Abstract
Understanding homeobox gene specificity and function has been hampered by the lack of proven direct transcriptional targets during development. Dlx genes are expressed in the developing forebrain, retina, craniofacial structures and limbs. Dlx1/Dlx2 double knockout mice die at birth with multiple defects including abnormal forebrain development and decreased Dlx5 and Dlx6 expression. We have successfully applied chromatin immunoprecipitation (ChIP) to identify a direct transcriptional target of DLX homeoproteins from embryonic tissues in vivo. We optimized cross-linking conditions to enrich for protein–DNA complexes, then using specific high affinity DLX antibodies captured immunoenriched DLX genomic DNA transcriptional targets. DLX homeobox proteins bind differentially to the Dlx5/Dlx6 intergenic enhancer in newborn retina (DLX2) and embryonic striatum (DLX1, DLX2) in situ. Reporter gene assays demonstrated the functional significance of the binding of DLX proteins to this regulatory element, confirmed in vitro by electrophoretic mobility shift assays, using tissue extracts or recombinant DLX proteins. ChIP provides the best approach to identify direct Dlx homeoprotein targets from developing tissues in situ. The use of this technology will advance our understanding of Dlx gene function in the vertebrate in vivo and can be applied to examine targets of other homeobox genes and other classes of transcription factors.
INTRODUCTION
Homeobox genes are transcription factors of fundamental importance during the development of evolutionarily diverse species. They have been implicated in many congenital malformations and developmental cancers (1,2). Insights into homeobox gene function in vivo have been gained through the mutagenesis, inactivation or ectopic expression of these genes in different animal models. However, there is little known about the identity of direct homeobox gene targets. It is critical to identify these ‘downstream’ genes to understand the genetic program used by homeobox genes to determine regional identity, patterning and cell-fate specification during vertebrate development.
Of six known mouse Dlx genes, four (Dlx1, 2, 5 and 6) are expressed in the developing forebrain and retina (3–6). The Dlx genes are arranged in bigenic clusters where each pair is linked to a different Hox cluster (Fig. 3a) (7). Mice lacking both Dlx1 and Dlx2 exhibit a time-dependent block in striatal neuronogenesis (8). Neurons born after embryonic day 12.5 (E12.5) do not fully differentiate and are blocked in their migration to the neocortex, olfactory bulb and hippocampus. In the Dlx1/2 mutants, a lateral tangential migration that appears to be the source for most neocortical GABAergic interneurons present in neonatal mice, is blocked (9,10). As well, we have demonstrated abnormal retinal development in these doubly null mice (de Melo,J., Du,G., Fonseca,M., Turk,W.J., Rubenstein,J.L.R. and Eisenstat,D.D., submitted).
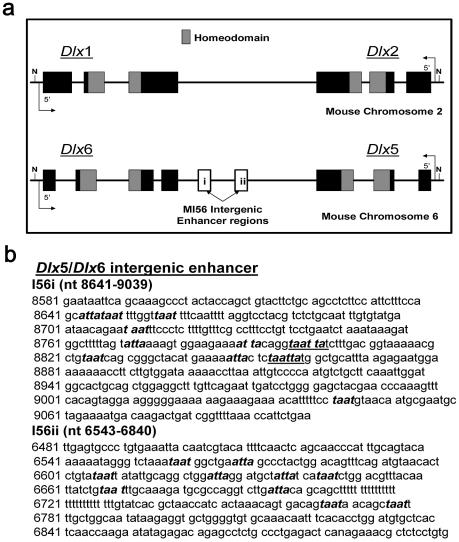
Figure 3.
(a) Genomic organization of Dlx1/Dlx2 and Dlx5/Dlx6 bigenic clusters in the mouse. Hox genes are sequentially oriented in a 3′–5′ direction from anterior to posterior, whereas the Dlx gene family is organized in three bigenic clusters with the genes facing one another as 5′–3′ 3′–5′ pairs. The Dlx5/Dlx6 intergenic region MI56 contains two enhancer regions, I56i and I56ii (13). The 60 amino acid homeodomain (shaded boxes) is highly conserved across Dlx family members. The Dlx3/Dlx7 cluster is not included (33). (b) Sequence and organization of candidate homeodomain DNA binding motifs in the Dlx5/Dlx6 intergenic enhancer. The MI56 sequence (GenBank, AY168010), is shown for two enhancer elements, I56i and i56ii, initially described by Zerucha et al. (13). Putative TAAT/ATTA homeodomain DNA binding motifs are highlighted in bold italics. The two underlined I56i sequences represent the 6th and 9th motifs critical for DLX1 and/or DLX2 binding and transcriptional activation demonstrated in Figures 5b and 6, respectively.
The redundant functions of the Dlx genes may be explained by their nearly identical homeodomains, whereas their unique functions may be due to the divergence of their amino acid sequences in other domains (3). In addition, while Dlx1, 2, 5 and 6 are localized to the same anatomical regions, they are expressed at different but overlapping stages of differentiation (3,4,6,8), suggesting that distinct Dlx genes are required at different stages of development. This temporal and spatial overlap could be the basis for the observed genetic redundancy between Dlx1 and Dlx2 (4,11,12).
A conserved enhancer located in the Dlx5/Dlx6 intergenic region (mouse Dlx5/6 intergenic enhancer; MI56) is an important site of cross-regulatory interactions between Dlx genes (13) and shows sequence similarity to an intergenic region of the Dlx3–7 cluster (14,15). In the Dlx1/Dlx2 knockout, activity of a reporter transgene regulated by MI56 is markedly reduced, analogous to decreased Dlx5 and Dlx6 expression in the Dlx1/Dlx2 double mutant striatum (8,13). Expression from MI56 marks adult mouse cortical GABAergic interneurons (16). However, these studies have not addressed whether the observed transcriptional events are due to the direct interaction of DLX homeoproteins with MI56 in vivo or via an indirect mechanism, such as regulation of other transcription factors. Here, we demonstrate for the first time that Dlx1 or Dlx2 regulate Dlx5 and/or Dlx6 expression in vivo by direct transcriptional regulation of MI56 by adapting chromatin immunoprecipitation (ChIP) to the developing vertebrate forebrain and retina.
MATERIALS AND METHODS
Animals
Timed pregnant CD1 mice were sacrificed by cervical dislocation following protocols approved by the University of Manitoba under the auspices of the Canadian Council on Animal Care. The presence of a vaginal plug after mating denoted embryonic day 0.5 (E0.5). Dlx5/Dlx6 intergenic enhancer-lacZ transgenic reporter mice (13) were obtained from M. Ekker (University of Ottawa, Canada), maintained in a CD1 background.
Chromatin immunoprecipitation
The medial and lateral ganglionic eminences (GE) and hindbrain tissue were dissected from embryonic day 13.5 (E13) CD1 mice and mechanically triturated to obtain single cells. Retinal tissue at birth (postnatal day 0; P0) was obtained by first removing the lens. Then 1–2 × 107 cells were cross-linked with 1% paraformaldehyde (PFA) for 2 h at room temperature (RT) in the presence of protease inhibitors. Cross-linked cells were resuspended in SDS lysis buffer (1% SDS, 50 mM Tris–HCl pH 8.1, 10 mM EDTA), then sonicated. Protein A-Sepharose beads (Pharmacia Biotech) were added to the supernatant to pre-clear the chromatin. DLX1 (4,9) or DLX2 antibodies (4,17) were repurified to obtain fractions with high-affinity binding, then added to the chromatin solution and incubated overnight at 4°C. Then Protein A and Protein G-Sepharose (Sigma) were added and incubated for 3 h. Pelleted beads were then sequentially washed with Low Salt Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl), High Salt Wash Buffer (with 500 mM NaCl), LiCl Buffer (0.25 M LiCl, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH 8.1, 1% NP-40) and TE buffer (10 mM Tris–Cl, 1 mM EDTA, pH 8.0). The enriched homeoprotein DNA complex was eluted twice using freshly prepared Elution Buffer (1% SDS, 0.1 M NaHCO3) and centrifuged. Then 4 M NaCl was added to the supernatant to reverse cross-linking, and incubated at 65°C for 4 h. Finally, proteinase K was added for 1 h at 45°C. Phenol/chloroform was used to extract residual proteins from the DNA.
cis-Diamminedichloroplatinum (cis-DDP) crosslinking
1–2 × 107 cells isolated from E13.5 GE were suspended with cold Hanks’ buffer containing 1 mM PMSF, centrifuged and washed, then resuspended in 1 mM cis-DDP solution (Sigma) and incubated at 37°C for 2 h. Then the cells were lysed using lysis buffer (200 mM K3PO4 pH 7.5, 5 M urea, 2 M guanidine–HCl, 2 M NaCl) with 1 mM PMSF and mixed with hydroxylapatite (HAP) resin (Bio-Rad) pre-equilibrated with lysis buffer for 1 h incubation at 4°C. Proteins cross-linked to DNA were released from the HAP-bound DNA by a 2 h incubation at 4°C with reverse lysis buffer (1 M thiourea is substituted for 5 M urea), dialyzed, lyophilized, then analyzed by SDS–PAGE and immunoblotted (18–21).
Polymerase chain reaction (PCR)
Oligonucleotide primers were designed according to two regions of the MI56 sequence (GenBank, AY168010), I56i and I56ii (13). For I56i, the sense primer was 5′-GACATTGGGGACAATTTA-3′ and the antisense primer was 5′-AATTTGTGTATGAATAAC-3′. For I56ii, the sense primer was 5′-GTTTGCACACCCCAGCACCTCT-3′ and the antisense primer was 5′-CAGCCATTATTTAGACCCTA-3′. PCR was carried out with genomic DNA derived from E13 mouse embryo used as a positive control. PCR products were separated by gel electrophoresis, purified then ligated into the pCR2.1 TOPO vector using a TOPO TA cloning kit (Invitrogen). Recombinant plasmid DNA was extracted using a Plasmid MiniPrep Kit (Qiagen Inc.) and M13 reverse universal primer was used for confirmation of sequence.
Electrophoretic mobility shift assays
The Dlx5/6 intergenic region I56i, nucleotides 8688–8917 (GenBank, AY168010), was excised from pCR2.1-TOPO vector (Invitrogen) with EcoRI, then the 5′ overhang was filled in with the large fragment of DNA polymerase I (Klenow) in the presence of radiolabeled [α32P]dATP. Purified recombinant protein or nuclear extracts from the E13 striatum were incubated with 1× binding buffer, poly(dI–dC), 1 mM PMSF and 32P-labeled DNA probes (2 × 104 c.p.m.). For ‘cold’ competition assays, double-stranded unlabeled fragments were added. For ‘supershift’ assays, the specific polyclonal DLX1 and DLX2 antibodies were added. A rabbit polyclonal antibody to human Secretory Component (Dako) was used as an ‘irrelevant’ antibody. Reactions were incubated for 30 min at RT. The DNA–protein complexes were resolved on a 4% non-denaturing polyacrylamide (37.5:1 acrylamide/bis-acrylamide) gel in 0.5× Tris–borate–EDTA solution. Gels were exposed to film and autoradiography was performed overnight at –70°C.
Sequences of four of 10 putative TAAT/ATTA homeodomain binding motifs (Fig. 3b) from the I56i region were used to generate individual synthetic oligonucleotides (16–20 bp). Double-stranded oligonucleotides were subjected to further electrophoretic mobility shift assay (EMSA) analysis. Motif 6: sense primer: 5′-CAAAGATAATTACCTG-3′, antisense primer: 5′-CAGGTAATTATCTTTG-3′; Motif 7: sense primer: 5′-CCCGCTGATTACAGCGT-3′, antisense primer: 5′-ACGCTGTAATCAGCGGG-3′; Motif 8: sense primer: 5′-AGTAATTTTTCATGGAGC-3′, antisense primer: 5′-GCTACATGAAAAATTACT-3′; Motif 9: sense primer: 5′-AAATGCAGCCATAATTAGAG-3′, antisense primer: 5′-CTCTAATTATGGCTGCATTT-3′. Putative homeodomain protein recognition sites as characterized by a TAAT/ATTA core sequence are in bold.
Constructs for reporter gene assays
Effector plasmids expressing the mouse Dlx1 and Dlx2 genes separately under control of a CMV promoter were constructed by inserting a PCR-amplified 790 bp Dlx1 cDNA and 1020 bp Dlx2 cDNA (gifts from J. Rubenstein, UCSF) into the pcDNA3 vector (Invitrogen). Reporter plasmids were constructed by inserting a 236 bp I56i intergenic enhancer fragment from position 8688–8917 (GenBank, AY168010) into the pGL3-promoter vector (Promega). Site-directed mutagenesis of putative DLX-binding sites was performed using the Quick Change kit (Stratagene). Three pGL3 promoter plasmids were generated containing deleted MI56i elements (ATAATTA). The first plasmid (Δ1) contained a deletion at nucleotides 8853–8860, the second plasmid (Δ2) contained a deletion at 8797–8804 and the third plasmid (Δ3) contained a deletion at both of these positions. Mutations were verified by DNA sequencing.
Reporter gene assays
Transient co-transfection experiments were performed in the P19 murine embryonic carcinoma (EC) cell line (a gift from M. McBurney, University of Ottawa, Canada). P19 cells were grown and maintained in alpha Dulbecco’s Modified Eagle’s medium (αDMEM) supplemented with 7.5% fetal bovine serum, 2.5% calf serum and 1% penicillin–streptomycin at 37°C with 5% CO2. Cells were seeded 24 h before transfection at a density of 1 × 107 per 36 mm2 dish. Cells were transiently transfected using Lipofectamine 2000 reagent (Invitrogen). All transfections contained pRSV-β-gal (Promega) as a control for transfection efficiency. Luciferase activity was measured using the Luciferase Reporter Assay System (Promega) and a standard luminometer.
Immunoblot, in situ hybridization, immunostaining, β-galactosidase staining
Immunoblot, in situ hybridization and immunostaining were performed on fixed, cryopreserved retinal tissues at P0 and in adult mice following established protocols (4,6,17). Primary antibodies were: αDLX1, rabbit polyclonal; αDLX2, rabbit polyclonal; anti-β-gal (ICN Pharmaceuticals), rabbit polyclonal; αHDAC1 (Affinity Bioreagents, Inc.), rabbit polyclonal. β-galactosidase staining was performed as described (13,16).
RESULTS
Dlx gene expression in embryonic forebrain and retina tissues
Both DLX1 and DLX2 are highly expressed in the anlagen of the embryonic striatum, the medial and lateral GE (Fig. 1a, A and B) (3,4,17), retinal neuroepithelium (Fig. 1a, C) (4,6) and the ventral thalamus (data not shown). Peak levels of expression occur in forebrain tissues before E14.5 (4).
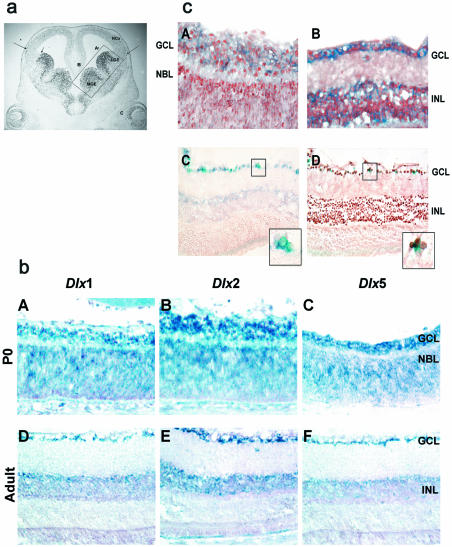
Figure 1.
(a) DLX2 expression in the developing forebrain and retina at E13.5. DLX2 is predominantly localized to the lateral (A) and medial (B) ganglionic eminences (LGE, MGE) as well as the retinal neuroepithelium (C) at this stage. There is a well-demarcated boundary of DLX2 expression at the GE-neocortical interface (small arrow). Migrating DLX2 immunoreactive cells, previously identified as GABAergic interneuron precursors, are visualized in the neocortex (NCx, long arrows). The boxed region outlines the regions of GE dissected for subsequent ChIP and EMSA. (b) Dlx isoform expression in the neonatal and adult retina. Dlx1 (panels A, D), Dlx2 (panels B, E) and Dlx5 (panels C, F) expression was determined at P0 (A–C) and in the adult (D–F) retina using digoxigenin-labeled RNA in situ hybridization. At P0, all three isoforms are expressed in the GCL, with Dlx2 expression predominating. In the adult retina, Dlx2 expression is evident in the GCL and inner nuclear layer (INL) (panel E), whereas Dlx1 and Dlx5 RNA expression are localized to the GCL more than the INL. DLX1 protein expression is not detected after P0 (6). (c) Dlx2 and Dlx5 are co-expressed with the Dlx5/6 intergenic enhancer in the retina. DLX2 protein (brown, nuclear staining) and Dlx5 RNA (purple, cytoplasmic staining) co-expression was established by combined immunohistochemistry and in situ hybridization in P0 (A) and adult (B) retinal tissue sections. Sections of adult retina stained for lacZ (blue) from the Dlx5/6 intergenic enhancer (MI56i) lacZ transgenic reporter mouse were probed with Dlx5 digoxigenin-labeled riboprobe (C, purple) and DLX2 antibody (D, brown), demonstrating co-localization of the MI56i enhancer element with Dlx5 and DLX2 in the GCL.
Dlx genes are co-expressed with the Dlx5/Dlx6 intergenic enhancer in the retina
Dlx1 and Dlx2 expression in the retina occurs after E12.5. Dlx5 is not expressed at this stage (4). We have completed a comprehensive analysis of Dlx1 and Dlx2 expression during retinal development. DLX1 expression is similar to DLX2 but is markedly reduced after birth (P0). In the adult, DLX2 is localized to retinal ganglion cells (RGC), amacrine cells, horizontal cells and a subset of bipolar cells, but not to photoreceptors (6). Digoxigenin RNA in situ hybridization studies demonstrated Dlx1, Dlx2 and Dlx5 retinal expression at P0 and in the adult. Dlx2 is expressed at higher levels than either Dlx1 or Dlx5 (Fig. 1b) consistent with its protein expression (6). Dlx5 is first expressed after E16, primarily in the ganglion cell layer (GCL, Fig. 1b, C and F) and DLX2 protein (brown) and Dlx5 RNA (blue purple) are co-expressed in the GCL at P0 (Fig. 1c, A) and in the GCL and inner nuclear layer (INL) in the adult retina (Fig. 1c, B) using combined immunohistochemistry with RNA in situ hybridization.
The Dlx5/6 intergenic enhancer/lacZ mouse expresses lacZ in anatomic locations where Dlx5, and to a lesser extent Dlx6, are expressed (13). Although Dlx1 and Dlx2 expression is significantly reduced in late gestation, the Dlx5/6 enhancer lacZ transgene may be expressed into adulthood (5,13,16,22). In the retina, lacZ expression was faintly detected at P0 (data not shown) but in the adult, lacZ was expressed in the GCL, containing RGCs and displaced amacrine cells (Fig. 1c). Adult retinal cells expressing lacZ (X-gal) co-expressed a digoxigenin-labeled Dlx5 riboprobe (Fig. 1c, C) and a subset were immunoreactive for DLX2 (Fig. 1c, D). Hence, DLX2 and MI56 are co-expressed in the GCL of the adult retina. These results support the Dlx5/6 intergenic enhancer as a candidate DLX1 or DLX2 target region in the developing retina in vivo.
Optimization of protein–DNA cross-linking in embryonic mouse forebrain and retina
We have optimized ChIP of embryonic striatum and retina, with respect to anatomic region, developmental stage, cell number, selection and concentration of a cross-linking reagent, and duration of cross-linking. We chose E13.5 GE and P0 retina due to the ease of dissection and the relative abundance of DLX1 and DLX2 expression at these time-points (Fig. 1a, A, B; Fig. 1b, A–C; Fig. 1c, A). An average litter of timed pregnant CD1 mice yielded 10–15 paired GE or retinas for ∼1–2 × 107 cells. We then cross-linked the tissues with PFA concentrations ranging from 1 to 4% with fixation times from 30 min to overnight. PFA cross-links protein to protein, protein to DNA and DNA to DNA depending on the substrate and binding conditions. 1% PFA was superior to 4% for cross-linking DLX proteins to nuclear DNA in GE (Fig. 2a).
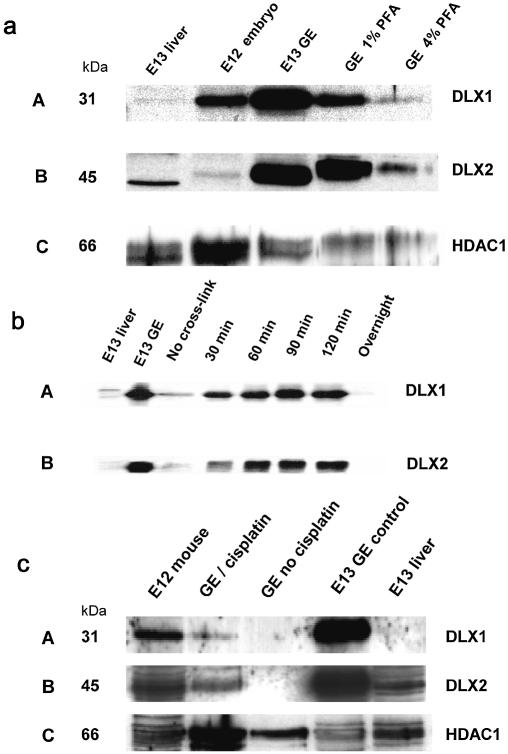
Figure 2.
(a) Paraformaldehyde cross-links DLX1 and DLX2 to nuclear DNA in situ. DLX1 and DLX2 are immunoenriched in E13.5 GE. 1% PFA improves cross-linking of DLX proteins compared with 4% PFA. Protein samples were obtained from embryonic liver (negative control), whole E12.5 embryo (positive control), E13.5 GE and E13.5 GE cross-linked with 4% or 1% PFA for 2 h. Blots shown in A, B and C were immunostained with αDLX1, αDLX2 and anti-histone deacetylase-1 (αHDAC1), respectively. (b) Duration of exposure to PFA affects the yield of homeoprotein–DNA cross-links in situ. Dissected GE at E13.5 were cross-linked with 1% PFA from 30 min to overnight. Peak levels of both DLX1 and DLX2 were detected at 90 min. Overnight exposure to PFA significantly abrogates detection of DLX proteins. (c) Cisplatin cross-links DLX1 and DLX2 to nuclear DNA in situ. Cisplatin (cis-DDP) was also used to cross-link DLX homeoproteins from E13.5 GE. Blots shown in Panels A, B and C were immunostained with αDLX1, αDLX2 and anti-histone deacetylase-1 (αHDAC1), respectively.
The duration of exposure to PFA was also critical for obtaining suitable yields of homeoprotein–DNA complexes. In GE, peak levels of DLX1 and DLX2 were obtained at 90 min, with levels rapidly falling off after 2 h and negligible amounts were present after an overnight fixation (Fig. 2b). Incubation time with the cross-linking reagent affects the quantity of protein–DNA complexes as well as the solubility of these complexes.
Cisplatin also cross-links DLX proteins to nuclear DNA in embryonic striatum
Cisplatin (cis-DDP) was tested as an alternative to PFA since it cross-links nuclear DNA at 4 Å rather than 2 Å and cross-links protein to DNA rather than to protein (18,20). Cisplatin preferentially cross-links nuclear matrix proteins, such as histone deacetylase-1 (HDAC1), to nuclear DNA in situ (18–20,23). Cisplatin cross-linked DLX1, DLX2 and HDAC1 to nuclear DNA from GE in situ (Fig. 2c) suggesting that DLX1 and DLX2 are nuclear matrix proteins. Cisplatin (1 mM) is sufficient to cross-link either DLX1 or DLX2 to nuclear DNA in situ. This cross-linking reagent was less efficient than 1% PFA for yields of DLX1 or DLX2, but relatively higher levels of HDAC1 were obtained. Since PFA produced a greater yield of DLX1 and DLX2 cross-linked products, we chose 1% PFA as the cross-linking reagent for subsequent ChIP experiments (Fig. 2a,b).
DLX proteins bind to the Dlx5/Dlx6 intergenic enhancer in embryonic striatum
Using this optimized ChIP procedure and specific polyclonal antibodies to DLX1 and DLX2 (4), DLX1 and DLX2 target genomic DNA sequences were isolated. Following sonication, the average fragment size, representing candidate regulatory sequences, was 300 bp (24). We selected the intergenic enhancer region (MI56) between Dlx5 and Dlx6 (Fig. 3a and b) (13). MI56 has two candidate homeodomain binding regions based upon groups of putative TAAT/ATTA DNA binding motifs: the I56i region (nucleotides 8641–9039) and I56ii (6543–6840) both have 10 motifs (13). We designed oligonucleotide primers to encompass both MI56 regions and performed PCR after ChIP of E13 GE fixed in 1% PFA for 2 h. Both DLX1 and DLX2 bound to I56i but not I56ii in GE (Fig. 4a). Mouse genomic DNA was used as a positive control for PCR. Negative controls included omission of the specific antibody from the IP (Fig. 4a, b and c) and use of embryonic hindbrain, a tissue that does not express any Dlx genes (Fig. 4c). Bands obtained from PCR were subcloned and sequence-verified.
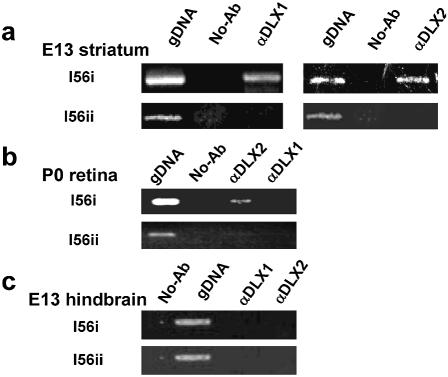
Figure 4.
(a) Chromatin IP: DLX1 and DLX2 bind to the Dlx5/6 intergenic enhancer in embryonic striatum in situ. E13.5 GE was cross-linked with 1% PFA and immunoprecipitated with specific DLX1 or DLX2 antibodies. After isolation of genomic DNA fragments bound to DLX1 or DLX2 homeoproteins, PCR using oligonucleotide primers to two regions (I56i and I56ii) of the Dlx5/6 intergenic region was performed. Total genomic DNA was used as a positive control. Both DLX1 and DLX2 bind to I56i but not to I56ii in situ. PCR products were subcloned and sequence-verified. (b) Chromatin IP: DLX2 but not DLX1 binds to the Dlx5/6 intergenic enhancer in neonatal retina in situ. ChIP was performed using P0 retina and specific DLX1 or DLX2 antibodies. In the P0 retina, only DLX2 binds to I56i; neither DLX1 nor DLX2 binds to I56ii in the retina in situ. (c) Chromatin IP: DLX1 and DLX2 do not bind to the Dlx5/6 intergenic enhancer in embryonic hindbrain in situ. E13.5 hindbrain was selected as a negative tissue control for ChIP with specific DLX1 or DLX2 antibodies. In embryonic hindbrain, neither DLX1 nor DLX2 bind to I56i or I56ii in situ.
DLX homeobox proteins differentially bind to the Dlx5/Dlx6 intergenic enhancer in the newborn retina
ChIP assays of retina tissue at P0 were performed (Fig. 4b). Similar to E13 GE, DLX homeoproteins bind only to the I56i region, not to I56ii in P0 retina in situ. However, unlike striatum, DLX2 but not DLX1 binds to the Dlx5/6 intergenic enhancer in retina at P0, probably reflecting the marked reduction in DLX1 protein in the neonatal retina (Fig. 4b).
Embryonic striatal nuclear extracts bind to specific homeodomain-DNA binding motifs of the Dlx5/Dlx6 intergenic enhancer
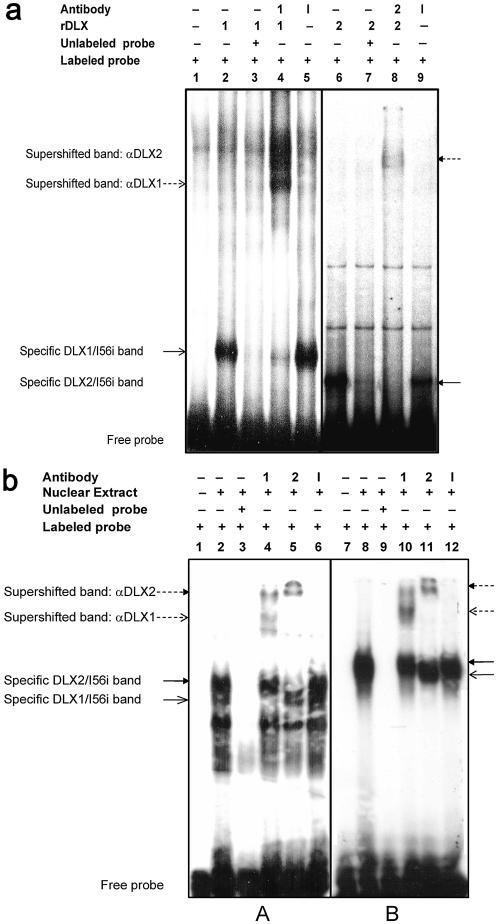
EMSA were used to provide in vitro evidence for the binding of DLX1 and/or DLX2 to MI56 isolated by ChIP in situ. We demonstrated specific DLX1– and DLX2–oligonucleotide complexes in vitro similar to DLX3–oligonucleotide complexes shown by Feledy et al. (25). Recombinant DLX1 and DLX2 bind to the I56i region and specific supershifted bands result on the addition of specific DLX1 or DLX2 antibodies. The addition of an irrelevant polyclonal primary antibody confirms the specific binding of homeoprotein–DNA complexes (Fig. 5a).
Figure 5.
(a) Electrophoretic mobility shift assays (EMSA) demonstrate recombinant DLX1 and DLX2 bind to the Dlx5/Dlx6 intergenic enhancer in vitro. EMSA was performed using recombinant DLX1 or DLX2 proteins and a radiolabeled I56i oligonucleotide probe, with cold competition (lanes 3 and 7) and specific DLX antibody ‘supershift’ assays (lanes 4 and 8). Lane 1: labeled I56i alone. Lanes 2–5: labeled I56i probes were incubated with rDLX1 (2), rDLX1 and unlabeled I56i (3), rDLX1 and anti-DLX1 (4), and rDLX1 and an irrelevant antibody (5). Lanes 6–9: I56i probes were incubated with rDLX2 (6), rDLX2 and unlabeled I56i (7), rDLX2 and anti-DLX2 (8), and rDLX2 and irrelevant antibody (9) (r: recombinant; 1: DLX1/anti-DLX1; 2: DLX2/anti-DLX2; I: irrelevant antibody). (b) EMSA of embryonic striatum demonstrates DLX1 and DLX2 binding to specific homeodomain binding motifs within the Dlx5/Dlx6 intergenic enhancer in situ. EMSA was performed using E13.5 GE tissue nuclear extracts and oligonucleotides for four TAAT/ATTA DNA binding motifs contained within I56i. Only the sixth and ninth motifs bound to E13.5 GE. (A) EMSA for homeodomain binding motif 9 (8853–8860 bp, lanes 1–6). (B) EMSA for motif 6 (8797–8804 bp, lanes 7–12). Cold competition assays are shown in lanes 3 and 9. Supershift assays with specific DLX antibodies are seen in lanes 4 and 10 (αDLX1) and lanes 5 and 11 (αDLX2).
We then developed EMSA for nuclear extracts enriched for DLX1 and DLX2 expression obtained from GE and retina tissues. Several of the 10 TAAT motifs located in the I56i DNA binding region (Fig. 3b) were previously shown by Zerucha et al. (13) to bind DLX2 in vitro; DLX1 binding to this region was not previously assessed. We sequentially deleted each region and determined that only the ninth (Fig. 5b, A) and sixth (Fig. 5b, B) homeodomain binding motifs were sufficient for binding both DLX1 and DLX2 in embryonic striatum in vitro corroborating the findings of Zerucha et al. (13). Supershift experiments confirm the specificity of each DLX protein–DNA complex. Hence, both recombinant DLX1 and DLX2 and striatal nuclear extracts bind to region 1 of the Dlx5/Dlx6 intergenic enhancer (I56i) in vitro.
The DLX2 homeoprotein preferentially activates reporter gene activity when bound to its target sequences in the Dlx5/Dlx6 intergenic enhancer
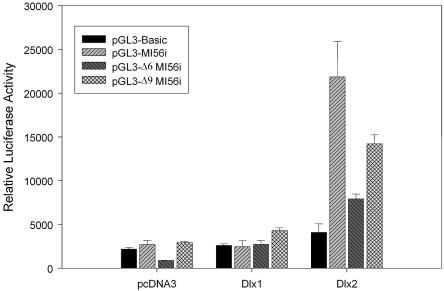
Although the ChIP assay was used to isolate mouse I56i as a DLX1 and DLX2 DNA target and EMSA confirmed protein–DNA interactions in vitro, it is important to demonstrate the functional significance of DLX1 or DLX2 binding to this enhancer element. P19 embryonal carcinoma cells are pluripotential cells that can differentiate into neuronal cells under specific conditions, such as treatment with retinoic acid (26). DLX2, but not DLX1, co-transfection activated luciferase expression. Western blotting confirmed that recombinant DLX1 and DLX2 proteins are both made in the transfected P19 cells (data not shown). Site-directed mutagenesis of the sixth or ninth TAAT motifs was completed prior to co-transfection with DLX expression constructs. Mutation of the sixth motif significantly reduced reporter gene activity mediated by DLX2, whereas mutation of the ninth motif reduced luciferase activity to a lesser extent following DLX2 co-transfection (Fig. 6). These results are similar to those obtained by Zerucha et al. using the zebrafish dlx4/dlx6 intergenic enhancer (13). Unlike DLX2, reporter gene expression was not affected by DLX1 co-transfection studies with the intact I56i enhancer element or mutated I56i enhancers containing mutations of the sixth and ninth TAAT motifs. Co-transfection of both DLX1 and DLX2 did not demonstrate any additive or synergistic effects compared with co-transfection of DLX2 alone (data not shown).
Figure 6.
DLX2 binding to the Dlx5/Dlx6 intergenic enhancer activates transcription of a luciferase reporter gene in vitro. In P19 embryonic carcinoma cells, a luciferase gene reporter construct containing I56i, or I56i with mutations of the 6th or 9th homeodomain DNA binding motifs, was co-transfected with expression constructs for DLX1 or DLX2. DLX2, but not DLX1, co-transfection activated reporter gene expression. Site-directed mutagenesis of the sixth TAAT motif significantly reduced reporter gene activity for DLX2 co-transfection.
DISCUSSION
We have optimized the ChIP assay, using our specific DLX1 and DLX2 antibodies, to demonstrate for the first time that both DLX1 and DLX2 bind to the Dlx5/Dlx6 intergenic enhancer in embryonic forebrain in vivo. Our results provide a direct mechanism to explain the marked reduction in both Dlx5 and Dlx6 expression in the embryonic striatum observed in the Dlx1/Dlx2 double knockout mouse (8). In addition, we have demonstrated similar transcriptional regulation of this enhancer by DLX2 but not DLX1 in the mouse retina, signifying a conservation of function of Dlx genes wherever these homeobox genes are expressed.
The subtle phenotypes observed in the developing forebrain of the Dlx1 and Dlx2 single knockouts, as well as the overlapping expression domains and co-expression in subsets of embryonic striatum, support functional redundancy of Dlx1 and Dlx2 (3–5,11,12). However, there are differences between Dlx1 and Dlx2 expression during early development of the GE and retina. For example, in the GE, more DLX2 than DLX1 expressing cells are cycling at E12.5 (4). Onset of DLX1 and DLX2 expression in the retina occurs by E12.5 but more DLX2 than DLX1 expressing cells are in M-phase (4). These differences suggest that DLX1 and DLX2 may have different yet overlapping gene targets in developing forebrain and retina.
DLX1 and DLX2 expression both significantly decrease by late gestation in the embryonic striatum. DLX1 and DLX2 bind to MI56 in E13.5 GE as demonstrated by ChIP assay. The activation of reporter gene expression by Dlx2 but not Dlx1 co-transfection with MI56 constructs suggests that DLX1 may require additional co-factors not present in the P19 embryonal carcinoma cells in vitro. However, luciferase and other reporter assays are in vitro ‘non-chromatin’ based methods. Hence, using ChIP, the investigator may detect binding without functional activation/repression of regulatory elements in vitro. The lack of demonstrated cooperation of DLX1 with DLX2 to activate the MI56 enhancer reporter gene suggests that DLX1 and DLX2 may have some non-overlapping functions during striatal neuronogenesis. These results support the functional differences observed between Dlx paralogs in Dlx1, Dlx2 and Dlx1/Dlx2 null mice (4,5,12).
In the retina, DLX2 and DLX1 expression extends throughout embryogenesis, with DLX2 expressed in the adult. DLX1 expression resembles DLX2 to P0 but is significantly decreased postnatally (6). At P0, the binding of DLX2 but not DLX1 to the MI56 may reflect decreased DLX1 protein levels in the neonatal retina. Alternatively, binding of DLX1 to the MI56 in vivo may not be detected if the specific DLX1 antibody is not sufficiently sensitive to detect low levels of DLX1 protein or able to immunoprecipitate small quantities of DLX1 homeoprotein–MI56 enhancer DNA complexes. β-gal antibody may be more sensitive than X-gal histochemistry in detecting lacZ expression since more cells are marked with the antibody at P0 and in the adult (data not shown). This could also reflect the possibility that the experimental conditions for tissue fixation and penetration of solutions may be less favorable for detection of the chromogenic substrate. The expression of lacZ in adult but minimally in neonatal retina is consistent with peak levels of Dlx5 and/or Dlx6 expression occurring after P0. Binding of DLX2 to the enhancer in retinal tissue at P0 may also reflect the relative sensitivity of the ChIP assay compared with routine histological detection, rather than false positive results. The ChIP protocol, which uses tissue from multiple embryonic or postnatal tissues to obtain 1–2 × 107 cells prior to processing and subsequent PCR amplification, is more likely to detect target sequences that are minimally expressed in developing tissues at a specific temporal or spatial coordinate. The retinal phenotype of the Dlx5/6 double knockout may be predicted to be more subtle than the Dlx1/2 null retina. Hence, unlike striatum, it may be helpful to perform ChIP at several time-points to obtain specific Dlx gene family targets in the developing and postnatal retina.
ChIP technology represented a major advance towards the identification of direct target genes of specific transcription factors, especially homeobox genes (23,27–29). The utilization of biochemical rather than genetic approaches, such as ChIP, provides several advantages. Identified target genes are directly downstream and derive from physiological homeodomain–DNA complexes obtained in vivo. Isolated gDNA fragments may be from regulatory elements of known or novel genes. ChIP may be applied to diverse species, including Drosophila and vertebrates. Cross-linking preserves naturally existing (in situ) protein–DNA interactions. Utilizing these methods, homeodomain targets have been isolated, including transcription factors, growth factors, adhesion molecules and secreted proteins (28,30,31). The search for homeobox targets in vertebrates has not yielded as many targets as in Drosophila. Although several candidates have been identified using in vitro and tissue culture methods, the significance of these interactions in vivo remains to be confirmed.
Tomotsune et al. (24) used ChIP to identify a tumor suppressor gene and putative adhesion molecule as a downstream target of Hox-C8 from mouse spinal cord, suggesting that it is possible to isolate other direct vertebrate homeobox gene targets. However, the ChIP methodology used by this group may have presented several technical obstacles for subsequent isolation of other homeobox gene targets. For example, cross-linking was performed overnight using 4% PFA and the entire spinal cord was used as a tissue source rather than a region expressing high levels of Hox-C8. It is unclear whether the specific antibody they used had high affinity for this homeobox protein. Hence, many investigators have eschewed ChIP in favor of cDNA microarray or other techniques to find genes potentially regulated by their transcription factor of interest. Recently, ChIP has been successfully coupled with DNA microarray analysis. Weinmann et al. (32) used human CpG microarrays probed with immunoprecipitated chromatin to rapidly identify target promoters for the transcription factor E2F. Hence, combining ChIP and microarray technologies may improve both the efficiency and scale of standard ChIP approaches.
We have optimized the ChIP approach by reducing the concentration of the cross-linking reagent PFA and incubation time to preferentially obtain homeoprotein–genomic DNA complexes from embryonic tissues in situ. The polyclonal antisera to DLX1 and DLX2 have been subjected to a rigorous affinity purification process and are sensitive and highly specific (4). In addition, chromatin is derived from nuclear extracts derived from tissues where peak developmental expression of DLX1 and DLX2 occurs in distinct anatomical regions, enriching for the selection of Dlx1 and Dlx2 specific DNA target fragments. The identification of multiple Dlx target sequences in vivo will facilitate the elucidation of a consensus Dlx DNA binding site initially derived in vitro for Dlx3 by Feledy et al. (25). Finally, the expression of novel Dlx gene targets identified by ChIP may be altered in the Dlx1/2 null mouse, providing further understanding of how Dlx genes function throughout development. By combining genetic and molecular approaches, we will identify other Dlx-dependent target genes that are critical effectors of forebrain and retinal development. In addition, this optimized ChIP approach can be used to isolate direct targets of other transcription factors during vertebrate development.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jeff Wigle and Spencer Gibson for critical review of the manuscript. We are grateful to Marc Ekker for providing the Dlx5/6 intergenic enhancer lacZ reporter transgenic mouse eye tissues used in this study. This work was supported by a Basil O’Connor Starter Scholar Award from the March of Dimes Birth Defects Foundation 5-FY00–615 (to D.D.E.), an operating grant from the CancerCare Manitoba Foundation (to J.R.D. and D.D.E) and an Establishment Grant from the Manitoba Health Research Council (to D.D.E.). T.N.L. was supported by a Canadian Institutes of Health Research Training Program Grant.
REFERENCES
- 1.Sorensen P.H., Lynch,J.C., Qualman,S.J., Tirabosco,R., Lim,J.F., Maurer,H.M., Bridge,J.A., Crist,W.M., Triche,T.J. and Barr,F.G. (2002) PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. J. Clin. Oncol., 20, 2672–2679. [DOI] [PubMed] [Google Scholar]
- 2.Downing J.R. and Shannon,K.M. (2002) Acute leukemia: a pediatric perspective. Cancer Cell, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- 3.Liu J.-K., Ghattas,I., Liu,S., Chen,S. and Rubenstein,J.L.R. (1997) Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev. Dyn., 210, 498–512. [DOI] [PubMed] [Google Scholar]
- 4.Eisenstat D.D., Liu,J.-K., Mione,M., Zhong,W., Yu,G., Anderson,S.A., Ghattas,I., Puelles,L. and Rubenstein,J.L. (1999) DLX-1, DLX-2 and DLX-5 expression define distinct stages of basal forebrain differentiation. J. Comp. Neurol., 414, 217–237. [DOI] [PubMed] [Google Scholar]
- 5.Panganiban G. and Rubenstein,J.L.R. (2002) Developmental functions of the distal-less/Dlx homeobox genes. Development, 129, 4371–4386. [DOI] [PubMed] [Google Scholar]
- 6.deMelo J., Qiu,X., Du,G., Cristante,L. and Eisenstat,D.D. (2003) Dlx1, Dlx2, Pax6, Brn3b and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J. Comp. Neurol., 461, 187–204. [DOI] [PubMed] [Google Scholar]
- 7.McGuinness T., Porteus,M.H., Smiga,S., Bulfone,A., Kingsley,C., Qiu,M., Liu,J.-K., Long,J.E., Xu,D. and Rubenstein,J.L. (1996) Sequence, organization and transcription of the Dlx-1 and Dlx-2 locus. Genomics, 35, 473–485. [DOI] [PubMed] [Google Scholar]
- 8.Anderson S., Qiu,M., Bulfone,A., Eisenstat,D.D., Meneses,J., Pedersen,R. and Rubenstein,J.L.R. (1997) Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron, 19, 27–37. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S., Eisenstat,D.D., Shi,L. and Rubenstein,J.L.R. (1997) Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science, 278, 474–476. [DOI] [PubMed] [Google Scholar]
- 10.Marin O. and Rubenstein,J.L.R. (2001) A long, remarkable journey: tangential migration in the telencephalon. Nature Rev. Neurosci., 2, 780–790. [DOI] [PubMed] [Google Scholar]
- 11.Qiu M., Bulfone,A., Martinez,S., Meneses,J.J., Shimamura,K., Pedersen,R.A. and Rubenstein,J.L.R. (1995) Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev., 9, 2523–2538. [DOI] [PubMed] [Google Scholar]
- 12.Qiu M., Bulfone,A., Ghattas,I., Meneses,J.J., Christensen,L., Sharpe,P.T., Presley,R., Pedersen,R.A. and Rubenstein,J.L. (1997) Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: Mutations of Dlx-1, Dlx-2 and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol., 185, 165–184. [DOI] [PubMed] [Google Scholar]
- 13.Zerucha T., Stühmer,T., Hatch,G., Park,B.K., Long,Q., Yu,G., Gambrotta,A., Schultz,J.R., Rubenstein,J.L. and Ekker,M. (2000) A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci., 20, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumiyama K. and Ruddle,F.H. (2003) Regulation of Dlx3 gene expression in visceral arches by evolutionarily conserved enhancer elements. Proc. Natl Acad. Sci. USA, 100, 4030–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghanem N., Jarinova,O., Amores,A., Long,Q., Hatch,G., Park,B.K., Rubenstein,J.L.R. and Ekker,M. (2003) Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res., 13, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stühmer T., Anderson,S.A., Ekker,M. and Rubenstein,J.L.R. (2002) Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development, 129, 245–252. [DOI] [PubMed] [Google Scholar]
- 17.Porteus M.H., Bulfone,A., Liu,J.-K., Puelles,L., Lo,L.C. and Rubenstein,J.L. (1994) DLX-2, MASH-1 and MAP-2 expression and bromodeoxyuridine incorporation define molecularly distinct cell populations in the embryonic mouse forebrain. J. Neurosci., 14, 6370–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuel S.K., Spencer,V.A., Bajno,L., Sun,J.M., Holth,L.T., Oesterreich,S. and Davie,J.R. (1998) In situ cross-linking by cisplatin of nuclear matrix-bound transcription factors to nuclear DNA of human breast cancer cells. Cancer Res., 58, 3004–3008. [PubMed] [Google Scholar]
- 19.Spencer V.A., Coutts,A.S., Samuel,S.K., Murphy,L.C. and Davie,J.R. (1998) Estrogen regulates the association of intermediate filament proteins with nuclear DNA in human breast cancer cells. J. Biol. Chem., 273, 29093–29097. [DOI] [PubMed] [Google Scholar]
- 20.Spencer V.A. and Davie,J.R. (2002) Isolation of proteins cross-linked to DNA by cisplatin. In Walker,J.M. (ed.), The Protein Protocols Handbook. Humana Press, Totowa, NJ, pp. 747–751. [Google Scholar]
- 21.Spencer V.A. and Davie,J.R. (2002) Isolation of proteins cross-linked to DNA by formaldehyde. In Walker,J.M. (ed.), The Protein Protocols Handbook. Humana Press, Totowa, NJ, pp. 753–757. [Google Scholar]
- 22.Stühmer T., Puelles,L., Ekker,M. and Rubenstein,J.L.R. (2002) Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb. Cortex, 12, 75–85. [DOI] [PubMed] [Google Scholar]
- 23.Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- 24.Tomotsune D., Shoji,H., Wakamatsu,Y., Kondoh,H. and Takahashi,N. (1993) The mouse homologue of the Drosophila tumour suppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature, 365, 69–72. [DOI] [PubMed] [Google Scholar]
- 25.Feledy J.A., Morasso,M.I., Jang,S.I. and Sargent,T.D. (1999) Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acids Res., 27, 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg R.W. and McBurney,M.W. (1990) Cell density and cell cycle effects on retinoic acid-induced embryonal carcinoma cell differentiation. Dev. Biol., 138, 123–135. [DOI] [PubMed] [Google Scholar]
- 27.Gould A.P., Brookman,J.J., Strutt,D.I. and White,R.A.H. (1990) Targets of homeotic gene control in Drosophila. Nature, 348, 308–312. [DOI] [PubMed] [Google Scholar]
- 28.Graba Y., Aragnol,D. and Pradel,J. (1997) Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays, 19, 379–388. [DOI] [PubMed] [Google Scholar]
- 29.Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- 30.Boudreau N. and Bissell,M.J. (1998) Extracellular matrix signaling: integration of form and function in normal and malignant cells. Curr. Opin. Cell Biol., 10, 640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manervik M. (1999) Target genes of homeodomain proteins. Bioessays, 21, 267–270. [DOI] [PubMed] [Google Scholar]
- 32.Weinmann A.S., Yan,P.S., Oberly,M.J., Huang,T.H. and Farnham,P.J. (2002) Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev., 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumiyama K., Irvine,S.Q., Stock,D.W., Weiss,K.M., Kawasaki,K., Shimizu,N., Shashikant,C.S., Miller,W. and Ruddle,F.H. (2002) Genomic structure and functional control of the Dlx3-7 bigene cluster. Proc. Natl Acad. Sci. USA, 99, 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]