Abstract
TAp73 is a structural homologue of the pre-eminent tumor suppressor p53. However, unlike p53, TAp73 is rarely mutated, and instead is frequently over-expressed in human tumors. It remains unclear whether TAp73 affords an advantage to tumor cells and if so, what is the underlying mechanism. Here we show that TAp73 supports the proliferation of human and mouse tumor cells. TAp73 activates the expression of the glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the pentose phosphate pathway (PPP). By stimulating G6PD, TAp73 increases PPP flux and directs glucose to the production of NADPH and ribose, for the synthesis of macromolecules and detoxification of reactive oxygen species (ROS). The growth defect of TAp73-deficient cells can be rescued by either enforced G6PD expression or the presence of nucleosides plus an ROS scavenger. These findings establish a critical role for TAp73 in regulating metabolism, and connect TAp73 and the PPP to oncogenic cell growth.
Introduction
p73 is a member of the p53 family1. While the importance of p53 in tumor suppression is firmly established2,3, p73 plays a complex role in tumorigenesis that is still not well-understood4-7. p73 is expressed in two major isoform classes (TAp73 and ΔNp73) with apparently distinct functions4-7 (Figure S1A). TAp73 isoforms, similar to p53, contain an N-terminal transactivation domain. TAp73 can activate p53-target genes and induce apoptosis or cell cycle arrest. In contrast, ΔNp73 lacks the transactivation domain but retains DNA-binding and oligomerization domains. ΔNp73 is able to exert a dominant negative effect on TAp73, as well as other p53 family members, through the formation of inactive hetero-oligomeric complexes or competition for promoter binding.
Unlike p53-deficient mice, which appear developmentally normal but highly prone to spontaneous tumors8,9, mice with total p73 loss have profound defects in the immune and nervous systems but no increases in tumor incidence10. Though total p73 loss cooperates with p53 loss to further promote tumor formation in a context-dependent manner11-13. TAp73-specific knockout mice exhibit partial embryonic lethality, infertility, and a marked increase in spontaneous and carcinogen-induced tumors14. These phenotypes are likely due, in part, to genomic instability in the absence of TAp7314,15. In contrast, ΔNp73 deficiency in mice leads to increased DNA damage signaling and p53-dependent apoptosis16, indicating a role for ΔNp73 in the suppression of the p53 response. These observations support a model in which TAp73, like p53, suppresses tumorigenesis, while ΔNp73 promotes it.
Nevertheless, in contrast to p53, which is the most frequently mutated gene in human tumors, TAp73 is rarely mutated in these tumors4,6,7. An analysis of ~1,500 human tumors indicated that less than 0.2% harbored a mutant p73 (either isoform class), as opposed to over 50% with a mutant p534. Instead, TAp73 is frequently over-expressed, along with ΔNp73, in a wide range of human cancers6,7. The conspicuous absence of TAp73 mutations and prevalence of TAp73 up-regulation suggest that TAp73 may afford proliferative advantages to tumor cells.
The metabolism in tumor cells is dramatically reprogrammed to enable robust biosynthesis and anti-oxidant defense17-19. While the generation of macromolecules is an intuitive requirement for tumor cell proliferation, recent evidence also supports the critical importance of ROS detoxification in oncogenic growth. Tumor cells commonly encounter high oxidative stress due to the effect of oncogenic mutations and their microenvironment18,20,21. While moderate and transient elevation in ROS is implicated in proliferation22,23, high and persistent elevation in ROS damages protein, DNA, and other cellular components and poses a continuous threat to the viability of tumor cells. The pentose phosphate pathway (PPP) is a major glucose metabolic pathway important for meeting the cellular demands of biosynthesis and anti-oxidant defense. It provides cells with ribose-5-phosphate (R5P) for de novo synthesis of RNA and DNA, and with the reducing equivalent NADPH for reductive biosynthesis (e.g., the synthesis of lipids and deoxyriboses) and anti-oxidant defense (Supplementary Fig. S2a). The pacesetter of the PPP is glucose-6-phosphate dehydrogenase (G6PD), which catalyzes the first committing step of this pathway. Here we investigate TAp73 in cell proliferation and identify a critical role for TAp73 in promoting biosynthesis and anti-oxidant defense via the induction of G6PD expression.
RESULTS
TAp73 supports tumor growth
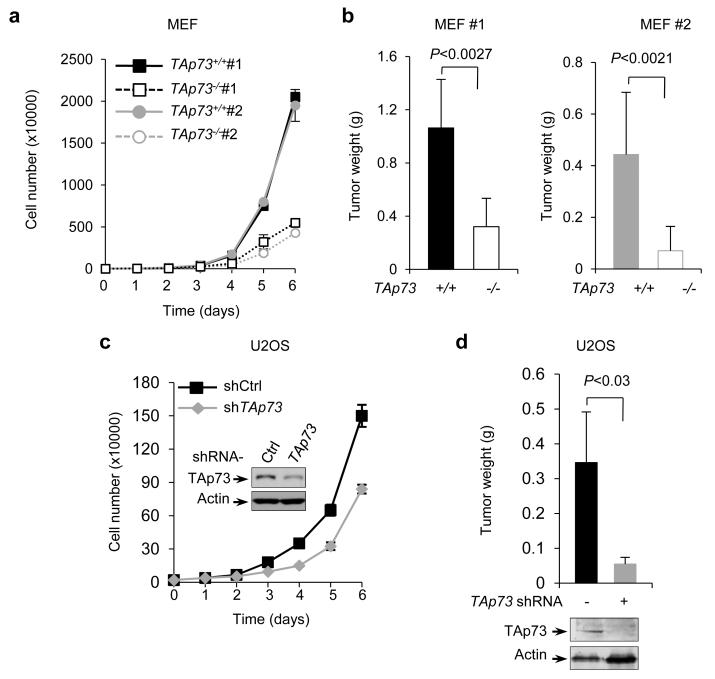
To investigate the role of TAp73 in tumor cell proliferation, we used E1A/RasV12-transformed mouse embryonic fibroblast cells (MEFs) with wild-type (+/+) or homozygous disruption of (−/−) TAp73 14. Interestingly, TAp73−/− MEF cells lines proliferated significantly slower than TAp73+/+ MEFs (Fig. 1a and Supplementary Fig. S1b). To investigate the role of TAp73 in tumor formation, we injected TAp73−/− and TAp73+/+ MEF cells into immuno-compromised mice. TAp73−/− MEF cells gave rise to much smaller tumors than TAp73+/+ MEF cells did (Fig. 1b and Supplementary Fig. S1c). To extend these analyses to human tumor cells, we stably knocked down TAp73 in human osteosarcoma U2OS cells using small hairpin RNA (shRNA). This significantly reduced proliferation of U2OS cells in culture and their growth in mouse xenograft tumors (Fig. 1c,d and Supplementary Fig. S1d). These observations suggest that TAp73 is required for optimal cell proliferation and tumor formation.
Figure 1.
TAp73 supports tumor cell growth
(a) Proliferation of two independent clones of TAp73−/− MEFs and the corresponding TAp73+/+ MEFs in culture. Data are means ± SD (n = 3 independent experiments)
(b) Independent TAp73+/+ and TAp73−/− MEF cells were injected subcutaneously (SC) in nude mice as described previously42. Shown are the average weights of the tumors (mean ± SD, n=3 mice in each group) three (left) or two (right) weeks later.
(c) Growth of U2OS cells stably expressing control or TAp73 shRNA. Protein expression in these cells is also shown. Data are means ± SD (n = 3 independent experiments)
(d) U2OS cells stably expressing control or TAp73 shRNA were individually injected in nude mice. Shown are tumor weights (mean ± SD, n=3 mice in each group) three weeks.
For comparison, we also tested the role of ΔNp73 in tumor cell proliferation using E1A/RasV12-transformed ΔNp73−/− MEFs and the matched ΔNp73+/+ MEFs16. We observed that loss of ΔNp73 resulted in a decrease in cell proliferation (Supplementary Fig. S1e, f), consistent with a role for ΔNp73 in tumor growth16.
TAp73 enhances the PPP
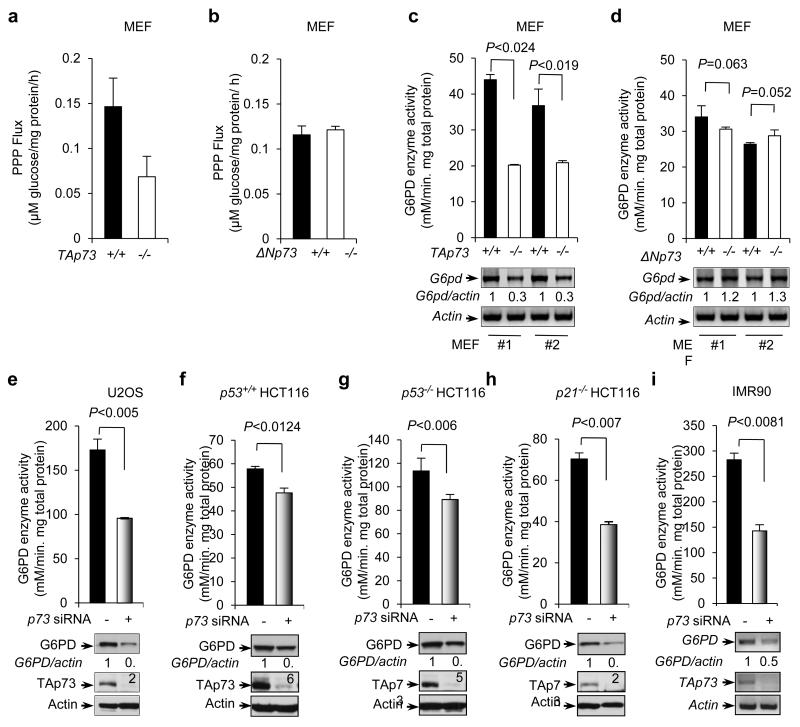
We investigated the mechanism by which TAp73 promotes proliferation. Our recent studies have shown that p53 inhibits the PPP24. We assessed whether TAp73 has any regulatory effects on the glucose flux through this pathway. TAp73+/+ and TAp73−/− MEF cells were cultured in medium containing [2-13C]glucose, and the PPP flux was measured based on the rate of glucose consumption and 13C labeling patterns in lactate determined by nuclear magnetic resonance (NMR) spectroscopy 24,25. Of note, TAp73 deficiency reduced the PPP flux by ~50%, while ΔNp73 deficiency had no effect (Fig. 2a,b), suggesting that TAp73, but not ΔNp73, enhances the PPP flux.
Figure 2.
p73 regulates PPP flux, G6PD activity and expression
(a,b) TAp73+/+ and TAp73−/− MEF cells (a), or ΔNp73+/+ and ΔNp73−/− MEF cells (b), were cultured in medium containing [2-13C]glucose. Oxidative PPP flux was measured based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 3 (indicating PPP) and carbon 2 (indicating glycolysis) of lactate determined by NMR spectroscopy. Data are means ± SD (n = 3 independent experiments).
(c,d) Top: G6PD activity in two independent clones of TAp73−/− MEFs (c), ΔNp73−/− MEFs (d), and the corresponding wild-type MEFs. Bottom: G6PD and β-actin mRNA levels, with the relative ratios of G6PD to β-actin indicated. Data are means ± SD (n = 3 independent experiments). Western blots represent two independent experiments.
(e-i) G6PD activity in U2OS cells (e), isogenic p53+/+ (f), p53−/− (g), or p21−/− (h) HCT116 cells, or IMR90 cells (i) that were treated with either control (−) or p73 siRNA. The expression of G6PD, TAp73, and actin was detected using Western blot (WB) (e-h) or RT-PCR (f). Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
The rate-limiting enzyme of the PPP is G6PD, which catalyzes the conversion of glucose-6-phosphate to 6-phosphate-gluconolactone (Supplementary Fig. S2a)26. We assayed G6PD activity in TAp73+/+ and TAp73−/− MEF cells. Importantly, TAp73−/− MEFs showed a strong reduction (~45-55%) in overall G6PD activity compared to TAp73+/+. However, no significant difference in G6PD activity was observed between ΔNp73−/− and ΔNp73+/+ MEFs (Fig. 2c,d). Correlating with these results, deficiency of TAp73, but not of ΔNp73, led to strong reduction in G6PD mRNA levels (Fig. 2c,d).
To ascertain the stimulatory effect of TAp73 on G6PD, we used small interfering RNA (siRNA) to knock down p73 in U2OS cells. This led to a noticeable decline in both the activity and expression levels of G6PD (Fig. 2e and Supplementary Fig. S2b). Introducing an siRNA-resistant form of TAp73 in p73 siRNA-treated cells largely restored G6PD expression (Supplementary Fig. S2b), underscoring the specificity of the siRNA. Likewise, upon p73 knockdown, G6PD activity and expression were reduced in three isogenic human colorectal HCT116 cell lines, which were wild type for p53 and its target gene p21 (p53+/+), deficient in p53 (p53−/−), or deficient in p21 (p21−/−) (Fig. 2f-h). A strong effect of p73 knockdown on G6PD was also observed in the normal human diploid fibroblast IMR90 cells (Fig. 2i).
G6PD is a target gene for TAp73
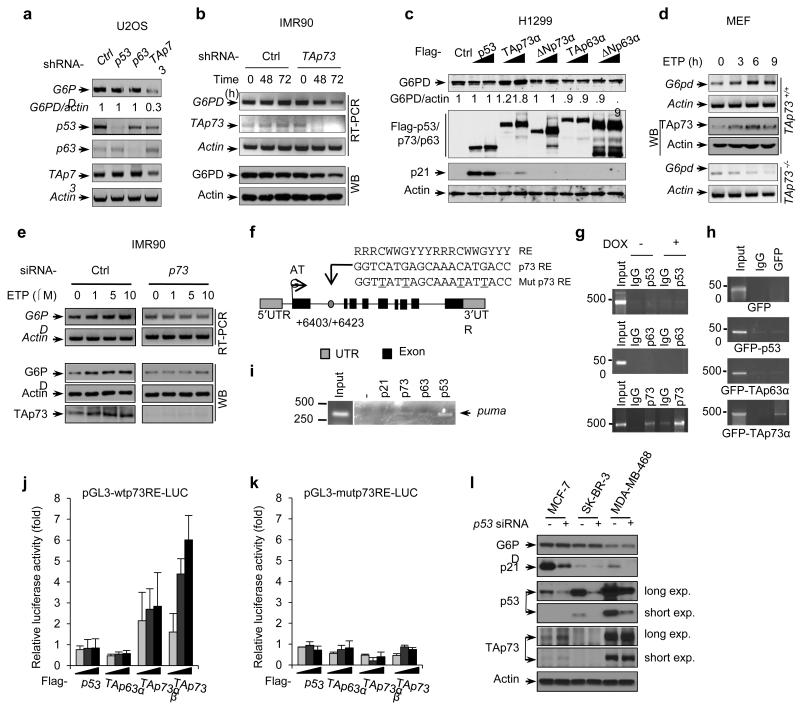
To examine the p73 isoform-specific effect on G6PD expression, we knocked down either total p73 or ΔNp73 using siRNAs in HeLa cells, which express relatively high levels of ΔNp73 compared to other cell lines tested (Supplementary Fig. S2c). Knockdown of total p73, but not ΔNp73, led to a noticeable reduction in G6PD levels (Supplementary Fig. S2d). We also knocked down TAp73 using shRNA, which strongly decreased G6PD expression in U2OS, IMR90, and human lung cancer H1299 cells (Fig. 3a,b and Supplementary Fig. S2e,f). Conversely, enforced expression of a TAp73 isoform (TAp73α), but not the corresponding ΔNp73 isoform (ΔNp73α), augmented G6PD expression (Fig. 3c and Supplementary Fig. S2g). Unlike TAp73, neither over-expression nor knockdown of p53, or the other p53 family member p63, affected G6PD levels (Fig. 3a,c and Supplementary Fig. S2f,g). Together, these results indicate that TAp73 specifically enhances the expression of G6PD.
Figure 3.
p73 regulates G6PD transcription
(a) mRNA levels in U2OS cells that were stably infected with lentiviruses expressing a control (Ctrl) shRNA or shRNA against the indicated p53 family gene.
(b) IMR90 cells were infected with lentiviruses expressing a control shRNA or a TAp73-specific shRNA for indicated durations. Protein and mRNA expression was detected using RT-PCR and WB, respectively. Data represent three independent experiments.
(c) H1299 cells were transfected with increasing amounts of control plasmid or plasmid expressing the indicated p53 family protein. Western blots represent two independent experiments, and the relative ratios of G6PD to β-actin are given. G6PD mRNA levels were detected by quantitative RT-PCR (Supplementary Fig. 2g).
(d) TAp73+/+ and TAp73−/− MEF cells were treated with ETP (20 μM) for the indicated durations and were analyzed by RT-PCR and WB.
(e) IMR90 cells transfected with p73 siRNA or control siRNA were treated with increasing amounts of ETP. G6PD expression was analyzed by RT-PCR and WB.
(f) Schematic representation of human G6PD genomic structure. Shown are the exon/intron organization, consensus p53 family protein RE, the potential p73 RE within the first intron, and the corresponding mutant p73 RE (with mutated nucleotides underlined). Arrows mark the positions of the primers used for PCR in the ChIP assay. UTR: un-translated region.
(g) A549 cells that were treated with and without doxorubicin (DOX, 1 μg/ml) were analyzed by ChIP assay with the indicated antibodies. Data represent two independent experiments.
(h) H1299 cells transfected with indicated GFP plasmids were analyzed by ChIP assay using anti-GFP antibody or normal mouse IgG and G6PD specific primers. The DNA size standard (in base pairs) is shown on the left. Data represent two independent experiments.
(i) Binding of endogenous p53, p63, and p73 to the puma genomic locus in A549 cells was analyzed by ChIP assay. Anti-p21 antibody and no antibody (−) were used as controls.
(j,k) Luciferase reporter constructs containing the wild-type (j) or mutant (k) p73 RE were transfected into H1299 cells together with increasing amounts of indicated plasmids. Renilla vector pRL-CMV was used as a transfection internal control. Relative levels of luciferase are shown. Data are means ± SD (n = 3 independent experiments).
(l) Protein expression of G6PD, TAp73, p53 and p21 in MCF-7 (wild-type p53), SK-BR-3 (p53 R175H), and MDA-MB-468 (p53 R273H) cells treated with p53 or control siRNA. Western blots represent three independent experiments.
p73 is activated by DNA damage signals27-30. When TAp73+/+ MEFs were treated with the genotoxic agent etoposide (ETP), TAp73 was stabilized and G6PD expression was elevated (Fig. 3d). In contrast, ETP reduced G6PD expression in TAp73−/− MEFs (Fig. 3d). When IMR90 cells were treated with ETP, TAp73 was also stabilized, and both the G6PD mRNA and protein were significantly increased. However, when p73 was knocked down, G6PD failed to accumulate upon DNA damage (Fig. 3e). Together, these results suggest that TAp73 stimulates the expression of G6PD both at basal levels in unstressed cells and when TAp73 is stabilized by DNA damage. In addition, a TAp73-independent mechanism might reduce G6PD expression upon DNA damage, but TAp73 can override this effect.
To determine whether TAp73 is a transcriptional activator for the G6PD gene, we analyzed the human G6PD gene sequence for potential p53 family protein response elements (REs), which share the consensus sequence of 5′-RRRCWWGYYY-(0-13bp spacer)-RRRCWWG YYY-3′ (where R is a purine, Y a pyrimidine, and W an A or T)31. One potential RE was identified in the second intron of G6PD (Fig. 3f). To investigate the binding of p73 to this RE, we performed chromatin immunoprecipitation (ChIP) assays. In the p53-wild-type A549 cells, endogenous p73 bound to the RE region of the G6PD gene; the binding was strengthened when cells were treated with the DNA damaging agent doxorubicin (DOX), which stabilized the p53 family proteins (Fig. 3g and Supplementary Fig. S2h). We observed no association between endogenous p53 or p63 and the G6PD gene under these conditions (Fig. 3g), even though p53 bound strongly to its target gene Puma as expected (Fig. 3i). Similarly, when H1299 cells were transfected individually with green fluorescence protein (GFP) fusions of p53 family members, GFP-TAp73α, but not GFP-p53 or GFP-TAp63α, associated with the G6PD genomic DNA (Fig. 3h).
To evaluate whether the RE within G6PD confers TAp73-dependent transcriptional activation, we cloned DNA fragments containing the wild-type RE or a mutant RE into the promoter region of a firefly luciferase reporter plasmid. Both TAp73α and TAp73β were able to induce luciferase expression from the wild-type reporter plasmid but not from the mutant reporter plasmid (Fig. 3j,k). Consistent with previous observations32,33, TAp73β had higher transactivation activity compared to TAp73α (Fig. 3j). In contrast, p53, TAp63a, and ΔNp73 failed to activate the wild-type RE-responsive luciferase (Fig. 3j and Supplementary Fig. S2i). These results suggest that TAp73, not p53 or TAp63, binds to the consensus p53 family RE within the second intron of G6PD and activates G6PD expression.
Tumor-derived p53 mutants may impair the function of TAp73 34. We tested two common p53 mutants, R175H and R273H. Over-expression of either mutant failed to suppress G6PD expression regardless whether TAp73 was co-expressed (Supplementary Fig. S2j). Also, knocking down these mutant forms of p53 did not affect the expression of G6PD (Fig. 3l). Thus, mutant p53 proteins may not impede the ability of TAp73 to stimulate G6PD expression.
p73 regulates NADPH homeostasis through G6PD
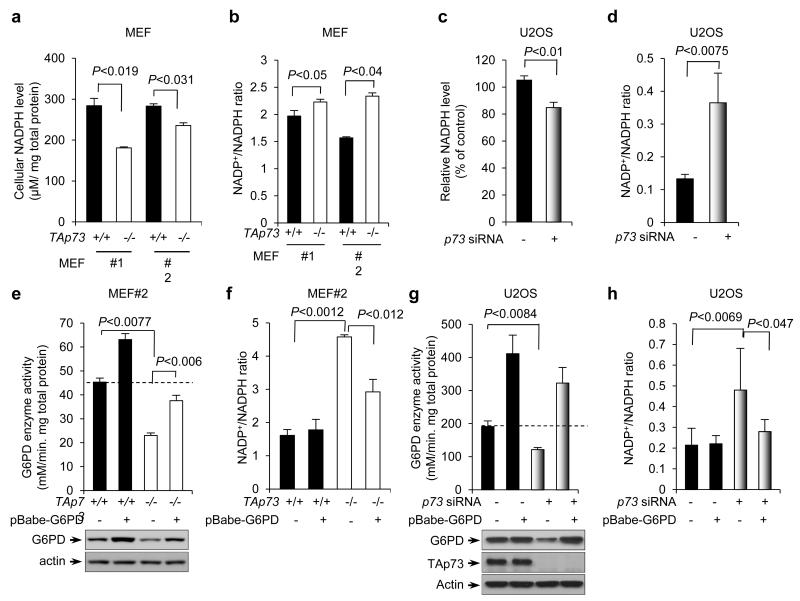
The PPP is a primary source of cellular NADPH35. In MEF cells, the lack of TAp73, but not ΔNp73, yielded noticeably reduced NADPH levels and increased NADP+/NADPH ratios (Fig. 4a,b and Supplementary Fig. S3a,b). Consistently with this finding, silencing p73 in U2OS and IMR90 cells lessened NADPH levels and raised NADP+/NADPH ratios (Fig. 4c,d and Supplementary Fig. S3c,d). Similar reductions in NADPH levels were also observed in p53+/+ and p53−/− HCT116 cells devoid of p73 (Supplementary Fig. S3e). To ascertain that the effect of TAp73 on NADPH is mediated by G6PD, we stably over-expressed G6PD in TAp73−/− MEF cells and p73-depleted U2OS cells with retrovirus-mediated gene transfer. This restored the levels of G6PD expression and activity and reduced the NADP+/NADPH ratios in these cells (Fig. 4e-h). Therefore, TAp73 maintains cellular NADPH content by stimulating the expression of G6PD.
Figure 4.
p73 regulates NADPH homeostasis.
(a,b) NADPH levels (a) and NADP+/NADPH ratios (b) in two independent clones of TAp73+/+ and TAp73−/− MEF cells are shown. Data are means ± SD (n = 3 independent experiments).
(c,d) NADPH levels (c) and NADP+/NADPH ratios (d) in U2OS cells transfected with control (−) or p73 siRNA. Data are means ± SD (n = 3 independent experiments).
(e,f) G6PD activity (e) and NADP+/NADPH ratios (f) in TAp73+/+ and TAp73−/− MEF cells stably overexpressing G6PD or vector control are shown. Protein expression was analyzed (e). Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
(g,h) G6PD activity (g) and NADP+/NADPH ratios (h) in U2OS cells stably overexpressing G6PD or vector control in the presence or absence of p73 siRNA are shown. Protein expression was analyzed (g). Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
A role of p73 in anti-oxidative defense
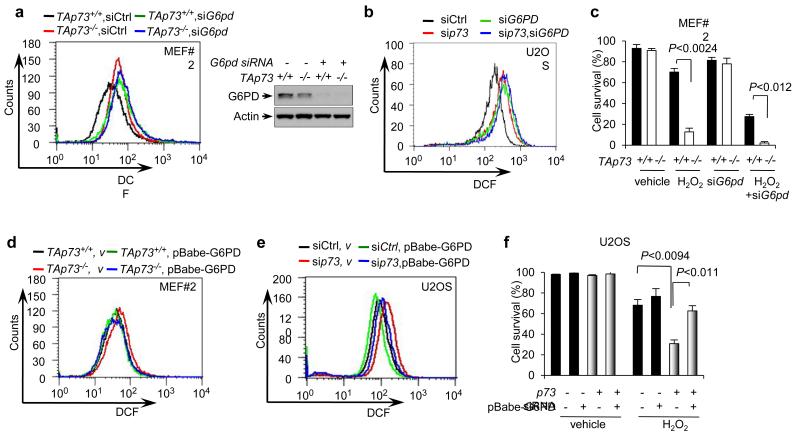
G6PD plays an important role in anti-oxidant metabolism by producing NADPH to regenerate reduced glutathione36,37. Treatment of TAp73+/+ MEFs with a G6PD-specific siRNA or the G6PD inhibitor dehydroepiandrosterone (DHEA) led to the accumulation of ROS (Fig. 5a and Supplementary Fig. S4a,b). Similarly, when G6PD was depleted in U2OS and IMR90 cells, ROS levels markedly rose (Fig. 5b and Supplementary Fig. S4c). Importantly, TAp73−/− MEFs also exhibited high ROS levels, similar to G6PD siRNA or DHEA-treated TAp73+/+ MEFs. In TAp73−/− MEFs, treatment with DHEA or G6PD siRNA only slightly increased ROS (Fig. 5a and Supplementary Fig. S4a,b). Similarly, U2OS and IMR90 cells devoid of p73 contained high levels of ROS, and knockdown of G6PD in these cells only raised ROS slightly higher (Fig. 5b and Supplementary Fig. S4c).
Figure 5.
A role of p73 in regulating ROS
(a) ROS levels in TAp73+/+ and TAp73−/− MEF cells treated with control or G6pd siRNA. Protein expression is shown (Right). Western blots represent three independent experiments.
(b) ROS levels in U2OS cells transfected with the indicated siRNA.
(c) TAp73+/+ and TAp73−/− MEF cells were treated with or without 150 μM H2O2 for 1 h and then cultured for 24 h in the presence or absence of G6pd siRNA. Cell viability was analyzed by trypan blue staining. Data are means ± SD (n = 3 independent experiments).
(d) ROS levels in TAp73+/+ and TAp73−/− MEF cells stably overexpressing G6PD or vector control. Protein expression is shown in Fig. 4e.
(e) U2OS cells stably overexpressing G6PD or vector control were transfected with p73 or control siRNA as indicated. ROS levels were analyzed.
(f) U2OS cells stably overexpressing G6PD or vector control were transfected with control siRNA (−) or p73 siRNA as indicated. Cells were treated with or without 100 μM H2O2 for 24 h and cell viability was analyzed by trypan blue staining. Data are means ± SD (n = 3 independent experiments).
We next examined the effect of TAp73 on cellular capacity to withstand oxidative stresses. Compared to TAp73+/+ cells, TAp73−/− MEFs were markedly more sensitive to hydrogen peroxide (H2O2) treatment (Fig. 5c and Supplementary Fig. S4d). Inhibition of G6PD by siRNA or DHEA reduced the survival of TAp73−/− cells and especially TAp73+/+ cells, minimizing their difference (Fig. 5c and Supplementary Fig. S4d). Knockdown of p73 in U2OS also sensitized these cells to H2O2 treatment, and silencing G6PD diminished the difference between these cells and the control cells (Supplementary Fig. S4e).
To ascertain that the sensitivity of TAp73-deficient cells to oxidative stresses is due to the reduction in G6PD expression, we tested the effect of G6PD expression in these cells. Forced expression of G6PD in TAp73−/−MEFs and p73-depleted U2OS cells reduced the ROS content (Fig. 5d,e). It also significantly enhanced the survival of p73-depeleted cells in the presence of H2O2 (Fig. 5f). Together, these data suggest that TAp73 plays a role in protecting against oxidative stress and that this effect of TAp73 is mediated, at least in part, by G6PD.
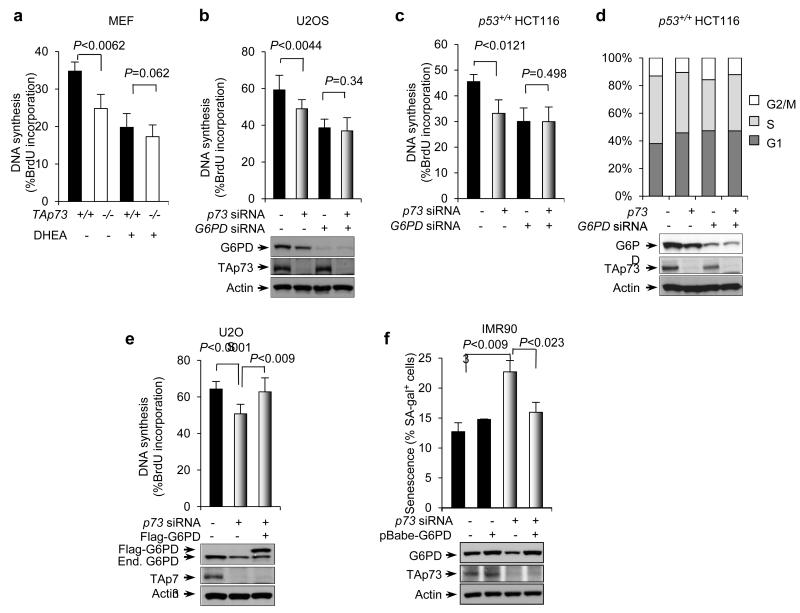
p73 regulates DNA biosynthesis and cell senescence
The PPP provides NADPH and ribose, both of which are important synthetic precursors for nucleic acids. We compared DNA synthesis in TAp73+/+ and TAp73−/− MEFs using 5-bromo-2′-deoxyuridine (BrdU) incorporation assays. DNA synthesis was sharply reduced in TAp73−/−MEFs compared to TAp73+/+ MEFs (Fig. 6a). Likewise, DNA synthesis slowed down when p73 was silenced in U2OS and various HCT116 cells (Fig. 6b,c and Supplementary Fig. S5a,b). In each cell type, treatment with DHEA or G6PD siRNA inhibited DNA synthesis in TAp73-depleted cells and especially in control cells, diminishing the difference between them (Fig. 6a-c and Supplementary Fig. S5a,b).
Figure 6.
p73 regulates DNA synthesis
(a) TAp73+/+ and TAp73−/− MEF cells were treated with or without DHEA. DNA synthesis was measured by BrdU incorporation. Percentages of BrdU-positive cells are shown as mean ± SD (n = 3 independent experiments).
(b, c) U2OS cells (b) and p53+/+ HCT116 cells (c) were transfected with control siRNA (−), p73 siRNA, and G6PD siRNA as indicated. Cells were assayed for BrdU incorporation. Protein expression is shown below. Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
(d) Cell-cycle profile of p53+/+ HCT116 cells transfected with p73 siRNA, G6PD siRNA, and control siRNA as indicated. Western blots represent three independent experiments.
(e) U2OS cells were transfected with control siRNA, p73 siRNA, and Flag-G6PD as indicated. BrdU incorporation (top) and protein expression (bottom) are shown. Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
(f) Percentage of SA-β-gal positive cells in IMR90 cells stably overexpressing G6PD or vector control in the presence or absence of p73 siRNA. Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
We also examined the effect of p73 on cell cycle progression. Knockdown of p73 led to a reduction in the number of cells in S phase and an increase in the number of cells in G1 and G2/M phases. This effect was independent of the status of p53 and the p53/p73-inducible cell cycle regulator p21. Knockdown of G6PD diminished the difference in DNA synthesis between p73-depleted cells and control cells (Fig. 6d and Supplementary Fig. S5c,d). Conversely, forced expression of G6PD almost completely restored DNA synthesis in p73-depleted U2OS cells (Fig. 6e). These results suggest that TAp73 stimulates DNA synthesis via G6PD.
The loss TAp73 in mouse cells accelerates cellular senescence38. We investigated the role of G6PD in this process using human IMR90 cells, a well-established model for senescence. Compared to cultures treated with control siRNA, IMR90 cultures treated with p73 siRNA showed a noticeable increase in the number of cells with senescence phenotypes, including flat morphology and the expression of the senescence-associated β-galactosidase (SA-β-gal) (Fig. 6f and Supplementary Fig. S5e). Knockdown of G6PD increased the number of senescent cells strongly in control cultures, but relatively moderately in p73-depleted cultures, reducing the difference between these two groups (Supplementary Fig. S5e). More importantly, over-expression of G6PD effectively reversed the induction of senescence in p73-depeleted cultures (Fig. 6f). These data suggest that the lack of TAp73 induces senescence in IMR90 cells due to the down-regulation of G6PD.
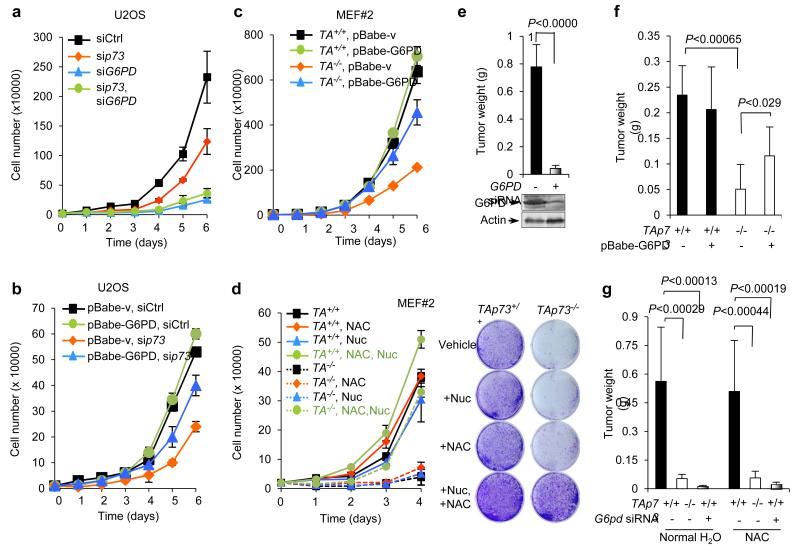
p73 enhances cell proliferation via G6PD
Knockdown of p73 strongly impeded cell growth in various cell lines, including MEFs, U2OS, and isogenic HCT116 cell lines (Fig. 1a,c, 7a, and Supplementary Fig. S1b,d, S6a-c). Growth delay due to p73 down-regulation also occurred in p53−/− HCT116 cells (Supplementary Fig. S6b), indicating a requirement for p73 in cell growth regardless of the status of p53. Depletion of G6PD reduced growth rate in control cells and especially in TAp73 knockdown cells (Fig. 7a and Supplementary Fig. S6a-c). Conversely, enforced expression of G6PD partially restored cell growth in p73 knockdown U2OS cells and TAp73−/− MEF cells but had a minimal effect on control cells (Fig. 7b,c and Supplementary Fig. S6d-h).
Figure 7.
p73 enhances cell growth through up-regulating G6PD
(a) Proliferation of U2OS cells transfected with control siRNA, p73 siRNA, and G6PD siRNA as indicated. Data are means ± SD (n = 3 independent experiments). Protein expression is shown in Fig. 6b.
(b) U2OS cells stably overexpressing G6PD or vector control were transfected with p73 or control siRNA as indicated. Cell proliferation is shown. Data are means ± SD (n = 3 independent experiments). Protein expression in is shown in Supplementary Fig. 4g.
(c) Growth of TAp73+/+ and TAp73−/− MEF cells stably overexpressing G6PD or vector control. Data are means ± SD (n = 3 independent experiments). Protein expression is shown in Fig. 4e.
(d) Growth of TAp73+/+ and TAp73−/− MEF cells medium containing 2 mM NAC and four ribonucleosides and four deoxyribonucleosides (Nuc) as indicated (left). Data are means ± SD (n = 3 independent experiments). Representative images of cells stained with crystal violet at day 4 (Right).
(e) U2OS cells expressing G6PD siRNA or control siRNA were individually injected subcutaneously (SC) in the dorsal flanks of the three nude mice. Tumor weights (mean ± SD, n=3 mice in each group) were measured 3 weeks after inoculation.
(f) TAp73+/+ and TAp73−/− MEF cells (2×106) stably overexpressing G6PD or vector control were individually injected subcutaneously (SC) in the dorsal flanks of the four nude mice. Tumor weights (mean ± SD, n=4 mice in each group) were measured 3 weeks after inoculation.
(g) TAp73+/+ and TAp73−/− MEF cells (1×106) expressing G6pd siRNA or control siRNA were individually injected subcutaneously (SC) in the dorsal flanks of the eight nude mice. Half were given water without NAC and the other half were given water with 40 mM NAC throughout the experiment. Tumor weights (mean ± SD, n=8 mice in each group) were measured 3 weeks after inoculation.
We examined whether the growth-stimulatory effect of TAp73 is related to both a decrease in ROS and an increase in intracellular nucleotides, two main outcomes of the PPP flux. We cultured TAp73+/+ and TAp73−/− MEF cells in medium containing the ROS scavenger N-acetyl-L-cysteine (NAC), four ribonucleosides (A, G, U, and C) plus four deoxynucleosides (dA, dG, dT, and dC), or both. Nucleosides in culture medium can be taken up by cells and converted to the corresponding nucleotides, thus bypassing the requirement for de novo ribose synthesis39,40. Importantly, in the presence of both NAC and nucleotides, but neither individually, TAp73−/− MEFs grew nearly as well as TAp73+/+ MEF cells (Fig. 7d). Moreover, combination treatment with NAC and nucleotides rescued the growth of TAp73−/− MEFs stably overexpressing G6PD or vector control, but to lesser extents compared to TAp73−/− MEFs (Supplementary Fig. S6e-h). These results suggest that TAp73 promotes cell proliferation at least partially through up-regulating G6PD, which leads to simultaneous reduction of ROS and enhancement of ribose synthesis.
Analysis of G6PD expression and metastasis status of a large number of primary breast tumors revealed that increasing G6PD expression was significantly correlated with the risk of metastasis (Supplementary Fig. S7a,b). To evaluate the role of G6PD in tumor formation, we injected immuno-compromised mice with U2OS cells or TAp73+/+ MEF cells expressing G6PD siRNA or control siRNA. G6PD siRNA-expressing cells generated much smaller tumors compared to control cells (Fig. 7e, g and Supplementary Fig. S7d). Conversely, reintroduction of G6PD partially restored tumor formation by TAp73−/− MEFs (Fig. 7f and Supplementary Fig. S7c). We also tested the effect of NAC on the growth of TAp73 and G6PD-deficient tumor cells. In agreement with cell proliferation results, NAC alone failed to reverse the inhibition of tumor growth derived from either TAp73−/− MEFs or G6PD-silenced MEFs (Fig. 7g and Supplementary Fig. S7d).
DISCUSSION
The present study indicates that TAp73 promotes G6PD expression and PPP flux, enhancing biosynthesis and ROS detoxification. Metabolic reprogramming in tumor cells is altered to meet the demand for robust production of macromolecules while minimizing oxidative damages17-19. Both require NADPH, and it has been proposed that NADPH production is a rate-liming step in cell proliferation17,18. Indeed, the production of NAPDH appears to be tightly controlled by oncogenes and tumor suppressors. For instance, the oncogene K-Ras stimulates NADPH production in pancreatic cancer cells by enhancing the flux through malic enzyme 1, a cytoplasmic isoform of the NADPH-generating malic enzymes41. In contrast, p53 inhibits the expression of all three malic enzymes that are localized either in the cytoplasm or in the mitochondria42. The PPP is not only a major source for NADPH but also highly responsive to oxidative stress37,43. G6PD is a common target for directly adjusting PPP flux. For example, it is allosterically activated by its substrate NADP+ and inhibited by its product NADPH. This homeostatic regulation ensures that the PPP is up-regulated when more NADPH is needed26. The stimulation of G6PD expression by TAp73 may represent a major mechanism that modulates G6PD expression. We found that G6PD over-expression partially restores the growth of TAp73−/− MEF cells and p73-depleted U2OS cells. This suggests that, at least in these cells, other TAp73 targets may contribute to its proliferative function. For example, a recent study revealed a role of TAp73 in regulating cytochrome c oxidase subunit 4 (Cox4i1), a subunit of the complex IV of the mitochondrial electron transport chain38. By maintaining levels of G6PD as well as Cox4il, TAp73 likely plays a pre-eminent role in anti-oxidant response in both unstressed and stressed cells.
Nevertheless, ROS scavengers alone cannot rescue the proliferative defects of TAp73-deficient cells. Rather, both ROS scavengers and nucleosides are needed (Fig. 7). These observations suggest that TAp73 promotes nucleotide synthesis as well as anti-oxidant defense, and that the PPP may be a main proliferative target of TAp73. Previous studies showed that upon over-expression, TAp73 could activate certain p53 target genes involved in apoptosis and cell cycle arrest. Yet, the physiological function of TAp73 is likely determined by its endogenous levels of expression, its interplay with the ΔN isoforms, and its regulation by upstream factors. Despite high similarity among p53 family members in the DNA-binding domain, TAp73 alone is able to induce G6PD expression, although the mechanism underlying this selectivity remains to be determined (Fig. 3). Moreover, ΔNp73 and tumor-associated p53 mutants do not appear to inhibit this function of TAp73 (Fig. 3). These observations support the notion that G6PD is a high-affinity, physiologically relevant target of TAp73, and that tumor cells benefit from TAp73 up-regulation even in the presence of elevated ΔNp73 or mutated p53 proteins.
The critical role of TAp73 in cell proliferation raises the question of why TAp73 deficient mice show increased tumor formation. An explanation may be that extensive genomic instability associated with congenital loss of TAp73 outweighs any proliferative defects in these mice by facilitating the accumulation of oncogenic mutations. It is noteworthy that the function of TAp73 in maintaining genomic stability appears to be cell type-specific, as it operates in lung fibroblasts but not in thymic cells, and that TAp73−/− mice primarily develop lung adenocarcinomas14. The role of TAp73 in G6PD expression, on the other hand, is observed in different cell types that have been tested in the current study. Rare mutations and prevalent up-regulation of TAp73 in a variety of human tumors suggest that human tumor cells opt for the powerful and probably ubiquitous proliferative advantages provided by TAp73.
Accumulating evidence indicates that PPP flux is enhanced in tumor cells either directly or indirectly. p53 suppresses the PPP through a direct interaction with G6PD. This activity is lost in tumor-derived p53 mutants24. PKM2, an isoform of pyruvate kinase that catalyzes the last step in glycolysis, is widely expressed in tumor cells. PKM2 has a low KM for its substrate, leading to the build-up of glycolytic intermediates for alternative pathways including the PPP44. The direct regulation of G6PD and PPP by two members of the p53 family, as well as indirectly by other mechanisms, underscores both their importance in tumorigenesis and their potential as therapeutic targets.
Methods
Antibodies and reagents
Antibodies against the following proteins/epitopes were used in this study with the company, catalog number, and dilution or concentration indicated: GFP (Clontech Laboratories, Mountain View, CA; 632381; 1:4,000), actin (Sigma-Aldrich, St Louis, MO; A2066; 1:4,000), Flag (Sigma, F3165, 1:4,000), p53 (DO-1) (Santa Cruz Biotechnology, Santa Cruz, CA; sc-126 HRP; 1:1,000), p73 (Imgenex Corp, San Diego, CA; IMG-259A; 1:1,000) (Bethyl Laboratories, Montgomery, TX; A300-126A; 1:1,000) (Sigma, SAB4300354, 1:1,000), mouse p73 (Santa Cruz, sc-7238, 1:500) (Cell Signaling Technology, Danvers, MA; 4300354; 1:1,000), G6PD (Sigma, HPA000834, 1:3,000), p21 (Cell Signaling, 2947, 1:1,000), and fluorescein isothiocyanate (FITC)-labeled anti-BrdU antibody (BD Bioscience Pharmingen; 556028; 10 μM). Anti-p63 antibody was kindly provided by Dr. C. Chen45.
The following reagents were purchased from Sigma: glucose-6-phosphate (G6P), 6-phosphogluconate (6PG), β-nicotinamide adenine dinucleotide 2′-phosphate (NADP+), β-nicotinamide adenine dinucleotide 2′-phosphate, reduced (NADPH), etoposide (ETP), doxorubicin (DOX), N-acetylcysteine (NAC), Propidium iodide (PI), dihydroepiandrosterone (DHEA), Crystal Violet (CV), Hydrogen peroxide (H2O2), 2′,7′-Dichlorofluorescin diacetate (DCF) and bromodeoxyuridine (BrdU).
Cell culture and gene knockdown with shRNA and siRNA
HCT116 cells and MEF cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Carlsbad, CA), and when indicated, in Minimum Essential Medium Eagle with ribonucleosides and deoxyribonucleosides (Sigma, M8042). TAp73−/−, ΔNp73−/−, and the corresponding wild-type MEFs have been previously described14,16. U2OS cells were maintained in McCoy’s 5A Medium, IMR90 cells in MEMα medium, and H1299 cells in RPMI1640 medium (Life Technologies, Carlsbad, CA). All mediums, if not specifically described, were supplemented with 10% fetal bovine serum (FBS).
Expression plasmids for shRNAs were made in a pLKO.1-puro vector. The targeted sequences were: p53, 5′-GACTCCAGTGGTAATCTAC-3′ 46; p63α, 5′-GGGTGAGCGTGTTATTGATGCT-3′ 47, and TAp73, 5′-GGATTCCAGCATGGACGTCTT-3′ 47 (all of human origin). The following siRNAs were purchased from Invitrogen (Carlsbad, CA) with the catalog number and sequences indicated: human G6PD, HSS103893, 5′-AAACCCACUCUCUUCAUCAGCUCGU-3′; human p73, HSS186396, 5′-GAGCUCGGGAGGGACUUCAACGAAG-3′; and rat/mouse G6PD, RSS302151, 5′-CCACCAAGCUGAUACACACAUAUUU-3′; human ΔNp73, 5′-UACGUCGGUGACCCCGCACGGU-3′.
siRNAs were transfected into cells using Lipofectamine RNAiMAX transfection Agent (Invitrogen, Carlsbad, CA) following the manufacturer’s instruction. Stable shRNA transfectants were selected in medium containing 1 μg/ml puromycin (Calbiochem, San Diego, CA, catalog No: 540222) as previously described48.
Semi-quantitative RT-PCR and quantitative RT-PCR.
Total RNA was isolated from cells by TRIzol® Reagent (Invitrogen, catalog No. 15596018) and 2 μg RNA of each sample was reversed to cDNA by First-strand cDNA Synthesis System (Marligen Biosciences, catalog No. 11801). 0.2 μg cDNA of each sample was used as a template to perform PCR. The primer pairs for human genes were: G6PD, 5′-ATGGCAGAGCAGGTGGCCCT-3′ and 5′-TCATGCAGGACTCGTGAATG-3′; β-actin, 5′-GACCTGACTGACTACCTCATGAAGAT-3′ and 5′-GTCACACTTCATGATGGAGTTGAAGG-3′; p53, 5′-CACGAGCTGCCCCCAGG-3′ and 5′-TCAGTCGACGTCTGAGT-3′; human p63, 5′-ACATGAATGGACTCAGCCCC-3′ and 5′-TCACTCCCCCTCCTCTTTGAT-3′; TAp73, 5′-ATGGCCCAGTCCACCGCCACC-3′ and 5′-TCGAAGGTGGAGCTGGG-3′. Primers for mouse genes were: G6pd, 5′-ATGGCAGAGCAGGTGG-3′ and 5′-CTCCTCCAGCTTAGGTC-3′; Actb, 5′-ACTACATTCAATTCCATC-3′ and 5′-CTAGAAGCACTTGCGGTG-3′.
G6PD enzyme activity
G6PD enzyme activity was determined as described49. The combined activity of G6PD and 6-phosphogluconate dehydrogenase (6PGD), the second enzyme of the PPP that also produces NADPH, was determined by the rate of conversion of NADP+ to NADPH in the presence of glucose-6-phosphate (G6P). 6PGD activity was measured by the conversion of NADP+ to NADPH in the presence of 6-phosphogluconate (6PG). G6PD activity was calculated by subtracting 6PGD from the combined activity. The reaction buffer contained 50 mM Tris (pH 8.1) and 1 mM MgCl2, and substrate concentrations were: G6P (200 μM) and 6PG (200 μM) and NADP+ (100 μM). Enzyme activities were normalized based on protein concentration, which was determined by a Bio-Rad protein assay kit (Bio-Rad, Richmond, CA, cat. no. 500-0006). The data are expressed in arbitrary absorption units.
Chromatin immunoprecipitation (ChIP) and Reporter assays
To identify potential p53 family protein response elements, we scan the G6PD gene using the Genomatix Promoter Inspector Program (Genomatrix Inc, Munich, Germany, software, http://www.genomatix.de). For ChIP assays, cells were cross-linked with 1% formaldehyde for 15 min at room temperature and cross-linking was stopped by the addition of 125 nM glycine (final concentration). Cell lysates were sonicated to generate DNA fragments with an average size below 1,000 bp and immunoprecipitated with indicated antibodies. Bound DNA fragments were eluted and amplified by PCR. Primer pairs were: g6pd, 5′-CCCTACTGTCCGTCTTACCTC-3′ (+6164/+6185) and 5′-CTCTCACTGCTGCACCCTTC-3′ (+6643/+6663); puma, 5-TGGCCTTGTGTCTGTGAGTAC-3; and 5-GGACAAGTCAGGACTTGCAGG-3′47.
For reporter assay, The G6PD genomic fragments (+6164 to +6663) containing either the wild-type (GGTCATGAGCAAACATGACC) or mutant (GGTTATTAGCAAATATTACC, with mutated nucleotides underlined) p73-binding region were cloned into pGL3-basic vector (Promega, Madison, WI, USA, catalog No: E1751). Luciferase reporter assays were performed as described previously42. Briefly, The reporter plasmids were transfected into p53-null H1299 cells together with a Renilla luciferase plasmid and increasing amounts of plasmids expressing p53 family proteins. 24 h after transfection, the luciferase activity was determined using a dual Luciferase Assay System (Promega, catalog No: E1910). Transfection efficiency was normalized on the basis of the Renilla luciferase activity.
NADPH and ROS levels
NADPH levels and NADP+/NADPH ratios were determined using the NADP+/NADPH Quantification kit (BioVision, Mountain View, CA, USA, catalog No: K347). ROS levels were analyzed as described50. Briefly, cells were incubated at 37°C for 30 min in phosphate buffered saline (PBS) containing 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2-DCFDA). Cells were then washed twice with PBS, treated with trypsin, and re-suspended in PBS. Fluorescence was immediately measured using a FACScan Flow Cytometer (Becton Dickinson, San Jose, CA).
PPP fluxes
The flux of the oxidative pentose phosphate pathway was measured based on the rate of glucose consumption and the ratio of 13C incorporated into carbon 2 (generated by glycolysis) and carbon 3 (generated by the PPP) of lactate determined by NMR spectroscopy as described previously24,51.
Analysis of cell cycle disruptions
Cells were washed with PBS twice and fixed with 75% ethanol overnight at −20 °C. cells were then treated with 0.1% tritonx-100 and 50 μg/ml RNase A for 30 minutes at 37 °C followed by PI staining. Cell cycle distribution was analyzed using a FACScalibur flow cytometer (BD Biosciences, CA). The data was analyzed using Flow Jo software (Tree Star, Ashland, OR).
BrdU incorporation assay
Cells were treated with G6PD siRNA or control siRNA. After indicated times, cells were pulse-labeled with BrdU (10 μM final concentration) for 1 h and analyzed by anti-BrdU immunostaining as described52. In brief, BrdU-labeled cells were washed with PBS and fixed sequentially with 2% paraformaldehyde and 70% ethanol at 4°C. Afterwards, cells were permeabilized with 0.2% TritonX-100 and washed with TBS. DNA was then denatured in 4N HCl-0.2% TritonX-100 for 2 h followed by sequential staining with anti-BrdU antibody and FITC-labeled BrdU specific antibody. The images were acquired with an Olympus DP71X microscope (Olympus, Center Valley, PA) and BrdU-positive cells were counted. The FITC signal was converted to a red color for better visualization.
Xenograft tumor models
Xenograft study was performed as described42. Briefly, cells were injected subcutaneously into the flanks of 4- to 5-wk-old athymic Balb-c nu/nu male mice (Taconic Farms, Germantown, NY). Three mice total were used for each treatment in Fig. 1b, 1d and Fig. 7e. Four mice (Fig. 7f) and eight mice (Fig. 7g) were used for each treatment. Tumor growth was evaluated at 2 or 3 weeks post-injection as indicated. All animal experiments were performed in accordance with relevant guidelines and regulations and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
Senescence-associated SA-β-gal activity
The SA-β-gal activity in cultured cells was determined using a Senescence Detection Kit (BioVision, Mountain View, CA, USA, catalog No: K320) following the manufacturer’s instructions. Percentages of SA-β-gal positive cells were calculated by counting more than 1,000 cells in random fields per cell line.
Cell proliferation assay and Crystal Violet (CV) staining of cells
Cell proliferation assay were performed as described42. Briefly, cells were transfected with siRNAs for 24 h and seeded in 6-well cell culture dishes in triplicates at a density of 5,000 or 20,000 cells as indicated per well in 2 ml of medium supplemented with 10% FBS. The medium was changed everyday. Cell number at the indicated time points was determined by counting using a hemocytometer. For Crystal Violet (CV) staining, cells were fixed with 10% Formalin for 5 minutes and stained with 0.05% CV for 30 minutes. After washed with distilled water, cells were photographed.
Statistical analysis
Statistical significance was analyzed by Student’s t-test and expressed as a P value.
Western blotting
Whole-cell lysates were made in modified RIPA lysis buffer (10 mM Tris–HCl at pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% NP-40, 1% Sodium deoxycholate, 0.025% SDS and complete protease cocktail) for 15 min on ice, and boiled in 2 × loading buffer. Protein samples were resolved by SDS-PAGE and transferred onto nitrocellulose membrane, which was blocked in 5% skim milk in TBST and probed with the indicated antibodies.
Univariable and multivariable survival analysis
We used a combined series of 871 breast cancer patients available through the GEO (accession numbers: GSE1456, GSE2990, GSE3494, GSE7390, GSE11121). All samples were profiled using the Affymetrix U133a microarray platform. Raw data were downloaded, processed using the RMA method, z-score transformed, and median centered. Probes mapping to the same gene were combined by using the probe with the highest variance. For Cox multivariable regression, G6PD and age were used as continuous variables, while other clinical and pathological factors were used as binary variables. Missing data were imputed. For Kaplan-Meier survival analysis, G6PD levels were discretized by using a mean cut-off. All survival analysis was performed using the R package “survival” version 2.37 and the R language and environment for statistical computing version 2.15.
Supplementary Material
Acknowledgements
We thank G. Melino, C. Chen, A. Tullo, U. M. Moll, M. C. Marín, B. Vogelstein, and W. El-Deiry for expression plasmids, antibodies, and/or cell lines; W. Wang, Y. Mei, N. Li, X. Wang, W. Tan, E. Witze, and K. E. Wellen for technical assistance; and C. O’Neill for help with manuscript preparation. Supported by China National Natural Science Foundation (30530200, 30871290 and 30728003), the Chinese Ministry of Science and Technology (2006CB910300 and 2010CB912804), and Chinese Academy of Sciences (KSCX1-YW-R-57) to M.W., and US National Institutes of Health (CA088868 and GM060911) and the Department of Defense (W81XWH-10-1-0468) to X.Y.
Footnotes
Author Contributions: W.D., P.J., X.Y. and M.W. designed the experiments and interpreted results. W.D. and P.J. performed all the experiments except those mentioned below. A.M. and P.J. analyzed the oxidative PPP flux. A.S. and M.B. helped with the FACS analysis and xenograft study, respectively. A.J.M. performed the breast cancer data analysis. T.W.M. supplied the MEF cells deficient of TAp73 and ΔNp73. W.D., P.J., and X.Y. wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Kaghad M, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 6.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 7.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 8.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 9.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 10.Yang A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 11.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Flores ER, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 13.Senoo M, Manis JP, Alt FW, McKeon F. p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell. 2004;6:85–89. doi: 10.1016/j.ccr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talos F, Nemajerova A, Flores ER, Petrenko O, Moll UM. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol Cell. 2007;27:647–659. doi: 10.1016/j.molcel.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 16.Wilhelm MT, et al. Isoform-specific p73 knockout mice reveal a novel role for {Delta}Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 19.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang P, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mancuso A, Sharfstein ST, Tucker SN, Clark DS, Blanch HW. Examination of primary metabolic pathways in a murine hybridoma with carbon-13 nuclear magnetic resonance spectroscopy. Biotechnol Bioeng. 1994;44:563–585. doi: 10.1002/bit.260440504. [DOI] [PubMed] [Google Scholar]
- 26.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 2007:577–589. [Google Scholar]
- 27.Gong JG, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 28.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 29.Yuan ZM, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 30.Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 32.Jost CA, Marin MC, Kaelin WG., Jr. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 33.De Laurenzi V, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Temple MD, Perrone GG, Dawes IW. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–326. doi: 10.1016/j.tcb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Pandolfi PP, et al. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filosa S, et al. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem J. 2003;370:935–943. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rufini A, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26:2009–2014. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dworkin CR, Gorman SD, Pashko LL, Cristofalo VJ, Schwartz AG. Inhibition of growth of HeLa and WI-38 cells by dehydroepiandrosterone and its reversal by ribo- and deoxyribonucleosides. Life Sci. 1986;38:1451–1457. doi: 10.1016/0024-3205(86)90557-6. [DOI] [PubMed] [Google Scholar]
- 40.Tian WN, et al. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998;273:10609–10617. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- 41.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anastasiou D, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15:1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 46.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 47.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 48.Godar S, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leopold JA, et al. Aldosterone impairs vascular reactivity by decreasing glucose-6-phosphate dehydrogenase activity. Nat Med. 2007;13:189–197. doi: 10.1038/nm1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cossarizza A, et al. Simultaneous analysis of reactive oxygen species and reduced glutathione content in living cells by polychromatic flow cytometry. Nat Protoc. 2009;4:1790–1797. doi: 10.1038/nprot.2009.189. [DOI] [PubMed] [Google Scholar]
- 51.Mancuso A, et al. Real-time detection of 13C NMR labeling kinetics in perfused EMT6 mouse mammary tumor cells and betaHC9 mouse insulinomas. Biotechnol Bioeng. 2004;87:835–848. doi: 10.1002/bit.20191. [DOI] [PubMed] [Google Scholar]
- 52.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.