Abstract
Objectives
To present a method for generating reference maps of typical brain characteristics of groups of subjects using a novel combination of rapid quantitative Magnetic Resonance Imaging (qMRI) and brain normalization. The reference maps can be used to detect significant tissue differences in patients, both locally and globally.
Materials and Methods
A rapid qMRI method was used to obtain the longitudinal relaxation rate (R1), the transverse relaxation rate (R2) and the proton density (PD). These three tissue properties were measured in the brains of 32 healthy subjects and in one patient diagnosed with Multiple Sclerosis (MS). The maps were normalized to a standard brain template using a linear affine registration. The differences of the mean value ofR1, R2 and PD of 31 healthy subjects in comparison to the oldest healthy subject and in comparison to an MS patient were calculated. Larger anatomical structures were characterized using a standard atlas. The vector sum of the normalized differences was used to show significant tissue differences.
Results
The coefficient of variation of the reference maps was high at the edges of the brain and the ventricles, moderate in the cortical grey matter and low in white matter and the deep grey matter structures. The elderly subject mainly showed significantly lower R1 and R2 and higher PD values along all sulci. The MS patient showed significantly lower R1 and R2 and higher PD values at the edges of the ventricular system as well as throughout the periventricular white matter, at the internal and external capsules and at each of the MS lesions.
Conclusion
Brain normalization of rapid qMRI is a promising new method to generate reference maps of typical brain characteristics and to automatically detect deviating tissue properties in the brain.
Introduction
Magnetic Resonance Imaging (MRI) is a very sensitive method for detecting focal changes in the brain. Using the common conventional imaging methods local tissue deviations are highlighted as darker or lighter areas depending on the choice of scanner parameters. Generally, however, no information is obtained on the absolute differences of the observed changes. This precludes objective measures that can determine the significance of deviating tissue properties. Particularly difficult is to detect diffuse changes throughout the brain since a slight change in signal intensity over a larger area may also be due to, for example, coil sensitivity differences or B1 field inhomogeneity. A direct comparison of changes across a group of subjects is virtually impossible, not only due to the differences in size and shape of the individual subjects, but also because the absolute image intensity in conventional MRI varies between subjects and examinations and thus cannot directly be compared.
Multi-parametric quantitative MRI (qMRI) can be used as a solution for objective measures of tissue properties. By qMRI, the physical properties that constitute an MR image, i.e. the longitudinal relaxation rate (R1), the transverse relaxation rate (R2), and the proton density (PD), are directly measured. The qMRI parameters provide an absolute scaling, independent of the MR scanner settings and imperfections [1]. Pathological processes such as axonal damage, gliosis, inflammation and edema are characterized by changes in relaxation behaviour and water content [2]–[6] and can therefore be detected as absolute differences in R1, R2 and PD values.
In order to generate reference maps of typical brain characteristics for a group, the quantitative maps of all subjects can be transformed into a standard stereotactic space. Spatial normalization to a common brain template [7] is a standard procedure, which has been used in many other applications, for example in studies on white and grey matter reduction in psychiatric and neurological disorders, e.g. Alzheimer’s disease and Multiple Sclerosis (MS) [8]–[10]. Another example is functional MRI (fMRI), where spatial normalization allows statistical comparisons of the blood oxygen level dependent (BOLD) time series in the participants during performance of cognitive tasks or during sensory stimuli [11]–[13]. In spatial normalization, the sum of the squared differences between the standard template and the images of individual subjects is minimized by spatial deformations and transformations of the intensity values in each image voxel. This technique is completely automatic and is therefore not sensitive to different experience and skill of the individual examiners. An additional advantage of normalization is that it enables the use of standardized atlases to define the various brain structures. By using this approach on qMRI, characteristic brain maps for an entire group of subjects can be generated, where for each voxel throughout the brain the average and standard deviation of the tissue properties are calculated. In previous work on normalized qMRI, various quantification methods were separately acquired and used for monitoring changes with aging or with diseases such as epilepsy and Multiple Sclerosis [14]–[20]. In this work a qMRI method was used that simultaneously measures all three MR tissue parameters R1, R2 and PD in a single acquisition within a clinically acceptable time, thereby providing both a rapid and objective method for tissue characterization.
There are two main applications of this technique: firstly, a single patient can be compared to a healthy reference group and specific deviations in the patient’s tissue values can be determined. Since the normal variation of the group is known, the significance of the individual differences can be calculated. This application may form a basis for objective pathology detection without user interaction. Secondly, two or more groups can be compared with each other, such that the individual (focal) changes average out, whereas common, more diffuse differences between brain tissue properties become emphasized. Depending on the size of the group, a high statistical power can be achieved. For both approaches atlas-based definitions can be used for the various brain structures, in order to avoid a subjective drawing of an area of interest.
The aim of this work was to propose a method of generating maps of typical brain characteristics in groups of healthy subjects and patients. To demonstrate the method, cerebral R1, R2 and PD maps were measured by rapid qMRI in a group of healthy subjects and normalized to a standard brain template. A standard brain atlas was used to calculate the mean qMRI values in a number of regions of interest. As an example of the usability of the method towards automatic pathology detection, the tissue characteristics of an elderly subject and a patient diagnosed with MS were compared to the reference values of the healthy group.
Methods
Ethics Statement
The study was approved by the Regional Ethics Committee in Linköping (Dnr. M88-07) and written informed consent was obtained from all participants.
Workflow
The workflow of the proposed method is schematically depicted in Fig. 1. The measured R1, R2 and PD maps were used to generate a T2-weighted image using the approach of synthetic MRI [21], [22]. The synthetic T2-weighted image was normalized to a standard T2-weighted brain template using affine transformation. The same transformation was used to normalize the separate maps. The normalized R1, R2 and PD maps were averaged to obtain mean values and standard deviations of the complete group. Finally, the differences between an individual and a reference group, or between two groups, can be determined. For a Gaussian distribution a significant (p<0.05) deviation of an individual compared with the mean of a reference group is found at a difference of, at minimum, 1.96 times the standard deviation. This threshold is generally somewhat higher due to the finite size of the reference group. For a comparison between two groups a two-sided t-test with assumed unequal variances is more appropriate, assuming that the measurement values for both groups have a normal distribution in R1, R2 and PD.
Figure 1. Schematic overview of the method.
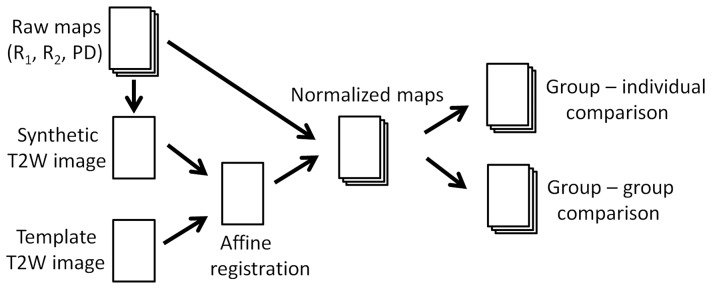
Magnetic Resonance quantification maps of R1 and R2 relaxation rates and proton density (PD) are used to synthesize a T2-weighted image. This image is normalized against a standard template T2-weighted image using affine registration. The same transformation is used to normalize the R1, R2 and PD maps. The individual quantitative maps from a group of subjects are averaged to obtain reference maps of typical cerebral tissue parameters. Such reference maps can either be compared to an individual brain or with another group.
Subjects
A group of 32 healthy subjects was examined using qMRI. Of these, 31 were used for the normalized reference group (14 male, 17 female, mean age 45±11 years, range 26–67 years). The remaining healthy subject, a female aged 72 years, was used as one example for the technique. The subjects were regarded as healthy based on an interview on disease history. Additionally, one patient (female, aged 40 years), diagnosed with Clinically Definite Multiple Sclerosis (Relapsing Remitting, disease duration 8 years, Expanded Disability Status Scale EDSS = 3.5) was used as a second example to demonstrate the usability of the method. All data was stored in the hospital’s research PACS.
Scanning Protocol
For quantification the QRAPMASTER (also ‘qMap’) sequence [23] was used. This is a multi spin-echo saturation recovery sequence with 4 saturation delays and 5 echoes. The saturation delays were at 100, 400, 1380 and 2860 ms with a repetition time (TR) of 2950 ms. The echo time (TE) was set to 14, 28, 42, 56 and 70 ms. Hence each acquisition leads to a matrix of 4×5 = 20 images per slice. The in-plane resolution was 1×1 mm2 over a field of view (FOV) of 210 mm; 30 axial slices of 4 mm thickness (no gap) were collected in a scan time of 8∶02 minutes. In addition,n T2-weighted (TR/TE = 6000/100 ms) and FLAIR (TR/TE/TI = 6000/120/2000 ms) images were acquired with the same FOV and number of slices in order to compare qMRI data with conventional imaging. The MR-scanner was an Achieva 1.5 T (Philips Healthcare, Best, The Netherlands).
Image Post-Processing
The raw image data were analyzed with the SyMRI Brain Studio software (SyntheticMR AB, Linköping, Sweden) to retrieve the R1, R2 and PD maps as described previously [23]. Using the same software the Brain Parenchymal Fraction (BPF), the total brain volume divided by the intracranial volume, was automatically measured. Additionally the lateral ventricles were manually segmented to obtain the Lateral Ventricle Fraction (LVF), defined as the total lateral ventricle volume divided by the intracranial volume.
Based the R1, R2 and PD maps a stack of T2-weighted images was synthesized using an TE = 100 ms and a TR = 4500 ms. These images were used as source images to calculate the transformation matrix to a standard stereotactic space in Montreal Neurological Institute (MNI) coordinates using SPM8 (Wellcome Department of Imaging Neuroscience, University College, London, UK). As a first step in this normalization process, the synthetic T2-weighted images were smoothed with an 8 mm Gaussian kernel to reduce the individual anatomical detail. This is required since the source and the MNI template should have approximately the same degree of smoothing. In this study we used the standard T2-weighted template available in SPM8. The synthetic T2-weighted images were transformed to match the size and position of the template by a 12-parameter (translation, rotation, shear, zoom) affine regularization. The same transformation matrix was then applied to the R1, R2 and PD maps. The resulting data was re-gridded to 2×2×2 mm3 to obtain an isotropic dataset. Mean values of R1, R2 and PD, standard deviations σ(R1), σ(R2) and σ(PD) and coefficients of variation (CoV = σ/mean) for all voxels in the brain were calculated.
In order to characterize different structures in the brain, we used the Wake Forrest University (WFU) PickAtlas [24] to define regions of interest (ROI). Six ROIs were predefined using the Automated Anatomical Labeling (AAL): the lateral ventricles, insula, the cingulate cortex, caudate nucleus, putamen, thalamus and five ROIs using the Talairach Daemon (TD): midbrain, pons and corpus callosum as well as regions representing whole brain grey matter (GM) and white matter (WM) Four ROIs were created by intersecting the WM ROI with corresponding cerebral lobes, thus obtaining frontal, parietal, occipital and sub-lobar WM. Mean value and standard deviation (SD) for R1, R2 and PD were calculated for all 15 predefined ROIs by taking the average R1, R2 and PD values within each individual ROI for the healthy group. Linear regression was performed on these values as a function of subject age.
To determine significant tissue deviations of the elderly subject and the MS patient, the differences between the quantitative tissue characteristics of the subject and the corresponding mean values of the reference group, ΔR1 = R1,s – R1,mean, ΔR2 = R2,s – R2,mean and ΔPD = PDs – PDmean, were normalized with the standard deviation σ, obtaining ΔR1/σ(R1), ΔR2/σ(R2) and ΔPD/σ(PD). Only significant differences were displayed by showing all data that was higher than a threshold of 2.04, corresponding to p = 0.05 for a reference group of 31 subjects. In order to combine all three qMRI maps the magnitude of the normalized vector sum S was calculated, defined as:
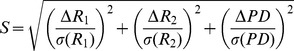 |
(1) |
In the visualization of the combined map all voxels that exceeded a value of 5 (corresponding to p<0.000001) were indicated with a color.
Results
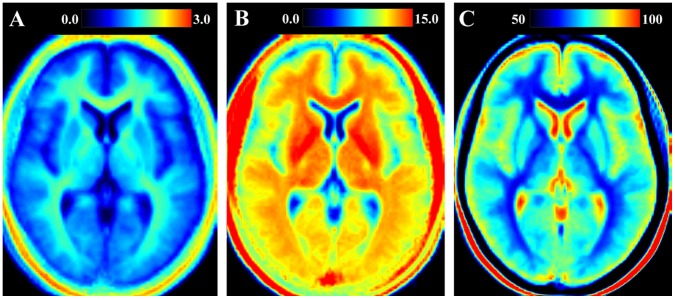
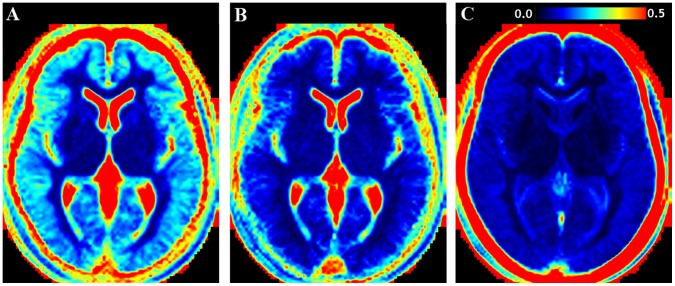
In Fig. 2 one axial slice of the normalized, mean R1, R2 and PD maps of the group of healthy subjects is shown. Clear differences in R1, R2 and PD values between the various brain structures are visible. Predefined, atlas-based ROIs that characterize the typical values of the healthy group are listed in Table 1. Also in Table 1 the change in qMRI values per year of subject age is displayed. Significant changes (written in bold face) were observed in all three qMRI parameters in the WM ROIs and in the caudate nucleus. In addition, significant correlation with age was observed for R1 in cortical GM and for R2 in the thalamus. In Fig. 3 the CoV maps of the three parameters are displayed. The CoV was especially high (>0.4, red in Fig. 3) at the edges of the ventricles and around the brain periphery. For R1 the CoV was also relatively high for the cortical grey matter, in the range 0.20–0.25 (light-blue), whereas it was approximately 0.10–0.15 for R2 and PD in the same area (dark blue). The CoV in white matter and in the deep grey matter structures such as the head of the caudate nucleus, the putamen and the hypothalamus was generally in the range 0.05–0.10 for R1, 0.03–0.07 for R2 and 0.02–0.05 for PD. Bands of higher CoV values at tissue interfaces, which might indicate problems in the normalization process, could not be observed.
Figure 2. Example of the normalized qMRI reference maps in a slice through the brain.
The figure shows the reference maps obtained from images pooled across a group of 31 healthy subjects with A: R1 relaxation rate on a scale 0–3 s−1, B: R2 relaxation rate on a scale 0–15 s−1 and C: proton density on a scale 50–100%, where 100% corresponds to pure water at 37°C.
Table 1. The mean value and standard deviation of the R1, R2 and PD values in various regions of interest, defined by using the PickAtlas.
| ROI | R1 | R2 | PD | R1 slope | R2 slope | PD slope |
| (s−1) | (s−1) | (%) | (s−1/year) | (s−1/year) | (%/year) | |
| Lateral ventricles | 0.38±0.32 | 3.89±2.35 | 94.3±8.8 | −0.0075 | −0.046 | 0.17 |
| Insula | 0.80±0.07 | 8.94±0.73 | 83.1±1.5 | −0.0004 | −0.006 | 0.00 |
| Cingulate cortex | 0.97±0.05 | 10.23±0.36 | 79.9±1.0 | −0.0007 | −0.005 | 0.01 |
| Caudate nucleus | 1.00±0.15 | 10.61±1.51 | 80.3±2.8 | −0.0088 | −0.091 | 0.15 |
| Cortical gray matter | 1.04±0.09 | 9.87±0.44 | 75.1±2.4 | 0.0031 | −0.006 | −0.03 |
| Pons | 1.24±0.07 | 11.34±0.50 | 73.5±1.5 | −0.0003 | 0.005 | 0.00 |
| Putamen | 1.25±0.05 | 13.07±0.42 | 75.2±1.6 | 0.0008 | 0.001 | −0.03 |
| Mid brain | 1.28±0.05 | 11.93±0.33 | 73.2±0.9 | −0.0010 | −0.007 | 0.01 |
| Thalamus | 1.33±0.07 | 12.81±0.21 | 72.5±1.9 | −0.0016 | −0.009 | 0.04 |
| Occipital white matter | 1.33±0.04 | 11.80±0.26 | 71.2±1.1 | −0.0005 | −0.004 | 0.00 |
| Frontal white matter | 1.33±0.06 | 11.77±0.32 | 71.1±1.4 | −0.0040 | −0.016 | 0.08 |
| Parietal white matter | 1.34±0.05 | 11.76±0.25 | 71.0±1.2 | −0.0002 | 0.000 | 0.01 |
| Sublobar white matter | 1.37±0.08 | 11.20±0.46 | 69.2±1.9 | −0.0043 | −0.026 | 0.10 |
| White matter | 1.38±0.05 | 11.79±0.26 | 70.0±1.0 | −0.0023 | −0.011 | 0.05 |
| Corpus callosum | 1.39±0.27 | 10.71±1.58 | 68.3±5.6 | −0.0084 | −0.057 | 0.16 |
Added are the linear regression slopes with subject age. Significant slopes (p<0.05) are displayed in bold face.
Figure 3. The coefficient of variation (CoV) of the reference maps.
The figure shows CoV in the same slice as visualized in Fig. 2, with A: CoV map of R1, B: CoV map of R2 and C: CoV map of PD. The same scaling of [0–0.5] was used in all CoV maps.
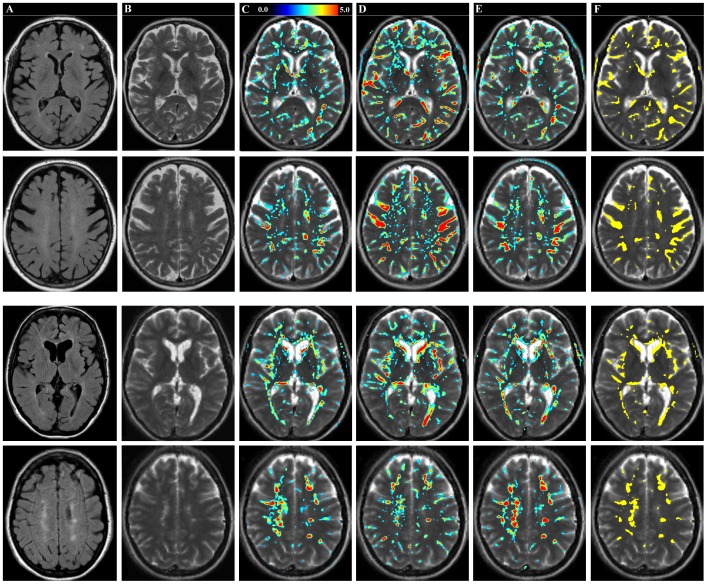
In Fig. 4 the normalized difference of the R1, R2 and PD maps of the elderly subject and the MS patient in comparison with the mean reference maps of the healthy group are displayed for two slices through the brain. Note that the scale for R1 and R2 relaxation is inverted compared to PD because typical differences between the examples and the group of healthy controls consisted of a decrease in R1 and R2 and an increase in PD. For the elderly subject clear changes were observed along the sulci. A number of focal changes were seen in the periventricular area and minor differences around the ventricles. For the MS patient the largest differences were observed as a rim along the ventricular system. Moreover moderate changes in the MS patient’s R1, R2 and PD are observed in the periventricular white matter, capsula interna, capsula externa and centrum semiovale. The MS lesions, visible as hyper-intensities in the FLAIR and T2-weighted images in Fig. 4A and B, also show a significant decrease in R1, R2 and increase in PD compared to healthy white matter at the same location. In the cortical grey matter no significant tissue deviations were observed. In Fig. 4F the vector sum is displayed where only differences are shown that exceed a threshold of 5. This map clearly displays the changes at the sulci of the elderly subject and at the edges of the ventricles, in the capsula externa and at all lesions of the MS patient.
Figure 4. Significant deviation of qMRI tissue properties in an elderly subject and an MS patient.
Shown are a FLAIR (A) and T2-weighted image (B) of two axial slices of the head of an elderly subject (female, 72 years old, top two rows) and an MS patient (female, 40 years old, bottom two rows). The color overlay in C corresponds to the normalized difference ΔR1/σ(R1) between the subject and the group of healthy controls on the same slices. Only values higher than a threshold of 2.04are shown. Synthetic T2-weighted images are calculated using the R1, R2 and PD maps and used as background images. Similar for ΔR2/σ(R2) in D and ΔPD/σ(PD) in E. In F the magnitude of the normalized vector sum of ΔR1/σ(R1), ΔR2/σ(R2) and ΔPD/σ(PD) is shown where voxels that exceed a threshold of 5 are indicated in yellow.
In order to verify the presence of brain atrophy the BPF and LVF were measured for all participants. The mean BPF of the reference group was 90.0±2.5%. Linear regression showed a decrease of BPF of 0.12% (p = 0.003) per year of age. The mean LVF was 1.1±0.5% in the reference group. Linear regression showed an increase of LVF of 0.025% (p<0.001) per year of age. The elderly subject had a BPF of 80.8% and an LVF of 1.7%. This BPF was rather small and significantly deviated from the reference group (−9.2%, p<0.001), although the subject was enlisted as healthy. The LVF difference was not significant (+0.6%, p = 0.2). The MS patient, on the other hand, had a BPF of 84.9% and an LVF of 3.6%. This situation was the reverse in comparison with the elderly subject, the BPF difference was just at the normal significance threshold (−5.1%, p = 0.05) whereas the LVF was significantly higher (+2.5%, p<0.001).
Discussion
A method is proposed to characterize brain tissue using a combination of rapid quantitative MRI and brain normalization to enable an automatic and objective comparison of tissue properties between groups and individuals. Using this method each voxel throughout the brain lies at the same stereotactic location for all subjects and the quantitative R1, R2 and PD values can be investigated for differences between groups and individuals. An atlas-based definition ensures an objective description of the various brain structures. The CoV is a good measure for the local sensitivity of the method since low CoV values are only possible when the tissue has low variation in qMRI values and normalization worked well for all subjects. The observed low CoV in white matter and deep grey matter structures of the group of healthy subjects indicates that the method is particularly suitable in these areas. Demonstration of the method on an elderly subject and an MS patient resulted in the observation of significant differences in all three normalized qMRI maps. Combining the maps into a normalized vector map clearly highlighted the subject-specific differences compared to a healthy reference group.
The qMRI values presented in Table 1 were similar to previously reported values with the same quantification method of the brain [23], [25] and close to values obtained by other methods [26]–[28]. The WM values were lower than previously reported values (e.g. T1 = 608 ms (R1 = 1.64 s−1) in [27] and T1 = 602 ms (R1 = 1.66 s−1) in [28]) but this seems mainly to be due to the rather wide region of interest defined in AAL, resulting is a minor overlap with GM areas for our method. A change of qMRI values with subject age was only observed in WM and the caudate nucleus. Although it seems likely that indeed changes in WM are larger than in GM in relation with age it must be noted that the sensitivity of the method was better in WM and the deep GM structures in comparison to cortical GM, which may partially obscure potential changes in cortical GM.
It is not straightforward to attribute the observed differences in the qMRI maps to specific causes regarding tissue microstructure. It seems likely that the changes in the elderly subject at the sulci indicate brain atrophy. This conclusion is supported by the measured low BPF inn the elderly subject. For the MS patient similar effects were observed at the edges of the brain stem and the ventricular system, supported by the large LVF observed in that patient. For results related to potential brain atrophy the decrease of R1 and R2 relaxation and increase in PD was caused by a partial volume effect where brain tissue was replaced by cerebrospinal fluid, since the total brain parenchymal volume decreased beyond the normal variation in the healthy subjects. The observed changes in the white matter and in the MS lesions, on the other hand, were more likely caused by actual changes in the brain tissue properties, which can be confirmed by the presence of the hyperintensities in the FLAIR and T2-weighted images. The significant deviations in the white matter in the vicinity of the capsula interna and externa, however, were not unambiguous to interpret. These R1, R2 and PD deviations could be caused by an actual change of the white matter tissue properties or rather a result of imperfect brain normalization. In a recent study by Van Hecke and co-workers [29] diffusion differences between MS patients and controls in this area was observed, indicating white matter aberrations in these regions. It is recognized that objective methods, such as qMRI, are important to quantify differences between patients and the healthy population [30], but more investigation is required to establish the distinction between different causes such as atrophy and actual tissue changes. A potential improvement would be the definition of a geometrical mask to exclude areas with high CSF content or tissue interfaces, to limit the effect of brain atrophy in the qMRI maps.
A limitation of the method is the potential inadequate ability of the geometrical morphing algorithm to successfully register the various brain structures. The natural variation of the size and shape of the brain and of the ventricular system resulted, even for the healthy population, in a high CoV at the interface between the brain and CSF. In addition, the complex shape of the cortical windings is different for each individual, resulting in a higher CoV in the entire cortex. This limits the statistical power in these areas. Resampling the qMRI maps to lower resolution will improve the registration but a trade-off must be made between maintaining anatomical detail and minimizing the CoV on tissue interfaces. It can be expected that these aspects will be particular challenging for patients with e.g. severe brain atrophy, hydrocephaly or very large brain pathologies. In future studies we will explore the possibilities for ameliorated normalization by using source images with higher resolution for the calculation of the transformation matrix and by using improved normalization algorithms that, besides a linear affine registration, also apply non-linear procedures.
An area of interest is the choice of the significance threshold. In many applications a significance level of p<0.05 is used, which also was applied in the present study for calculating the tissue maps in Fig. 4C–E. For a normal t-distribution this choice implies that there may be 5% of all pixels in an image that appear to deviate significantly purely by chance. For an image matrix of 256×256 pixels statistically this corresponds to over 3000 pixels. Increasing the threshold to t = 5 decreases this number to less than 0.5 pixels, i.e. it is not likely that any pixel in the image will appear to deviate by chance. This threshold was used for the combined quantitative map displayed in Fig. 4F. Further validation is required, however, to define a threshold which constitutes an optimal balance between false positives and false negatives that might be appropriate for clinical applications.
In the future we will aim for automatic pathology detection based on this method. Besides the potential for brain atrophy and MS lesion detection, the proposed method could be applied to objectively measure the volume of brain tumors or the edemic area after stroke [31]–[33]. A significant advantage of the method is that individual segmentation of the brain is no longer required since all brains are mapped to the same standard brain template. Using a standard atlas removes ambiguity concerning the definitions of regions of interest. An inverse transformation of the resulting pathology maps can be co-registered to the conventionally acquired images to indicate the findings and obtain the true volumes of the ROIs. In this work it has been shown that tissue deviations in individual patients can automatically be found. For pathology detection, however, it is required that the method is validated by confirmation of a neuroradiologist. Validation can be combined with a more thorough analysis of the observed qMRI differences using more sophisticated methods such as multivariate or principle component analysis, rather than the example of the vector sum.
The alternative application of the method, to compare two or more groups of subjects to find group-specific features, was not investigated here. Analyzing groups, rather than individuals, will increase the statistical power substantially. However, the method will then be less sensitive to individual, focal changes but will highlight more diffuse differences, common for the whole group. Therefore the proposed method is expected to be suitable for the investigation of diffuse white matter changes such as in Normal Appearing White Matter or Dirty Appearing White Matter (NAWM/DAWM) in MS. It is known that there is only a moderate correlation between the focal changes in the brain of MS patients and clinical disability [34], [35]. We believe that this technique can be applied to shed more insights into the simultaneous diffuse global changes [36]–[39].
In conclusion, we have shown that it is possible to characterize tissue properties for each voxel throughout the brain for a group of subjects using brain normalization of multi-parametric qMRI data. A standard brain atlas can be used to define regions of interest for objective comparison. The method allows automatic determination of significant tissue differences, exemplified with and elderly subject and a patient with MS in comparison to a healthy reference group. The results from this study are promising since the method is automatic, objective and may lead to robust pathology detection.
Acknowledgments
Dr. Anne-Marie Landtblom is acknowledged for patient recruitment.
Funding Statement
The study was supported by grants from The National Research Council (VR/NT), the University Hospital of Linköping and the County Council of Östergötland. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tofts P (2003) Quantitative MRI of the Brain. Wiley.
- 2. Kumar R, Delshad S, Woo MA, Macey PM, Harper RM (2012) Age-related regional brain T2-relaxation changes in healthy adults. J Magn Reson Imaging 35: 300–308. [DOI] [PubMed] [Google Scholar]
- 3. Neema M, Stankiewicz J, Arora A, Dandamudi VS, Batt CE, et al. (2007) T1- and T2-based MRI measures of diffuse gray matter and white matter damage in patients with multiple sclerosis. J Neuroimaging 17: 16S–21S. [DOI] [PubMed] [Google Scholar]
- 4. Oh J, Cha S, Aiken AH, Han ET, Crane JC, et al. (2005) Quantitative apparent diffusion coefficients and T2 relaxation times in characterizing contrast enhancing brain tumors and regions of peritumoral edema. J Magn Reson Imaging 21: 701–708. [DOI] [PubMed] [Google Scholar]
- 5. Larsson HB, Frederiksen J, Petersen J, Nordenbo A, Zeeberg I, et al. (1989) Assessment of demyelineation, edema and gliosis by in-vivo determination of T1 and T2 in the brain of patients with acute attack of Multiple Sclerosis. J Magn Reson Med 11: 337–338. [DOI] [PubMed] [Google Scholar]
- 6. Vymazal J, Righini A, Brooks RA, Canesi M, Mariani C, et al. (1999) T1 and T2 in the brain of healthy subjects, patients with Parkinsons disease and patients with Multiple System Athrophy: Relation to iron content. Radiology 211: 489–495. [DOI] [PubMed] [Google Scholar]
- 7. Friston KJ, Ashburner J, Frith CD, Poline J-B, Heather JD, et al. (1995) Spatial registration and normalization of images. Hum Brain Mapp 3: 165–189. [Google Scholar]
- 8. Selvaraj S, Arnone D, Job D, Stanfield A, Farrow TF, et al. (2012) Grey matter differences in bipolar disorder: a meta-analysis of voxel-based morphometry studies. Bipolar Disord 14: 135–145. [DOI] [PubMed] [Google Scholar]
- 9. Li J, Pan P, Huang R, Shang H (2012) A meta-analysis of voxel-based morphometry studies of white matter volume alterations in Alzheimer’s disease. Neurosci Biobehav Rev 36: 757–763. [DOI] [PubMed] [Google Scholar]
- 10. Riccitelli G, Rocca MA, Pagani E, Martinelli V, Radaelli M, et al. (2012) Mapping regional grey and white matter atrophy in relapsing-remitting multiple sclerosis. Multiple Sclerosis J 18: 1027–1037. [DOI] [PubMed] [Google Scholar]
- 11. Engström M, Vigren P, Karlsson T, Landtblom AM (2009) Working Memory in 8 Kleine-Levin Syndrome Patients: An fMRI Study. Sleep 32: 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Ettinger-Veenstra HM, Ragnehed M, Hällgren M, Karlsson T, Landtblom AM, et al. (2010) Right-hemispheric brain activation correlates to language performance. NeuroImage 49: 3481–3488. [DOI] [PubMed] [Google Scholar]
- 13. Larsson MBO, Tillisch K, Craig AD, Engström M, Labus J, et al. (2012) Brain Responses to Visceral Stimuli Reflect Visceral Sensitivity Thresholds in Patients With Irritable Bowel Syndrome. Gastroenterology 142: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pell GS, Briellmann RS, Waites AB, Abbott DF, Jackson GD (2004) Voxel-based relaxometry: a new approach for analysis of T2 relaxometry changes in epilepsy. Neuroimage 21: 707–713. [DOI] [PubMed] [Google Scholar]
- 15. Aubert-Broche B, Grova C, Pike GB, Collins DL (2009) Clustering of atlas-defined cortical regions based on relaxation times and proton density. Neuroimage 47: 523–532. [DOI] [PubMed] [Google Scholar]
- 16. Hasan KM, Walimuni IS, Abid H, Wolinsky JS, Narayana PA (2012) Multi-modal quantitative MRI investigation of brain tissue neurodegeneration in multiple sclerosis. J Magn Reson Imaging 35: 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hasan KM, Walimuni IS, Abid H, Datta S, Wolinsky JS, et al. (2012) Human brain atlas-based multimodal MRI analysis of volumetry, diffusimetry, relaxometry and lesion distribution in multiple sclerosis patients and healthy adult controls: implications for understanding the pathogenesis of multiple sclerosis and consolidation of quantitative MRI results in MS. J Neurol Sci. 313: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saito N, Sakai O, Ozonoff A, Jara H (2009) Relaxo-volumetric multispectral quantitative magnetic resonance imaging of the brain over the human lifespan: global and regional aging patterns. Magn Reson Imaging 27: 895–906. [DOI] [PubMed] [Google Scholar]
- 19. Walimuni IS, Hasan KM (2011) Atlas-based investigation of human brain tissue microstructural spatial heterogeneity and interplay between transverse relaxation time and radial diffusivity. Neuroimage 57: 1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alfano B, Brunetti A, Larobina M, Quarantelli M, Tedeschi E, et al. (2000) Automated segmentation and measurement of global white matter lesion volume in patients with multiple sclerosis. J Magn Reson Imaging 12: 799–807. [DOI] [PubMed] [Google Scholar]
- 21.Riederer SJ, Lee JN, Farzeneh F, Wang HZ, Wright RC (1986) Magnetic resonance image synthesis: Clinical implementation. Acta Radiol Suppl 369: 466–468. [PubMed]
- 22. Bobman SA, Riederer SJ, Lee JN, Suddarth SA, Wang BP, et al. (1985) Cerebral magnetic resonance image synthesis. Am J Neuro Rad 6: 265–269. [PMC free article] [PubMed] [Google Scholar]
- 23. Warntjes JBM, Dahlqvist Leinhard O, West J, Lundberg P (2008) Optimization for Clinical Usage of Rapid Magnetic Resonance Quantification on the brain. Magn Reson Med 60: 320–329. [DOI] [PubMed] [Google Scholar]
- 24. Maldjian JA, Laurienti PJ, Kraft RA, Burdettea JH (2003) An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage 19: 1233–1239. [DOI] [PubMed] [Google Scholar]
- 25. West J, Warntjes JBM, Lundberg P (2012) Novel whole brain segmentation and volume estimation using quantitative MRI. Eur Radiol 22: 998–1007. [DOI] [PubMed] [Google Scholar]
- 26. Deoni SCL, Peters TM, Rutt BK (2005) High-Resolution T1 and T2 Mapping of the Brain in a Clinically Acceptable Time with DESPOT1 and DESPOT2. Magnetic Resonance in Medicine 53: 237–241. [DOI] [PubMed] [Google Scholar]
- 27. Neeb H, Zilles K, Shah NJ (2006) A new method for fast quantitative mapping of absolute water content in vivo. Neuroimage 31: 1156–1168. [DOI] [PubMed] [Google Scholar]
- 28. Deichmann R (2005) Fast high-resolution T1 mapping of the human brain. Magn Reson Med 54: 20–27. [DOI] [PubMed] [Google Scholar]
- 29. Van Hecke W, Nagels G, Leemans A, Vandervliet E, Sijbers J, et al. (2010) Correlation of Cognitive Dysfunction and Diffusion Tensor MRI Measures in Patients With Mild and Moderate Multiple Sclerosis. J Magn Res Imag 31: 1492–1498. [DOI] [PubMed] [Google Scholar]
- 30. Filippi M, Agosta F (2010) Imaging biomarkers in multiple sclerosis. J Magn Reson Imaging 31: 770–788. [DOI] [PubMed] [Google Scholar]
- 31. Loubinoux I, Volk A, Borredon J, Guirimand S, Tiffon B, et al. (1997) Spreading of vasogenic edema and cytotoxic edema assessed by quantitative diffusion and T2 magnetic resonance imaging. Stroke 28: 419–426. [DOI] [PubMed] [Google Scholar]
- 32. Verhaert D, Thavendiranathan P, Giri S, Mihai G, Rajagopalan S, et al. (2011) Direct T2 quantification of myocardial edema in acute ischemic injury. JACC Cardiovasc Imaging 4: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sbarbati A, Reggiani A, Nicolato E, Arban R, Lunati E, et al. (2002) Regional changes in the contralateral “healthy” hemisphere after ischemic lesions evaluated by quantitative T2 parametric maps. Anat Rec 266: 118–122. [DOI] [PubMed] [Google Scholar]
- 34. Barkhof F (2002) The clinico-radiological paradox in multiple sclerosis revisited. Curr Opin Neurol 15: 239–245. [DOI] [PubMed] [Google Scholar]
- 35. Chard D, Miller D (2009) Is multiple sclerosis a generalized disease of the central nervous system? An MRI perspective. Curr Opin Neurol 22: 214–218. [DOI] [PubMed] [Google Scholar]
- 36. Filippi M, Tortorella C, Bozzali M (1999) Normal-appearing white matter changes in multiple sclerosis: the contribution of magnetic resonance techniques. Mult Scler 5: 273–282. [DOI] [PubMed] [Google Scholar]
- 37. Vrenken H, Geurts JJ, Knol DL, van Dijk LN, Dattola V, et al. (2006) Whole-brain T1 mapping in multiple sclerosis: global changes of normal-appearing gray and white matter. Radiology 240: 811–820. [DOI] [PubMed] [Google Scholar]
- 38. Stevenson VL, Parker GJ, Barker GJ, Birnie K, Tofts PS, et al. (2000) Variations in T1 and T2 relaxation times of normal appearing white matter and lesions in multiple sclerosis. J Neurol Sci 178: 81–87. [DOI] [PubMed] [Google Scholar]
- 39. Neema M, Goldberg-Zimring D, Guss ZD, Healy BC, Guttmann CR, et al. (2009) 3T MRI relaxometry detects T2 prolongation in the cerebral normal-appearing white matter in multiple sclerosis. Neuroimage 46: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]