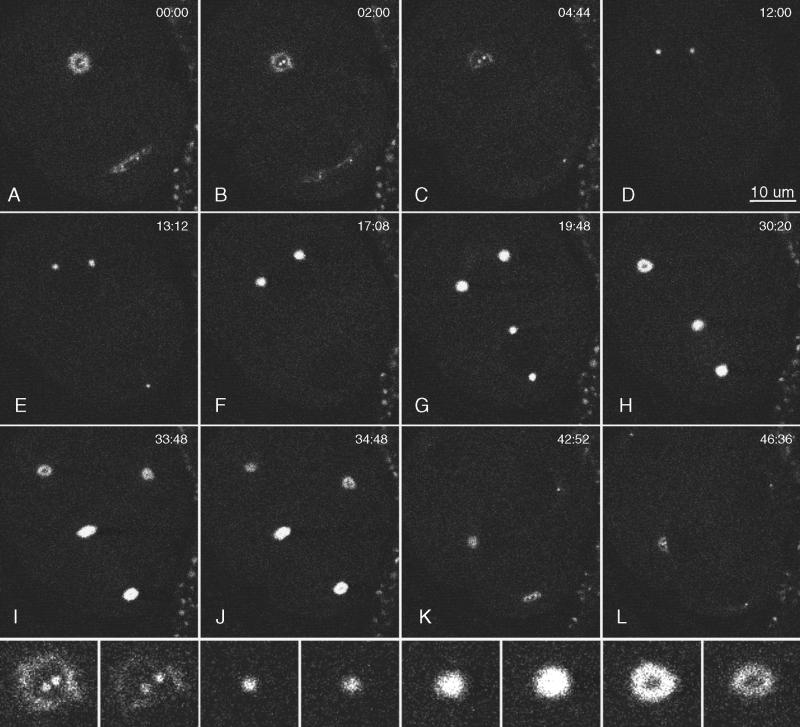
Figure 4.
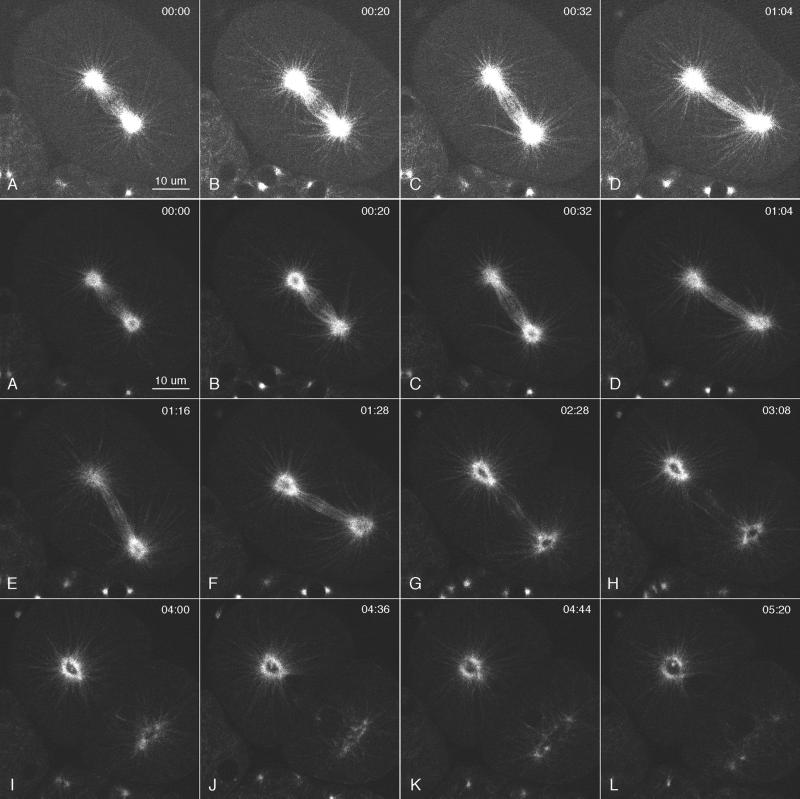
Multiphoton images of GFP::γ-tubulin in a live wild-type embryo. Anterior is top left. Images were collected in a single focal plane (∼0.5 μm thick), and the elapsed time from the first frame is shown in minutes:seconds. (A and B) One-cell embryo during telophase. The anterior centrosome was a hollow sphere (a ring in cross section), and the posterior centrosome was flattening. Within each ring, 2 small dots of GFP::γ-tubulin were seen that grew into the centrosomes of the next cell cycle. (C) The flattened posterior centrosome dispersed before the anterior centrosome. In this frame and others, some daughter centrosomes are not visible, because they moved out of the focal plane. (D–F) In the anterior daughter cell (AB), the new centrosomes enlarged and migrated to opposite sides of the nucleus. (G) In the posterior daughter cell (P1), the centrosomes enlarged, migrated to opposite sides of the nucleus, and underwent the typical 90-deg rotation to align on the anterior–posterior axis. Mitosis in the anterior AB cell preceded that in P1. (H–L) Although more difficult to see in the second round of mitosis, the GFP::γ-tubulin signal was lost from the middle of each centrosome, and new daughter centrosomes appeared within the dark centers. The posterior centrosome in P1 repeated the pattern of flattening and early disappearance observed during the division of P0 (K and L). Bottom row, enlarged views of the top left centrosome from the period covered in B–I. Note the dispersal of the “mother” centrosome, appearance of daughter centrosomes within the mother ring, increase in centrosome size during mitosis, and reappearance of a dark center during the second anaphase.
Analysis of GFP::γ-Tubulin and GFP::β-Tubulin in Living Embryos
To study the dynamic behavior of γ-tubulin and other elements of the spindle, we devised a method to express GFP-tagged versions of C. elegans γ-tubulin, β-tubulin, and histone H2B in the germ line. The pie-1 gene, a maternal-effect lethal locus, is transcribed in the germ line of adult hermaphrodites. A vector that would express GFP fusion genes was constructed with the use of the promoter, 3′ sequences, and a large intron of pie-1 (for details, see Figure 1 and MATERIALS AND METHODS). To generate transgenic worms, a mixture of the fusion construct, a dominant marker gene, and complex worm genomic DNA was microinjected into wild-type hermaphrodites to generate complex extrachromosomal arrays (Kelly et al., 1997). For each of the fusion genes, transformed lines were established that produced a GFP signal in the germ line and in early embryos. At typical culture temperatures (16–20°C), GFP fusion expression in the germ line diminished and was silenced after 1 or 2 generations. However, the expression did not diminish and was heritable for many (>100) generations if worms were maintained at an elevated temperature (25°C). We made 1 line containing GFP::β-tubulin integrated into the genome (see Movie 4); in this line, GFP expression was not dependent on culturing worms at elevated temperature.
Two issues are of particular concern in studying fluorescent proteins in live C. elegans embryos. First, because the embryos are relatively thick, even when partially flattened under a coverslip, out-of-focus fluorescence can reduce image quality. Second, because we wanted to observe the embryos for extended periods, photobleaching could also cause problems, both by dimming the GFP signal and by causing free radical–induced physiological damage. To minimize these problems, time-lapse movies of GFP-tagged proteins were made with the use of a prototype multiphoton microscope built and maintained by J. White and D. Wokosin (University of Wisconsin). The narrow plane of fluorescence excitation produced by the multiphoton illumination system substantially reduced both out-of-focus fluorescence and photobleaching. Although useful image series could be collected by conventional scanning confocal fluorescence microscopy, the signal-to-noise ratio decreased rapidly, and image quality was inferior.
Time-lapse movies of GFP::γ-tubulin in living embryos revealed elements of γ-tubulin and centrosome behavior that were not evident in fixed, immunostained embryos (Figure 4, Movie 1). During anaphase, the centrosomal spheres of γ-tubulin became hollow, and 2 dots of γ-tubulin appeared within each ring (Figure 4, A–C). After anaphase, the γ-tubulin rings disappeared, and the dots grew into the next pair of centrosomes. Thus, the dots represent the recruitment of γ-tubulin by separated and perhaps duplicated centrioles within the expanding and disassembling mother centrosome. The behavior of the posterior centrosome differed from that of the anterior centrosome during anaphase B spindle elongation. As described previously, unlike the anterior pole, which remained relatively stationary, the posterior pole swung from side to side and moved toward the posterior end of the embryo (Albertson, 1984; Figures 4, G–J, and 5, C–H). After the posterior migration had ceased, the γ-tubulin ring at the posterior pole flattened laterally, decreased in brightness, and appeared to fragment, as if subject to forces directed toward the lateral cortex (Figure 4, A, B, and K; see Movie 1). In contrast, the γ-tubulin ring at the anterior pole persisted longer and showed little flattening or fragmentation as it decreased in brightness (Figure 4C). In subsequent rounds of mitosis, this pattern of centrosomal γ-tubulin dynamics was repeated on a progressively smaller scale.
The dynamic behavior of centrosomes seen with GFP::β-tubulin was similar to that seen with GFP::γ-tubulin (Figure 5, Movie 2). Centrosomes recruited GFP::β-tubulin and enlarged from prophase through metaphase (see Movie 3 and Figure 8, A and B). They showed ring-like profiles with darkened centers, and during anaphase–telophase, the posterior ring underwent swinging, posterior migration, flattening, and fragmentation (Figure 5, C–J, Movies 2 and 3). Two small dots of GFP::β-tubulin, presumably the daughter centrosomes, could be seen within the expanding rings during anaphase–telophase (Figure 5, I–L). An interesting contrast in the behaviors of centrosomal γ- and β-tubulins was seen in the timing of the appearance of darkened centers. With GFP::β-tubulin, ring profiles could be seen in prometaphase and metaphase (Figure 5A). With anti-γ-tubulin and GFP::γ-tubulin, they were not seen until anaphase (Figures 3D and 4H, respectively). This suggests that despite the presence of γ-tubulin throughout the centrosome during spindle assembly, most recruitment and polymerization of α/β-tubulin dimers occurs in the outer layers.
Figure 5.
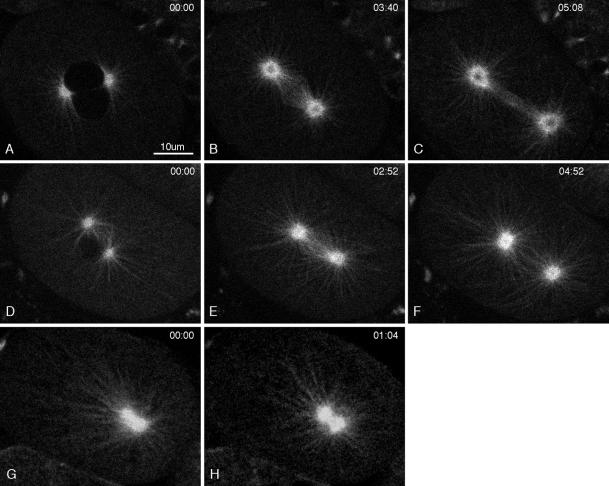
Multiphoton image series of GFP::β-tubulin in a live wild-type embryo. Elapsed time from the first frame is shown in minutes:seconds. Brightness and contrast were adjusted to better reveal single microtubules in the top row and centrosome dynamics in the bottom 3 rows. Microtubule dynamics can be seen best in Movies 2 and 3. (A–C) Metaphase and early anaphase. (D–L) Anaphase B and telophase. The posterior spindle pole underwent dramatic side-to-side swinging during spindle elongation (C–G). Many astral microtubules extended to near the cortex. They usually appeared “swept back” on the side from which the spindle pole was moving (D). Some microtubules bent on the side to which the spindle pole was moving (see Movies 2 and 3). Note the bright area at the equator (D–F), which likely represents the region of interdigitation of interpolar microtubules. (G–L) Telophase and cytokinesis. The dark centers of the centrosomes enlarged, and the posterior mother centrosome flattened and fragmented. Two daughter centrosomes appeared within each dark center.
Figure 8.
Multiphoton movies of GFP:: β-tubulin in a wild-type embryo (A–C), in a moderate γ-tubulin(RNAi) embryo (D–F), and in a severe γ-tubulin(RNAi) embryo (G–H). Elapsed time from the first frame is shown in minutes:seconds. (A and D) Prophase. The stage in D was further advanced than in A, and the 2 pronuclei were stacked one above the other. (B, E, and G) Metaphase, although the precise stage in the RNAi embryos (E and G) is difficult to determine. Both E and G were immediately before the spindle gyrations seen in RNAi embryos that we interpret as anaphase. (C and F) Anaphase B spindle elongation. Notice that an ordered bundle of interpolar microtubules is not seen in the moderate RNAi embryo (F). (H) In the severe embryo, the poles of the collapsed spindle did not separate. Comparison of G and H shows that the microtubule pattern changed with time, suggesting that even in severe γ-tubulin(RNAi) embryos, centrosomal microtubules are dynamic (also see Movie 4).
The behavior of GFP astral microtubules suggests a mechanism for generating the forces that drive the swinging, posterior migration and flattening of the posterior centrosome during anaphase B and telophase. Best seen in multiphoton movies, GFP microtubules between the posterior centrosome and the lateral cortex were abundant during migration and swinging of the posterior pole (Movies 2 and 3). During each lateral swing, astral microtubules trailing the pole were uniformly arranged in a gentle arc, appearing to be pulled by pole movement. Those leading the pole were often bowed in various directions. Thus, force for the lateral motion did not appear to be generated by microtubule polymerization on the lagging side or by synchronous microtubule depolymerization on the leading side. We speculate that the force for swinging and for centrosome flattening stem from microtubule capture by a small number of lateral–posterior cortical sites containing minus-end–directed microtubule motors. Those tethered motors would generate force by binding to and pulling on a few centrosomal microtubules.
γ-Tubulin Is Required for Normal Spindle Assembly and Chromosome Segregation
To study the effects of inhibiting γ-tubulin function in early embryos, we used RNAi (Fire et al., 1998). RNAi has been shown to induce loss-of-function phenotypes for many genes (Rocheleau et al., 1997) and is particularly effective at depleting maternal proteins in early embryos. Hermaphrodites injected with dsRNA corresponding to the γ-tubulin gene gave rise to 100% dead embryos starting at ∼18 h after injection. Examination of such RNAi embryos by means of GFP::γ-tubulin (our unpublished data) or by anti-γ-tubulin antibody staining (Figure 3F) revealed that γ-tubulin was reduced below detectability. Furthermore, we did not observe a more severe RNAi phenotype when γ-tubulin was depleted from hermaphrodites with only 1 copy of the γ-tubulin gene (see MATERIALS AND METHODS). Thus, it is likely that the most severe RNAi embryos displayed defects caused by a nearly complete loss of γ-tubulin function.
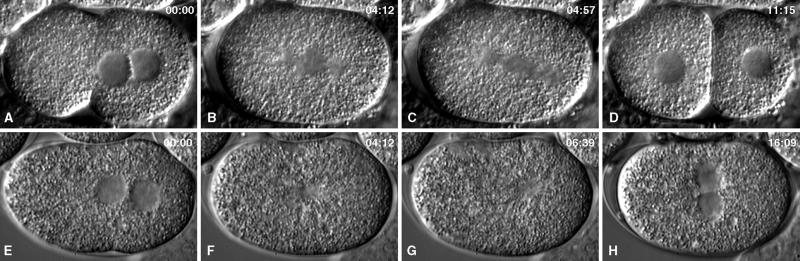
In γ-tubulin(RNAi) embryos observed by Nomarski differential interference contrast (Figure 6), the earliest phenotype seen was in female pronuclear migration. In wild-type embryos, the female pronucleus moves slowly from the anterior pole to the center of the embryo and then moves fast and directly to the male pronucleus. Our tracking measurements yielded velocities of 0.06 μm/s for the slow phase and 0.28 μm/s for the fast phase (n = 4 embryos at 22–23°C). Severe depletion of γ-tubulin did not detectably alter the slow phase of migration. However, the fast phase was absent or greatly reduced (0.08 μm/s; n = 5 embryos at 22–23°C).
Figure 6.
Comparison of a wild-type embryo (A–D) and a severely affected γ-tubulin(RNAi) embryo (E–H) during the first cell cycle. In these Nomarski images, nuclei (A, D, E, and H) and spindles (B, C, F, and G) are seen as granule-free zones. The top row shows a wild-type embryo at pronuclear meeting (A), prometaphase–metaphase (B), anaphase (B), and after cytokinesis (D). The bottom row shows corresponding stages in a γ-tubulin(RNAi) embryo. RNAi prevented the formation of a bipolar spindle, prevented normal chromosome segregation, and prevented cytokinesis.
After the pronuclei met (Figure 6E) and nuclear envelopes broke down, subsequent spindle assembly and function were aberrant. Bipolar spindles were not observed. Instead, a clear zone formed in the center of the embryo (Figure 6F). At a time when one would expect anaphase, this apparent monoaster and associated cytoplasm went through a brief period of gyrations, primarily back and forth along the anterior–posterior axis (Figure 6G). After the gyrations stopped, multiple nuclei of various sizes formed in the center of the embryo, and cytokinesis did not occur (Figure 6H). It was apparent that depletion of γ-tubulin inhibited spindle assembly and, perhaps as a consequence, prevented cytokinesis.
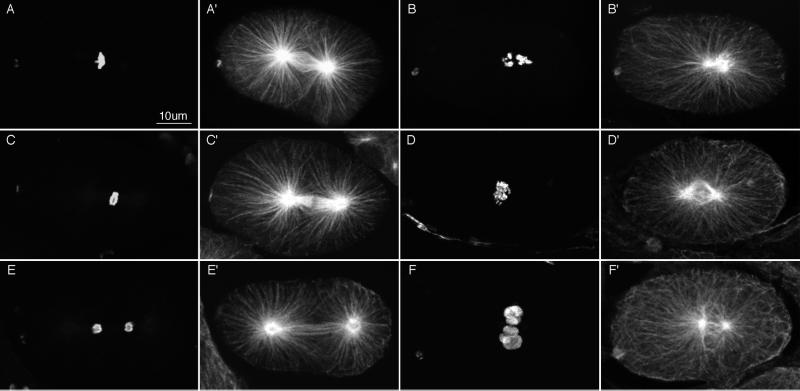
The Nomarski movies suggested that γ-tubulin depletion blocked some, but not all, microtubule-dependent processes. To investigate the effects of γ-tubulin depletion on microtubules and chromosomes, we used immunofluorescence microscopy. RNAi embryos were fixed and stained with anti-tubulin and anti-histone antibodies (Figure 7). All mitotic embryos contained stunted spindles. Some spindles appeared completely collapsed with condensed chromosomes collected near a central diaster (Figure 7, B and B′). Other spindles were bipolar, but the poles were abnormally close to one another (Figure 7, D′ and F′); kinetochore and interpolar microtubules were either absent or poorly organized. In these cases, condensed chromosomes were often collected in the foreshortened interpolar region (Figure 7D) but were never seen organized into a tight metaphase plate. Decondensed chromosomes were also seen between the poles (Figure 7F), probably representing failed chromosome segregation and aberrant telophase. Given the expectation that γ-tubulin would be required for nucleating microtubules from centrosomes, it was striking that in all mitotic embryos observed, astral arrays of microtubules were present. Most studies to date suggest that astral microtubules are dynamic and less stable than the kinetochore microtubules, which attach to kinetochores and drive chromosome movements, or the interpolar microtubules, which interact with microtubules from the opposite pole and help push spindle poles apart (Mitchison and Kirschner, 1985; Kirschner and Mitchison, 1986; Mitchison et al., 1986; Saxton and McIntosh, 1987; Mastronarde et al., 1993). Our studies reveal that kinetochore and interpolar microtubules are more sensitive to a loss of γ-tubulin than are astral microtubules.
Figure 7.
Anti-tubulin and anti-histone immunofluorescence images of wild-type embryos (left 2 columns) and γ-tubulin(RNAi) embryos (right 2 columns). Fixed embryos were stained with rabbit anti-acetylated histone H4 antibodies to reveal chromatin (A–F) and mouse anti-α-tubulin antibodies to reveal microtubules (A′–F′). Each image is a projection of 5–7 sequential optical sections collected with a scanning confocal fluorescence microscope. Wild-type embryos are shown in prometaphase (A and A′), early anaphase (C and C′), and telophase (E and E′). (B and B′) Severely affected γ-tubulin(RNAi) embryo. Numerous long astral microtubules emanate from a collapsed spindle; chromosomes were not organized. (D and D′) Less severely affected RNAi embryo with a partially collapsed and poorly organized central spindle. Chromosomes appeared to be in early anaphase and were not well organized. (F and F′) RNAi embryo at telophase. The spindle had not segregated the chromosomes and showed little sign of interpolar microtubule bundles or spindle elongation. Chromatin was decondensing in micronuclei at the center of the embryo.
Effects of γ-Tubulin Depletion on Microtubules and Chromosomes in Living Embryos
To eliminate the uncertain temporal sequence inherent in studying microtubule and chromatin phenotypes in fixed embryos, we monitored events in living embryos containing GFP fusion proteins. To analyze the effects of γ-tubulin depletion on microtubule organization and dynamics, we compared wild-type and γ-tubulin(RNAi) embryos expressing GFP::β-tubulin (Figure 8, Movies 2–5). In all γ-tubulin(RNAi) embryos examined, it was clear that γ-tubulin depletion did not prevent the nucleation of astral microtubules (Figure 8, D–H). As in wild-type embryos, astral arrays appeared in prophase and persisted through anaphase. Furthermore, the changing patterns of astral microtubule distribution suggested that they were dynamic. Two interesting features of RNAi embryos were the absence of a distinct darkened zone of depleted GFP::β-tubulin in the center of the centrosome and the failure of the posterior centrosome to flatten and fragment; instead, RNAi embryos displayed a more uniform distribution of GFP::β-tubulin throughout the centrosome, and the posterior centrosome remained spherical (Figure 8, D–H).
In the most severe RNAi embryos, the mitotic spindle consisted of a collapsed diastral array of microtubules that persisted through telophase (Figure 8G, Movie 4). There was little or no evidence of kinetochore or interpolar microtubules. This diaster often failed to rotate onto the anterior–posterior axis and usually underwent several dramatic excursions toward the posterior pole of the embryo (Movie 4). In less severely affected embryos (termed “moderate RNAi embryos”), spindles were bipolar but stunted, with disorganized microtubules between the poles (Figure 8, D–F, Movie 5), suggesting again poor development of kinetochore and interpolar microtubules. During anaphase, moderate RNAi embryos failed to form a dense bundle of parallel microtubules between the spindle poles (compare Figure 8F with wild-type embryos in Figures 5, B–F, and 8C), and the spindle poles failed to separate. The poles were sometimes pushed apart during telophase by the formation of multiple micronuclei (Movie 5). Kinetochore and interpolar microtubules arise through the capture and stabilization of astral microtubule plus ends by either kinetochores or antiparallel plus ends emanating from the opposing pole (Mitchison and Kirschner, 1985; Kirschner and Mitchison, 1986; Mastronarde et al., 1993). Our results suggest that γ-tubulin somehow influences this capture of astral microtubule plus ends or subsequent stabilization of the captured microtubules.
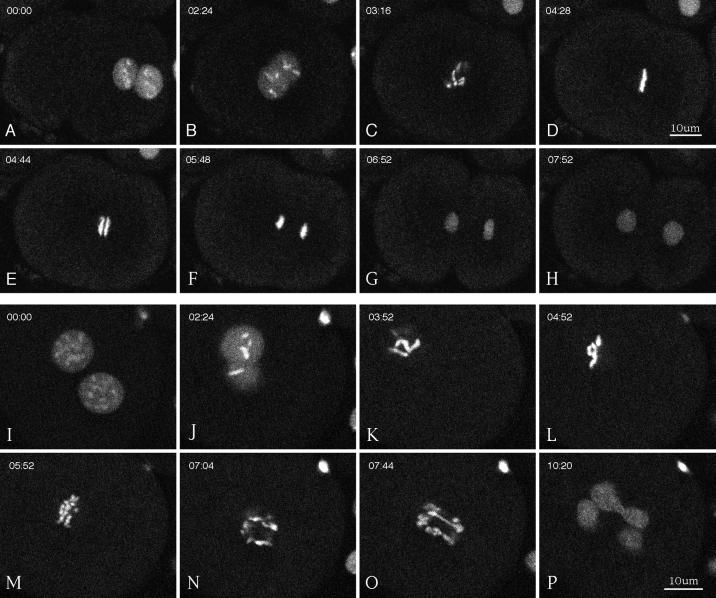
To investigate the ability of the defective spindles described above to organize and separate chromosomes during mitosis, we analyzed wild-type and γ-tubulin(RNAi) embryos expressing GFP::histone (Figure 9, Movies 6–8). In newly fertilized wild-type embryos, GFP::histone appeared on oocyte chromosomes as they completed meiosis. Fluorescence appeared on sperm chromatin during formation of the sperm pronucleus, presumably reflecting the replacement of sperm-specific chromatin proteins by maternal histones. Thereafter, during mitosis GFP::histone was clearly associated with condensed chromosomes, and during interphase it was distributed diffusely throughout the nucleus (Figure 9, A–H, Movie 6). In severe γ-tubulin(RNAi) embryos, chromosomes condensed during pronuclear migration, as in wild-type embryos. After nuclear envelope breakdown, they clustered together loosely in the center of the embryo. At the time when anaphase should have occurred, chromosomes did not segregate; instead they decondensed and formed multiple micronuclei (see Movie 7). In moderate RNAi embryos, chromosomes often congressed together but did not organize into a tight metaphase plate (Figure 9, K–L). Anaphase was defective, as evidenced by chromosome bridges, poor segregation, and the consequent formation of multiple micronuclei (Figure 9, M–P, Movie 8).
Figure 9.
Multiphoton movies of GFP::histone in a wild-type embryo (A–H) and in a moderate γ-tubulin(RNAi) embryo (I–P). Elapsed time from the first frame is shown in minutes:seconds. (A, B, I, and J) Meeting, centering, and rotation of the pronuclei and prophase chromosome condensation. (C and K) Breakdown of the nuclear envelopes. (D and L) Metaphase. Tight alignment of chromosomes is shown on the metaphase plate in a wild-type embryo (D) versus loose alignment in the γ-tubulin(RNAi) embryo (L). (E and M) Early anaphase. (F and N) Late anaphase. (G, H, O, and P) Telophase. Note the poor segregation of chromosomes in the γ-tubulin(RNAi) embryo (N–P). Cytokinesis occurred in the wild-type embryo (H) but not in the RNAi embryo (P).
DISCUSSION
γ-Tubulin, Microtubule Nucleation, and Microtubule Function
Since its discovery (Oakley and Oakley, 1989), γ-tubulin has emerged as a conserved and essential component of many forms of microtubule-organizing centers, including centrosomes. There is strong support for the view that γ-tubulin, in the form of γ-TuRCs, stimulates microtubule growth from microtubule-organizing centers by providing a stable seed for α/β-tubulin polymerization (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000). However, our studies of C. elegans embryos depleted of γ-tubulin suggest that γ-tubulin and γ-TuRCs are not essential for microtubule growth from centrosomes. Although our RNAi approach may not have generated a true null phenotype, our findings are consistent with results from flies and yeast that demonstrate that microtubule functions are not completely eliminated in γ-tubulin null mutants (Horio et al., 1991; Sobel and Snyder, 1995; Wilson and Borisy, 1998).
The results from γ-tubulin mutant analysis, as well as recent demonstrations that functional mitotic spindles can form in the absence of centrosomes (de Saint Phalle and Sullivan, 1998; Khodjakov et al., 2000; Megraw et al., 2001), point to the existence of backup systems for organizing spindle microtubules. First, there appears to be a γ-tubulin–independent pathway for microtubule nucleation from centrosomes. Centrosome components other than γ-tubulin must be able to recruit sufficiently high concentrations of α/β-tubulin to create stable seeds that can support extensive polymerization. Second, there is a centrosome-independent pathway for organization of bipolar spindles. As seen with meiotic spindles in several systems and with spindles assembled around DNA-coated beads, this pathway relies on the ability of chromatin to nucleate microtubules and on the ability of microtubule motors to organize those microtubules into a bipolar spindle (Heald et al., 1996; Matthies et al., 1996).
It is interesting that microtubules emanating from γ-tubulin–depleted centrosomes in C. elegans embryos were sufficient for some processes but not for others. For example, slow migration of the female pronucleus occurred, but fast pronuclear migration was inhibited. Both phases of pronuclear migration depend on microtubules (Strome and Wood, 1983). However, slow migration may depend on the meshwork of interphase microtubules, whereas directed fast migration may use microtubules emanating from the centrosomes of the male pronucleus. When sperm–aster microtubule plus ends contact minus-end–directed microtubule motors on the female pronucleus, the motors could then pull the pronuclei directly together. The inhibition of fast migration suggests that γ-tubulin is important for the generation or function of sperm–aster microtubules that can support such force production.
Depletion of γ-tubulin also appears to affect the functions of a subset of mitotic spindle microtubules. Duplicated centrosomes in severe γ-tubulin(RNAi) embryos could generate astral arrays of microtubules, but the asters were not properly separated, resulting in collapsed spindles. This suggests that γ-tubulin is needed for stable interpolar connections, which are normally established by lateral interactions of microtubule plus ends from opposite poles (Mastronarde et al., 1993). Spindles with separated poles often did form in less severely affected γ-tubulin(RNAi) embryos. In these cases, chromosome congression to the metaphase plate and anaphase chromosome segregation failed, suggesting that the accumulation of stable kinetochore microtubules requires γ-tubulin. Combining our results with the previous observations that mitotic spindles without functional centrosomes can mediate normal chromosome segregation (de Saint Phalle and Sullivan, 1998; Khodjakov et al., 2000; Megraw et al., 2001), it appears that the stabilizing influence of γ-tubulin does not require that it reside in a centrosome. At least, it seems evident that γ-tubulin function goes beyond the simple nucleation of microtubules.
There are various ways to explain the effects of γ-tubulin depletion on microtubule-dependent processes. We will focus on 3.
Altered Templating.
Centrosomal γ-TuRCs may serve as structural templates for the growth of microtubules, with the number of protofilaments of α/β-tubulin defined by the number of γ-tubulins per ring (Li and Joshi, 1995). As discussed above, centrosomes depleted of γ-tubulin appear to be able to nucleate microtubules with the use of alternative mechanisms. However, those alternatives may produce microtubules with an aberrant number of protofilaments and, hence, altered tubulin lattice characteristics. In this case, the differential sensitivity of the 3 classes of spindle microtubules to γ-tubulin depletion would imply that although structural abnormalities do not interfere with simple growth of astral microtubules, they do interfere with the higher-order interactions required to convert astral microtubules to kinetochore and interpolar microtubules. In support of this possibility, it has been shown elegantly with Drosophila axonemes that higher-order microtubule interactions are quite sensitive to subtle perturbations of tubulin sequence and, presumably, tubulin lattice characteristics (Raff et al., 1997). Future high-resolution ultrastructural studies may resolve the question of whether γ-tubulin depletion affects protofilament number.
Altered Plus Ends.
Most evidence points to an exclusive association of γ-TuRCs with minus ends (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000). However, it is possible that γ-tubulin also interacts with plus ends. Cell fractionation suggests that the majority of γ-tubulin is not associated with centrosomes (Moudjou et al., 1996). Furthermore, γ-tubulin can interact with β-tubulin (Leguy et al., 2000), the subunit exposed at the plus ends of microtubules (Nogales et al., 1999). If γ-tubulin can bind plus ends, then γ-tubulin depletion could affect plus-end dynamics or other properties that are necessary for proper interactions with kinetochores and interpolar microtubules. Indeed, Paluh et al. (2000) proposed that the defects observed in γ-tubulin–mutant yeast cells might be due to hyperstabilization of microtubules. Our movies of GFP::β-tubulin in embryos do not reveal the actual growth or shortening of single microtubule ends. We expect that future improvements in live GFP imaging technologies will allow the spatial and temporal resolution required to measure such microtubule plus-end dynamics in C. elegans.
Altered Minus Ends.
Centrosomal γ-TuRCs in the pericentriolar matrix provide for controlled nucleation of microtubules. They may also be important for stabilization of microtubules after they are nucleated. In keeping with this, it has been shown that γ-TuRCs block minus-end polymerization–depolymerization dynamics in vitro (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000). The alternate nucleation mechanisms, which we hypothesize are active in γ-tubulin–depleted embryos, may not provide proper minus-end stabilization. Because such “free” minus ends are likely to lose α/β subunits (Rodionov and Borisy, 1997), nucleation and polymerization of microtubules from centrosomes lacking γ-TuRCs would be followed by slow minus-end depolymerization. This condition could still allow the growth of astral microtubules with fast-growing plus ends. However, capture and stabilization of those plus ends by kinetochores or by plus ends from the opposite pole would be fruitless. Minus-end depolymerization would disconnect them from the spindle pole and render them too short lived to produce the forces required for proper spindle assembly and function.
Centrosome Dynamics
Keating and White (1998) have described the dynamics of C. elegans centrosomes in 1-cell embryos seen with rhodamine–α/β-tubulin. Our observations of the dynamic behavior of GFP::γ- and GFP::β-tubulin confirm their results and provide additional insights. In particular, we documented that first β-tubulin and then γ-tubulin appear to shift from even distributions throughout the centrosome to expanding hollow spheres. β-Tubulin shows the hollow distribution as early as prometaphase, whereas γ-tubulin shows hollowing later, during anaphase. With both GFP fusion proteins, daughter centrosomes are seen within the mother spheres during anaphase. Depletion of γ-tubulin reduces or eliminates the centrosome hollowing and expansion seen with GFP::β-tubulin. This suggests that the alternative α/β recruitment–nucleation mechanism is distributed evenly throughout the centrosome and cannot support the forces that cause outward centrosome expansion during anaphase.
It is interesting that the anterior and posterior mother centrosomes behave differently, especially in light of the importance of first-division asymmetries in establishing developmental lineages. Most of anaphase B spindle elongation is accomplished by movements of the posterior centrosome (Albertson, 1984; Kemphues et al., 1988). It swings from side to side as it moves toward the posterior end of the embryo. After swinging, the posterior ring of γ- and β-tubulin flattens and appears to be pulled apart laterally. The anterior sphere does not flatten. Perhaps the forces that generate the posterior pole's swinging motions, its flattening, and its lateral dispersion are all generated by minus-end–directed microtubule motors concentrated at a few sites in the posterior–lateral cortex. When an astral microtubule plus end from the posterior centrosome is first captured by one of those sites, the spindle pole is drawn toward it, in a lateral and slightly posterior direction. Subsequent microtubule contacts with sites on the opposite side swing the centrosome back and further toward the posterior. Eventually, multiple sites are engaged on both sides; the centrosome is held in balance between them near the posterior end; and continued force production stretches the centrosome apart laterally. In support of the existence of sites in the posterior cortex that “pull” astral microtubules toward them, Grill et al. (2001) recently observed that, after removal of the spindle midzone, the posterior spindle pole moved rapidly and with transverse oscillations to the posterior cortex.
Expression of GFP Fusion Proteins in the Germ Line and Early Embryos
Achieving transgene expression in the C. elegans germ line has been problematic. Transgenic animals are typically generated by injecting DNA into the syncytial germ line of adult hermaphrodites. Injected DNA molecules recombine to form relatively large extrachromosomal arrays, which are segregated with variable efficiency during mitosis and meiosis. In general, transgenes present in such arrays are expressed efficiently in the somatic cells of offspring but are silenced in the germ line (Kelly et al., 1997). Germ-line expression of some transgenes has been achieved by increasing the DNA complexity of extrachromosomal arrays (Kelly et al., 1997). However, not all transgenes tested could be desilenced (W. Kelly, personal communication), and germ-line silencing of transgenes has remained a formidable barrier to expressing tagged versions of proteins in germ cells and early embryos. We report here a successful approach for overcoming germ- line silencing. This was accomplished by combining the high DNA complexity approach with a vector that contains regulatory sequences from a gene that is normally expressed in the germ line (pie-1). Germ-line expression of multiple genes was robust. They include fusion genes encoding GFP-tagged versions of γ-tubulin, β-tubulin, and histone H2B (reported here) as well as actin, PGL-1, and IFE-1 (M. Dunn, K. Reese, and G. Seydoux, unpublished data; A. Amiri, B. Keiper, I. Kawasaki, Y. Fan, Y. Kohara, R. Rhodes, S. Strome, unpublished data). All of these GFP constructs are available on request.
Several observations on expression of GFP fusion proteins from the extrachromosomal arrays we generated are worth noting. First, the pie-1 promoter successfully drove expression of transgenes in the germ line but often also drove ectopic expression in a subset of somatic cells. Ectopic expression may be due to position effects in the complex arrays. Second, when worms were maintained at 16–20°C, germ-line expression of the transgenes was generally restricted to the F2 generation after transformation. Maintaining worms at 25°C allowed robust expression in the germ line for many generations (>100). Third, even in worms maintained at 25°C, there was some stochastic loss of transgene expression in the germ line. This loss was most noticeable in early generations. After many generations of selecting GFP-positive worms for propagation, the lines became more stable. Fourth, none of the GFP lines we generated by microinjection has integrated the transgenic array into a chromosomal site. However, integration of the GFP::histone-expressing construct has been achieved by γ irradiation of array-bearing worms (Kaitna et al., 2000). Also, Praitis et al. (2001) recently reported integration of our GFP::histone construct by bombarding worms with DNA-coated gold beads. We used their method to generate a strain with integrated GFP::β-tubulin. Hermaphrodites bearing these integrated GFP constructs express GFP fusion protein in the germ line stably and do not require high-temperature culture for robust expression. The availability of these lines should facilitate the type of live-embryo analysis of protein dynamics we have described here.
Supplementary Material
ACKNOWLEDGMENTS
We thank Judith Kimble for encouragement and sharing facilities with S.S. and W.S. during a wonderful sabbatical at the University of Wisconsin; Jayne Squirrell, Bill Mohler, Kevin Eliceiri, and David Wokosin for advice and help with multiphoton imaging; Laurel Bender, Diane Schmidt, and Lisa Misner for help generating 2 GFP strains; Yuji Kohara for cDNA clones; David Allis, Margaret Fuller, and Marc Monestier for antibodies; and members of the White, Kimble, Seydoux, Strome, and Saxton laboratories for discussions and ideas. This research was supported by National Institutes of Health grants GM58811 (to W.S. and S.S.), GM34059 (to S.S.), and GM52454 (to J.W.), a Guggenheim fellowship (to S.S.), grants from the Packard and Searle Foundations (to G.S.), and an established investigatorship from the American Heart Association (with funds contributed in part by the American Heart Association, Indiana Affiliate; to W.S.).
Abbreviations used:
- dsRNA
double-stranded RNA
- γ-TuRC
γ-tubulin ring complex
- GFP
green fluorescent protein
- RNAi
RNA-mediated interference
Footnotes
REFERENCES
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode Caenorhabditis elegans. Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Fukuda M, Nishida E. Identification and characterization of Caenorhabditis elegansgamma-tubulin in dividing cells and differentiated tissues. J Cell Sci. 2000;113:3747–3759. doi: 10.1242/jcs.113.21.3747. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T.C.e.S. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- de Saint Phalle B, Sullivan W. Spindle assembly and mitosis without centrosomes in parthenogenetic Sciaraembryos. J Cell Biol. 1998;141:1383–1391. doi: 10.1083/jcb.141.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of gamma-tubulin recruitment in Xenopussperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Furuta T, Tuck S, Kirchner J, Koch B, Auty R, Kitagawa R, Rose AM, Greenstein D. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol Biol Cell. 2000;11:1401–1419. doi: 10.1091/mbc.11.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Steizer EHK, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegansembryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Gunawardane RN, Lizarraga SB, Wiese C, Wilde A, Zheng Y. Gamma-tubulin complexes and their role in microtubule nucleation. In: Palazzo RE, Schatten GP, editors. Current Topics in Developmental Biology: The Centrosome in Cell Replication and Early Development. San Diego: Academic Press; 2000. pp. 55–73. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegansembryos, encodes a putative ser/thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopusegg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. Incenp and an Aurora-like kinase form a complex that is essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Keating HH, White JG. Centrosome dynamics in early embryos of Caenorhabditis elegans. J Cell Sci. 1998;111:3027–3033. doi: 10.1242/jcs.111.20.3027. [DOI] [PubMed] [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegansgene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegansembryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Leguy R, Melki R, Pantaloni D, Carlier MF. Monomeric gamma-tubulin nucleates microtubules. J Biol Chem. 2000;275:21975–21980. doi: 10.1074/jbc.M000688200. [DOI] [PubMed] [Google Scholar]
- Li Q, Joshi HC. Gamma-tubulin is a minus end-specific microtubule binding protein. J Cell Biol. 1995;131:207–214. doi: 10.1083/jcb.131.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Leone JW, Cook RG, Allis CD. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies HJG, McDonald HB, Goldstein LSB, Theurkauf WE. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–27. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monestier M, Novick KE, Losman MJ. d-Penicillamine- and quinidine-induced antinuclear antibodies in A.SW (H-2s) mice: similarities with autoantibodies in spontaneous and heavy metal-induced autoimmunity. Eur J Immunol. 1994;24:723–730. doi: 10.1002/eji.1830240335. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA. Three-dimensional structural characterization of centrosomes from early Drosophilaembryos. J Cell Biol. 1995a;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995b;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. Gamma-tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR. Gamma-tubulin. In: Palazzo RE, Schatten GP, editors. Current Topics in Developmental Biology. The Centrosome in Cell Replication and Early Development. San Diego: Academic Press; 2000. pp. 27–54. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux AL, Cande WZ. A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol Biol Cell. 2000;11:1225–1239. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in C. elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff EC, Fackenthal JD, Hutchens JA, Hoyle HD, Turner FR. Microtubule architecture specified by a beta-tubulin isoform. Science. 1997;275:70–73. doi: 10.1126/science.275.5296.70. [DOI] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegansis mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegansembryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rodionov VI, Borisy GG. Microtubule treadmilling in vivo. Science. 1997;275:215–218. doi: 10.1126/science.275.5297.215. [DOI] [PubMed] [Google Scholar]
- Saxton WM, McIntosh JR. Interzone microtubule behavior in late anaphase and telophase spindles. J Cell Biol. 1987;105:875–886. doi: 10.1083/jcb.105.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg BJ, Khodjakov A, Rieder CL, Palazzo RE. The disassembly and reassembly of functional centrosomes in vitro. Proc Natl Acad Sci USA. 1998;95:9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. Asymmetric movements of cytoplasmic components in Caenorhabditis elegansembryos. J Emb Exp Morphol. 1986;97:15–29. [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegansembryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Gomes R, Sampaio P, Perdigao J, Gonzalez C. Gamma-tubulin is required for the structure and function of the microtubule organizing center in Drosophilaneuroblasts. EMBO J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Wilson EB. The Cell in Development and Heredity. 3rd ed. New York: MacMillan; 1925. [Google Scholar]
- Wilson PG, Borisy GG. Maternally expressed gamma Tub37CD in Drosophilais differentially required for female meiosis and embryonic mitosis. Dev Biol. 1998;199:273–290. doi: 10.1006/dbio.1998.8900. [DOI] [PubMed] [Google Scholar]
- Wokosin DL, Centonze VE, White JG, Hird SN, Sepsenwol S, Malcolm GPA, Maker GT, Ferguson AI. Multi-photon excitation imaging with an all-solid-state laser. Proc Soc Photo-Optical Instrum Eng. 1996;2678:38–49. [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.