Abstract
The phage-shock protein PspE and GlpE of the glycerol 3-phosphate regulon of Salmonella enterica serovar Typhimurium are predicted to belong to the class of thiosulfate sulfurtransferases, enzymes that traffic sulfur between molecules. In the present study we demonstrated that the two genes contribute to S. Typhimurium virulence, as a glpE and pspE double deletion strain showed significantly decreased virulence in a mouse model of systemic infection. However, challenge of cultured epithelial cells and macrophages did not reveal any virulence-associated phenotypes. We hypothesized that their contribution to virulence could be in sulfur metabolism or by contributing to resistance to nitric oxide, oxidative stress, or cyanide detoxification. In vitro studies demonstrated that glpE but not pspE was important for resistance to H2O2. Since the double mutant, which was the one affected in virulence, was not affected in this assay, we concluded that resistance to oxidative stress and the virulence phenotype was most likely not linked. The two genes did not contribute to nitric oxid stress, to synthesis of essential sulfur containing amino acids, nor to detoxification of cyanide. Currently, the precise mechanism by which they contribute to virulence remains elusive.
Introduction
The Gram-negative bacterium Salmonella enterica serovar Typhimurium (S. Typhimurium) is a major pathogen of both animals and humans. It invades epithelial cells of the small intestine and causes inflammation of this organ, usually leading to a self-limiting gastroenteritis [1]–[2]. In mice, the bacterium causes a typhoid-like systemic disease. Important features of this manifestation include the ability to invade in the intestine, to infect and kill macrophages, to survive and replicate within dendritic cells and macrophages and to spread to the reticulo-endothelial system of organs such as liver and spleen [1]–[2]. In order to do so, S. Typhimurium possesses several virulence factors that are often encoded as gene clusters on so called Salmonella pathogenicity islands (SPIs). Two of the major SPIs, SPI-1 and SPI-2, encode type three secretion systems (T3SSs) that inject effector molecules into the host cell to mediate the invasion process and intracellular survival [3].
Salmonella has to cope with several stress conditions during the infection process [4]–[5]. Nitric oxide (NO) stress caused by the release of NO and reactive nitrogen species (RNS) is one such stress factor [6]–[10]. NO and RNS nitrosylate and inactivate reactive metal centers and iron-sulfur clusters, thereby inhibiting the functionality of key bacterial enzymes, such as metabolic, respiratory and DNA synthesizing proteins [6]–[10]. Thus to carry out the infection, Salmonella has to activate several defense mechanisms to detoxify NO and RNS, and to repair the damages that they cause [6], [10]–[11]. Oxidative stress is another host defense mechanism that Salmonella has to overcome during infection, and several stress systems specifically deal with detoxification of oxygen radicals [4].
Sulfurtransferases shuffle sulfur between molecules [12]–[13]. The enzymes have a carboxy-terminal domain carrying an active-site cysteine, which is important for the sulfur transport [12]. Among other functions, sulfurtransferases are believed to be involved in sulfur metabolism [14]–[15], cyanide detoxification [16]–[17] and the repair and assembly of the iron-sulfur clusters mentioned above as targets for the damaging effects of NO and RNS [13], [18]–[21].
PspE and GlpE are single-domain thiosulfate sulfurtransferases (TSTs), which have been demonstrated in vitro to have rhodanese activity in Escherichia coli [22]–[24]. The enzymes can detoxify cyanide; however, other substrates such as dithiols may also be utilized by these enzymes [23]–[24]. Recently PspE has been categorized in E. coli as a periplasmic rhodanese. It was shown to contribute to the restoration of disulphide bond formation in proteins in the cell envelope in a DsbA mutant in conjunction with the protein DsbC [25]. No function has yet been attributed to these two proteins in Salmonella.
PspE is a member of the phage-shock protein (Psp) system that responds to membrane stress (reviewed in [26]–[27]). Expression of pspE in S. Typhimurium occurs together with the other genes of the pspABCDE operon from the σ54-dependent psp promoter [28]. However, pspE expression is likely to happen from an intrinsic, pspE-specific σ70-dependent promoter [29], as shown for E. coli [30]. Expression of pspE is highly induced during infection of eukaryotic cells [31]–[32], which indicates a role in host pathogen interaction.
glpE is a member of the glpEGR operon [33]. Transcription in E. coli has been shown to occur from a cyclic AMP-cAMP receptor protein (cAMP-CRP) complex-dependent promoter, generating one polycistronic glpEGR mRNA [34]. Furthermore, glpG and/or glpR genes are transcribed from three additional promoters [33]. GlpR is a repressor of the glycerol 3-phosphate regulon, thus involved in the metabolism of glycerol 3-phosphate and its precursors [35]. However, glpE might function independent of the other members of the glp regulon, as it does not contribute to the metabolism of glycerol 3-phosphate in E. coli [22], [24]. In E. coli the cytoplasmic protein GlpE and the periplasmic protein PspE show functional redundancy and together they are responsible for 95% of the thiosulfate sulfurtransferase activity [24].
Given that pspE is highly expressed in S. Typhimurium during infection [31]–[32], and given that GlpE and PspE seem to have overlapping functions in E. coli [24], we hypothesized that their combined activity might be important for virulence of S. Typhimurium. We demonstrated that this is indeed the case, and that virulence in a mouse model was affected when both genes were inactivated, but not when single genes were knocked-out. Despite further studies using cell culture models and different in vitro growth and survival assays, however, we failed to identify the mechanism by which these proteins contribute to virulence.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacteria used in this study are listed in Table 1. Deletion of single genes with parallel insertion of a resistance cassette in S. Typhimurium 4/74 was performed using the Lambda Red recombination system as described [38]. Sequences of oligonucleotides used for Lambda Red mediated mutagenesis and PCR verifications are listed in Table 2. Insertions were confirmed by PCR and sequencing, using standard procedures. Phage P22HT105/int−201-mediated transduction was performed as described previously [41] to transfer mutations to a clean 4/74 background and to generate double knockout mutants.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype | Reference or source |
| JEO3774 | S. Typhimurium 4/74 wild type | [36] |
| JEO4560 | S. Typhimurium 4/74 ΔglpE::Kan (kanr) | This work |
| JEO4865 | S. Typhimurium 4/74 ΔglpE::Kan+pINS05 (kanr ampr) | This work |
| JEO4835 | S. Typhimurium. 4/74 ΔpspE::Cm (cmr) | This work |
| JEO4867 | S. Typhimurium 4/74 ΔpspE::Cm+pINS02 (cmr, ampr) | This work |
| JEO4836 | S. Typhimurium 4/74 ΔglpE::Kan/ΔpspE::Cm (kanr, cmr) | This work |
| JEO4869 | S. Typhimurium 4/74 ΔglpE::Kan/ΔpspE::Cm +pINS04 (kanr, cmr, ampr) | This work |
| JEO3775 | S. Typhimurium 4/74 ΔinvH201::TnphoA | [37] |
| pKD3 | Red template for amplification of Cm resistance cassette (ampr, cmr) | [38] |
| pKD4 | Red template for amplification of Kan resistance cassette (ampr, Kanr) | [38] |
| pKD46 | Vector for Lambda Red mediated mutagenesis; λ-Red expression from arabinose-inducible promoter; temperature sensitive (ampr) | [38] |
| pACYC177 | Cloning vector (ampr, kanr) | [39] |
| pINS02 | pspE in pACYC177 (ampr) | This work |
| pINS04 | glpE and pspE in pACYC177(ampr) | This work |
| pINS05 | glpE in pACYC177 (ampr, kanr) | This work |
| E. coli TOP10 | Chemically competent E. coli cloning strain | Invitrogen |
| KP1274 | S. Typhimurium LT2 (restriction-deficient) | [40] |
Table 2. Oligonucleotide sequences for PCR based amplification and sequencing.
| Primer | Sequence | Application |
| glpE | for: 5′GCCTTAATTGGCTTCACCGGCAATAATGAAAGAGCGATTCTGTGTAGGCTGGAGCTGCTTC′3 | Lambda Red recombination |
| rev: 5′ CCCTTCTCGTCGGAGGCTGCATCTTTATGGGATTACGCGCATATGAATATCCTCCTTAG′3 | ||
| pspE | for: 5′TGCGTTAGCGTTATTCGTAGCCATGCCGCTTTATGCCGCATGTGTAGGCTGGAGCTGCTTC′3 | Lambda Red recombination |
| rev: 5′CGGCATATCAAGACGACTGATGCCGCCCATATTCATCGCGCATATGAATATCCTCCTTAG′3 | ||
| glpE_C | for: 5′ACGCGCTATTGGTAACATCG′3 | Proof of Lambda Red mutation |
| rev: 5′CCTGGGTGGCCATGTAATCA′3 | ||
| pspE_C | for: 5′CTGGAGCCAGTACTTCGTAAG′3 | Proof of Lambda Red mutation |
| rev: 5′GCAGCCTGCGTTATAGATATGTGC′3 | ||
| glpE_BamHI | for: 5′ATGGATCCCAGACGGAACTCGTAGTGCT′3 | Complementation |
| rev: 5′ATGGATCCGAGGATTCGCAAAGGAAGTA′3 | ||
| pspE_HindIII | for: 5′ACTGAAGCTTGCTACGTCACGTCAGATACG′3 | Complementation |
| rev: 5′ACTGAAGCTTACTACCCGGAGAGATCAACA′3 | ||
| pACYC177_BamHI | for: 5′CGGTTCGGTTTATTGACGAC′3 | Proof of insertion |
| rev: 5′ACCAGTGCCCTTCTGATGAA′3 | ||
| pACYC177_HindIII | for: 5′CGTATTTCGTCTCGCTCAG′3 | Proof of insertion |
| rev: 5′CGACTCGTCCAACATCAATA′3 | ||
| hilA _q | for: 5′AACACTGTACGGACAGGGCTATCGG′3 | qPCR [42] |
| rev: 5′TACCATCGGGTATCATCTGCCCGGA′3 | ||
| invG_q | for: 5′GGAACAATGATCCGAGTGCT′3 | qPCR |
| rev: 5′ACGCATCTACGGAGGTGGTA′3 | ||
| prgH_q | for: 5′GTCCTGTGCGGTAATCTGCT′3 | qPCR |
| rev: 5′ATGGAAACTCACAGCCGTTC′3 | ||
| sopB_q | for: 5′ACTCAGCAGCAGGATGGCTTACCTG′3 | qPCR [42] |
| rev: 5′TCATGCACACTCACCGTGGACATCC′3 | ||
| pspE_q | for: 5′GCCGCAGAATACTGGATAGATG′3 | qPCR |
| rev: 5′TCCTGTCTGGAACTACCGTTTC′3 | ||
| glpE_q | for: 5′AGAGGCGTATCAGAAACTGCAC′3 | qPCR |
| rev: 5′CCAGCGTATCGTTAGTCAAGTG′3 | ||
| rsmC_q2 | for: 5′GAAAAGCAGCCGCAGTTTAG′3 | qPCR |
| rev: 5′CAGTTGGCTACCAACATCCA′3 | ||
| nusG_q2 | for: 5′GTCCGTTCGCAGACTTTAAC′3 | qPCR |
| rev: 5′GCTTTCTCAACCTGACTGAAG′3 |
Strains were maintained in LB-Lennox broth (LB). For growth on solid media, LB was enriched with 1.5% agar producing LB agar plates. If not stated otherwise, bacteria were grown in M9 minimal salt medium (containing per liter: 12.8 g Na2HPO4-12H20, 3.0 g KH2PO4, 0.5 g NaCl, 1.0 g NH4Cl, 0.1 mM CaCl2, 2 mM MgSO4 and 0.4% Glucose) at 37°C, 200 rpm for 16–18 h. When necessary, media was supplemented with antibiotics at the following concentration: 100 µg ampicillin ml−1, 50 µg kanamycin ml−1 and 10 µg chloramphenicol ml−1.
Construction of Complementation Plasmids
glpE and pspE-specific PCR products plus their upstream located promoter regions (approx. 400 bp) were cloned into pACYC177 [39] following standard procedures. Oligonucleotides used for construction of complementation plasmids and verificatoin of insertions are listed in Table 2. The constructs were transformed into One Shot® E. coli TOP10 chemically competent cells following the recommendations given by the supplier (Invitrogen). Insertion of glpE and pspE was confirmed by PCR and sequencing. The plasmids were further transformed into KP1274 [40], a restriction-deficient Salmonella strain, and finally to glpE and pspE mutant strains to test for genetic complementation. Expression of glpE and pspE genes from the complementation plasmids was confirmed by qPCR (see method below).
RNA Extraction and qPCR
Bacteria were grown to logarithmic phase in M9 (OD600nm = 0.4±0.01). RNA was isolated from 1.5 ml aliquots by mechanical disruption with the FastPrep system (Bio101; Q-biogene) and help of the RNeasy mini kit (Qiagen). Quantity and quality of total RNA was checked with the NanoDrop 1000 spectrophotometer (Thermo Scientific) and on a 1.5% (w/v) agarose gel. All enzymatic steps described below were performed according to the supplier’s recommendation (Fermentas). The RNA was DNase treated with the RNase free DNaseI kit and reverse transcribed with the RevertAid H minus reverse transcriptase kit. The qPCR was done with the Maxima SYBR Green/Rox qPCR Master Mix and gene specific oligonucleotides (Table 2) in a MxPro3000 cycler (Stratagene). qPCR was performed in parallel in samples of the wild type and mutant strains. Data were normalized against two reference genes, rsmC and nusG [32], [43], producing similar results. Relative gene expression (change fold = CF) in mutant strains compared to the wild type, was calculated by help of the 2−ΔΔCT method [44] corrected by the different primer efficiencies [45].
The Contribution of the Gene Products to Sulfur Metabolism
The ability to grow in M9 media which contains MgSO4 as the only sulfur source was measured. Bacteria from exponentially growing cultures in LB were collected by centrifugation, washed in PBS and re-suspended in the M9 medium at an OD600 value of 0.005. The contribution of the enzymes to metabolism of thiosulfate and sulphite was investigated by parallel incubation of wild type and mutated strain in TSI-agar (Oxoid CM0277, Thermo Scientific) and Iron-sulfite agar (Oxoid CM0079, Thermo Scientific) for 24 hours at 37°C.
Induction of Membrane Stress by SDS
Growth was determined in presence of 0.01% (w/v) and 0.1% (w/v) SDS. Bacteria were grown to stationary phase in LB medium and adjusted to the same number. Bacteria were spotted on LB agar plates as previously described [46]. Prior to spotting the plates were adjusted to the test condition by addition of SDS, and growth was evaluated after 16–18 h of incubation at 37°C. As a control, growth on LB agar plates without SDS was followed in parallel. For control of plate assay, a broth assay was also performed with one of the concentrations. 20 ml of M9 media in 100 ml test tubes with 0.01% SDS was inoculated with colony material of each strain from an LB plate (OD600 value of 0.05), and growth of the bacteria at 37°C with shaking was followed.
Resistance Towards Cyanide
The ability to detoxify cyanide was determined by growth in basis medium (in 1 liter containing 10 g peptone; 5 g NaCl; 0.225 g KH2PO4; 5.64 g Na2HPO4; pH = 7,4) supplemented with potassium cyanide (KCN) at the following concentrations: 0,3 mg/l; 0,6 mg/l; 3 mg/l; 15 mg/l; 75 mg/l. Media was inoculated with a colony of bacteria grown on LB agar, and incubated at 37°C. Growth was evaluated after two days of incubation. As a control, growth of each strain in non-supplemented basis medium was investigated in parallel.
Resistance Towards H2O2
Strains were inoculated in LB media and incubated overnight at 37°C. The next day a dilution with PBS was made to OD600 = 0.2. The exact CFU (T0) was determined by plating in duplicate on LA agar using the glass bead method. H2O2 was added to a concentration of 10 mM, and CFU was determined at times T1, T2, T3 and T6 hours as mentioned for T0.
Resistance to NO Stress
Resistance to NO stress was tested in growth experiments in the presence of S-Nitrosoglutathione (GSNO; Sigma-Aldrich) and in survival experiments after exposure to peroxynitrite (Caymen Chemicals). To determine the exact concentration of peroxinitrite, absorbance at 302 nm (A) was measured and the concentration C (C = A/E) was calculated based on the extinction coefficient ε = 1670 M−1 cm−1.
For growth experiments in the presence of GSNO, stationary phase bacterial cultures were adjusted to an optical density (OD) at 600nm of 0.005 in fresh M9 medium supplemented with 0.1 mM, 0.25 mM, 0.5 mM and 1 mM GSNO. Growth was performed in 96-well plates and followed over a period of 20 h by OD600nm measurements in a microplate spectrophotometer (PowerWave XS, Biotek) with intermediate shaking of the plate.
For survival studies, logarithmically grown cultures in M9 (OD600nm = 0.04±0.01) were treated with 360 µM peroxynitrite and samples were taken after 0 and 15 min. To determine the number of bacteria in the samples, serial dilutions were plated on LB agar plates. Survival of bacteria was determined by calculating the number of bacteria as colony forming units (CFU) per ml (CFU ml−1) after 15 min in relation to the number of bacteria at the beginning of the experiment. In growth and survival experiments in the presence of NO stress, two non-treated controls were investigated in parallel: one in non-supplemented medium and one in medium diluted with the corresponding compound solvent (0.3 M NaOH for peroxynitrite and distilled H20 for GSNO experiments = 1∶10 diluted M9).
Epithelial Cell Infection
Invasion of cultured epithelial INT-407 cells was investigated using a Gentamicin protection approach as described previously [47] with few modifications. In brief INT-407 cells were grown in MEM+Glutamax™-I, Earles, 25 mM HEPES (Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Invitrogen) at 37°C in an atmosphere containing 5% CO2. 18–20 h prior to infection, cells were seeded in 24-well plates at 1×105 cells per well. Bacteria grown to stationary phase were re-grown to logarithmic phase in M9 (OD600nm = 0.4±0.01) and were added to INT-407 cells at a multiplicity of infection (MOI) of 100. After 15 min of infection at 37°C and 5% CO2, cells were treated with 100 µg ml−1 gentamicin for 1 h to kill extracellular bacteria and dissolved in 0.1% (v/v) Triton X-100. Serial dilutions of the lysates were spread on LB agar plates to determine the number of invaded bacteria as CFU ml−1. To adjust for day to day variation, values were adjusted against CFU ml−1 of the wild type. In parallel bacterial enumerations of the inoculum was determined to ensure equal starting numbers.
Infection of Macrophages
Cell culture experiments with J774.1A macrophages were performed essentially as previously described [48]. In brief J774.1A cells were grown in RPMI1640+Glutamax™-I, 25 mM HEPES (Gibco), supplemented with 10% (v/v) heat-inactivated FBS at 37°C and 5% CO2. 20–24 h prior to infection, cells were seeded in 24-well plates at 4×105 cells per well. Bacteria grown to stationary phase in M9 were harvested at 8000 rpm for 5 min, re-suspended in 0.9% (w/v) NaCl, complement-opsonized and added to the cells at a MOI of 10∶1. A separate experiment was set up to demonstrate that the assay was capable of showing differences in survival capability between strains. This assay employed the wild type strain 14028 and an isogenic strain with insertion mutation of the SPI-2 gene ssaV, which causes attenuation for intra cellular propagation [49]. After 25 min of infection at 37°C, 5% CO2 (designated time zero p.i.), cells were further incubated in the presence of 100 µg ml−1 gentamicin for 1 h. The cells were either dissolved in 0.1% (v/v) Triton-X 100 (intracellular survival 1 h p.i.), or further incubated in the presence of 25 µg ml−1 gentamicin for a total incubation time of 4 h and 24 h (intracellular survival/replication 4 h and 24 h p.i.), and then dissolved as described above. In the control experiment, survival/replication was only followed for 20 hours. Serial dilutions were plated on LB agar plates to determine bacterial numbers of the inoculum (to ensure equal starting numbers) and the lysates as CFU ml−1. Values determined at T4 and T24 were expressed relatively to CFU for the WT determined at T1.
Cytotoxicity Towards Macrophages
Cytotoxicity of bacteria towards macrophages was determined by measuring release of the lactate dehydrogenase (LDH). Infection studies with J774.1A macrophages and bacteria grown to stationary phase in M9 were performed as described above. 24 h p.i., supernatants were collected and LDH release was determined using the CytoTox 96®Non-Radioactive Cytotoxicity Assay kit (Promega) following the supplier instructions. Cytotoxicity was calculated as the percentage of LDH release in infected cells in relation to LDH release in non-infected, enzymatically lysed cells (maximum release).
Mouse Infection
The ability to infect female six-week old C57/BL6 mice was assed as described previously [42]. Essentially mice were inoculated i.p. with a 50∶50 mixture of wild type and mutant bacteria at a challenge dose of 5×103 CFU of each strain. The number of bacteria recovered from the spleen after 4 to 6 days of infection (variation of up to 30 hours was introduced between the time where mice were sacrificed because some animals had to be sacrificed earlier than day 6 due to severe illness to comply with the ethical clearance) was between 1×107 CFU ml−1 and 5×108 CFU ml−1. A few samples showed counts below 106 CFU ml−1 and these were not included in the analysis. The competitive method used has been reported to be more sensitive for testing of virulence than individual challenge of strains, since the mouse-to-mouse variation is eliminated [50]. This lowers the need for many mice in each group. Competition indices were calculated based on input (CFU ml−1 of inoculum) and output (CFU ml−1 of spleen sample) numbers of wild type versus mutant bacteria as described previously [42]. Mice infection studies were performed with permission to John Elmerdahl Olsen from the Danish Animal Experiments Inspectorate, license number 2009/561-1675.
Statistical Analyses
Statistical significance of the differences between wild type and mutant strains was determined using GraphPad Prism®, version 5.0 (GraphPad software) with one-sample t-test analysis. Grubb’s outlier test was performed to exclude outliers with a significance of 0.05.
Results and Discussion
PspE is a member of the Psp system, which helps S. Typhimurium to cope with membrane stress (reviewed in [27]–[28], [51]). Recently, the first protein encoded in the pspABCDE operon, PspA, was demonstrated to be required for virulence in the mouse model of systemic infection [52]. However, pspE expression is predicted to occur independently from a pspE-specific promoter [29] and it is highly expressed during cell infection [31]–[32]. Therefore, we wanted to investigate the role of PspE in virulence in S. Typhimurium independent from the contribution of the remaining part of the Psp system. Furthermore, in order to investigate whether it has functional overlap with another TST, GlpE, as it does in E. coli [24], we also investigated the role this single domain TST in parallel. We generated glpE and pspE single and double knockout mutant strains in S. Typhimurium 4/74 as well as in trans complemented strains (Table 1) and characterized all strains. qPCR was used to measure expression of the two genes in wild type, mutated and complemented strains. Wild type and complemented strains expressed the genes in the expected way, while no expression was observed in mutated strains (data not shown).
GlpE and PspE have a Role in Systemic Disease in the Mouse Model
In order to investigate whether GlpE and PspE have a role in virulence, we tested glpE and pspE mutant strains in competition with the wild type strain in a mouse model of systemic infection. Deletion of pspE increased virulence in C57/BL6 mice slightly (CI: 1.14; p<0.01). Introduction of the gene to the mutant in trans on the plasmid pINS02 removed significant differences between the two strains, suggesting that the increase in virulence was indeed caused by the pspE mutation. In contrast, deletion of glpE did not cause a significant change in virulence (Table 3). Interestingly, the ΔglpE/ΔpspE double mutant strain showed significantly decreased virulence (CI: 0.69±0.10; p<0.01), and introduction of the plasmid pINS04, encoding cloned copies of the two genes restored virulence to wild type level. Thus, the combined lack of GlpE and PspE decreased the ability of S. Typhimurium to carry out systemic disease in the mouse model, suggesting a role for TST in the infection process and complementary function of the two TSTs during the infection.
Table 3. Competitive indices of S. Typhimurium 4/74 wild type bacteria relative to mutant bacteria in the mouse spleen.
| 4/74 wild type versus | C.I. ± STD |
| ΔglpE (6) | 0.80±0.25 |
| ΔpspE (4) | 1.14±0.03** |
| ΔpspE+pINS02 (6) | 0.72±0.47 |
| ΔglpE/ΔpspE (4) | 0.69±0.10** |
| ΔglpE/ΔpspE+pINS04 (4) | 0.97±0.25 |
C57/BL6 mice were infected i.p. with equal numbers of mutant and wild type bacteria (each 5×103 CFU). After 4 to 6 days, mice were sacrificed and the spleen was removed. Serial dilutions were spotted on LB agar plates and number of wild type and mutant bacteria in a total of 100 colonies was further determined by selection of the resistance marker. Competitive indices (C.I.) were calculated as previously described [42]. The results are shown as mean values ± STD based on the number of mice tested as indicated in brackets. Significant differences from 1.0 (**p<0.01) were determined by one-sample t-test analysis.
The method used for virulence testing, i.e. competitive testing of wild type and mutant in the same animal was developed by Beuzon et al. [50] as a way to increase sensitivity in virulence testing. Since both wild type and mutant strain are tested in the same mice, the mouse-to-mouse variation in susceptibility to Salmonella infection is eliminated. As a consequence, fewer mice are needed to obtain the same statistical power as testing of each strain individually, which is in line with the international agreement to reduce the number of experimental animals used for research.
Growth Phenotypes of GlpE and PspE Mutants of S. Typhimurium
To rule out that the virulence phenotype was caused by a simple growth defect, we investigated whether GlpE and PspE mutants grew similar to the wild type strain in rich (LB) (data not shown) and minimal (M9+glucose) medium (Figure 1). Single as well as the double mutant showed similar growth curves as the wild type strain. TSTs are believed to assist in sulfur metabolism [13]. This also tested whether glpE and pspE are needed for the synthesis of sulfur containing amino acids, since the M9 medium contained MgSO4 as the only sulfur source, i.e. mutants were not affected in growth with MgSO4 as the only sulfur source. We also tested the ability of the strains to reduce thiosulfate and sulfite and also here we did not observe difference between the wild type and mutated strains, i.e all strains showed the typical ability of Salmonella to reduce these substances (data not shown), indicating that glpE and pspE are dispensable for the synthesis of essential sulfur containing amino acids in S. Typhimurium and for metabolism of thiosulfate and sulfite.
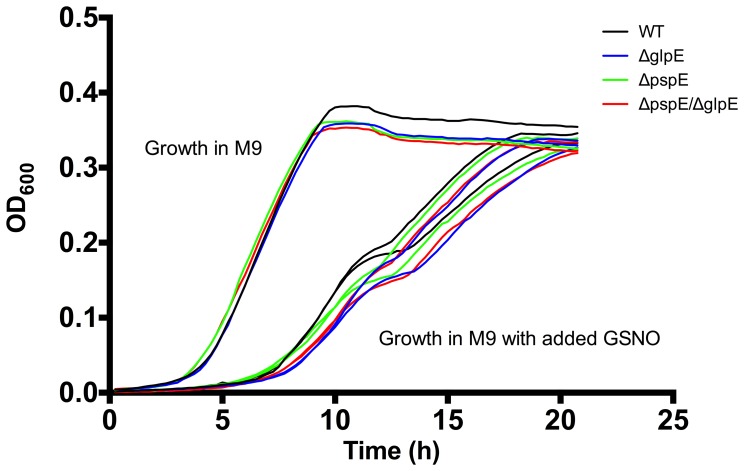
Figure 1. Growth of wild type and glpE and pspE mutant strains in M9 medium with and without GSNO.
Bacteria grown to stationary phase in M9 were re-grown in M9 medium without supplement and in media with varying concentrations of GSNO. The figure shows results for growth in M9, M9+0.5 mM GSNO and M9+1 mM GSNO. Growth of the wild type (black), ΔglpE (blue), ΔpspE (green) and ΔglpE/ΔpspE (red) strains was monitored for 20 h at 37°C with intermediate shaking of the plate. The data show representative results of two independent replicates. Growth experiments showed similar growth of ΔglpE, ΔpspE and ΔglpE/ΔpspE strains compared to their isogenic wild type strain.
GlpE and PspE Mutants are Dispensable During SDS Induced Membrane Stress
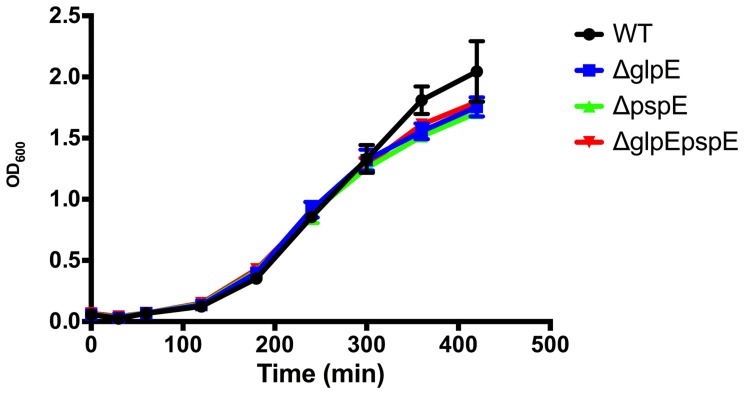
Since the Psp system is believed to aid in counteracting membrane stress [26]–[27], we investigated whether the mutants would show increased sensitivity to SDS, which is the prototype stress factor for detergent shock proteins [53]. The 4/74 ΔpspE, ΔglpE and ΔpspE/glpE strains grew equally well as the wild type strain in the presence of 0.01% and 0.1% SDS (data not shown). Growth control on plates without SDS was included, and comparison to this showed that the conditions tested affected the growth of the wild type strain, showing that it indeed experienced a stress. The plate assay used was less sensitive than comparative growth experiments, and to further substantiate our conclusions, growth in the presence of 0.01% SDS was also performed in M9 media in 100 ml flasks. As seen in Figure 2, no difference was observed between wild type and mutated strains.
Figure 2. Growth of wild type 4/74 and ΔpspE, ΔglpE and ΔpspE/glpE strains in the presence 0.01% SDS.
Strains were grown in 100 ml flasks containing 20 ml M9 supplemented with 0.01% SDS. The growth of wild type and mutated strains was similar.
GlpE and PspE are Dispensable for Resistance Towards NO stress in vitro
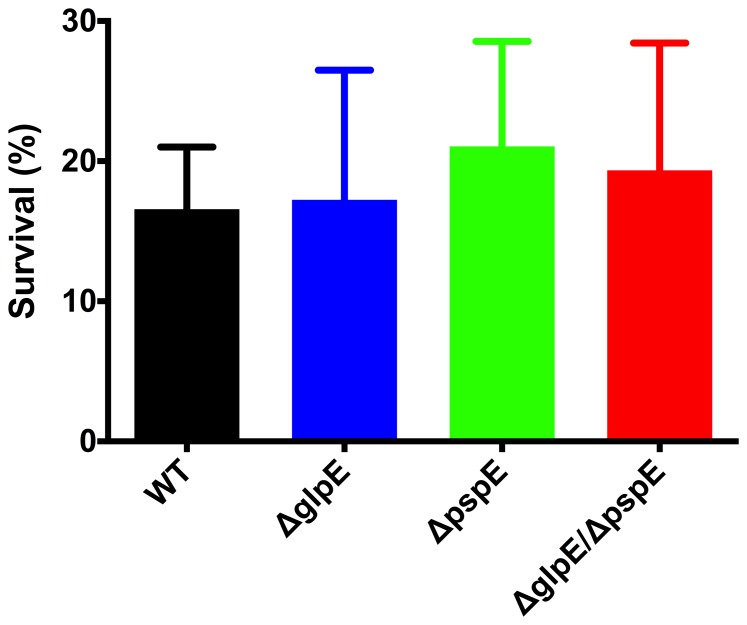
Nitric oxide compounds produced by the host are believed to interfere with important iron-sulfur complexes in bacteria [6]. S. Typhimurium has been reported to contain four iron-storage proteins, of which ferritin B encoded by ftnB and regulated by the Fur-system [54] has been identified as important for repair of iron-sulfur clusters [55]. Sulfur transferases are believed to be involved in the synthesis and repair of iron-sulfur clusters [13] as the bovine liver rhodanese is able to re-constitute iron-sulfur clusters of various enzymes in vitro [18]–[21]. We hypothesized that GlpE and PspE might be important for growth and survival in the presence of NO and RNS. To test this, we performed growth experiments of 4/74 wild type and mutant strains in the presence of GSNO and survival experiments after exposure to peroxinitrite. GSNO is a NO donor that primarily reacts with thiols, causing nitrosylation of proteins [9]. Peroxynitrite is a RNS that usually is formed in the cell by the reaction of NO with superoxide anion and which reacts with metal centers. Addition of GSNO at concentrations ranging from 0.1 to 1 mM lowered growth of the wild type strain in M9 compared to growth in the control medium without GSNO addition (Figure 1), showing that the growth reducing effect was indeed a result of GSNO addition. Growth inhibition was, however, similar between the wild type and the glpE and pspE mutant strains. From the experiments performed we cannot rule out that mutant specific responses would have been observed at higher concentrations. Furthermore, survival of the ΔglpE, ΔpspE and ΔglpE/ΔpspE strains in the presence of 360 µM peroxinitrite for 15 min likewise was similar to survival of the wild type strain (Fig. 3). Altogether, GlpE and PspE were concluded not to be required for resistance of S. Typhimurium to NO stress in vitro within the concentration range tested.
Figure 3. Survival of wild type 4/74 and ΔpspE, ΔglpE and ΔpspE/glpE strains in the presence of peroxynitrite treatment. Bacteria grown to logarithmic phase (OD600nm = 0.04±0.01) were treated with 360 µM peroxynitrite for 15 min.
Survival of bacteria was determined by calculating the number of bacteria after 15 min in relation to the number of bacteria at the beginning of the experiment. Results show mean values of at least three independent experiments ± SEM.
glpE but not pspE Contributes Significantly to Resistance Towards H2O2
Like nitric oxide, H2O2 also affects the cell through damage of iron- clusters [56], and we found it indicated to investigate the role of GlpE and PspE in the protection against this oxidative stress molecule. We grew our mutants in the presence of 5 mM and 10 mM H2O2 in LB and M9 media and observed that the wild type strain was slightly affected in growth and that the ΔglpE, but not the ΔpspE mutant, was severely affected in growth under this condition in both media (growth in 10 mM H2O2 shown in Figure 4). The phenotype was fully complemented by addition of the wild type gene in trans. The role of GlpE in oxidative stress adaptation has not previously been investigated, and this observation is the first clear phenotype associated with GlpE in S. Typhimurium. Unexpectedly, the double ΔglpEpspE mutant was not affected (Figure 4). The reason for this remains elusive, but the observation was very reproducible and may indicate that the lack of GlpE is only critical for resistance to H2O2 in the presence of a fully functional PspE.
Figure 4. Growth of WT (4/74) and pspE and glpE mutated and complemented strains in M9 medium with 10 mM H2O2.
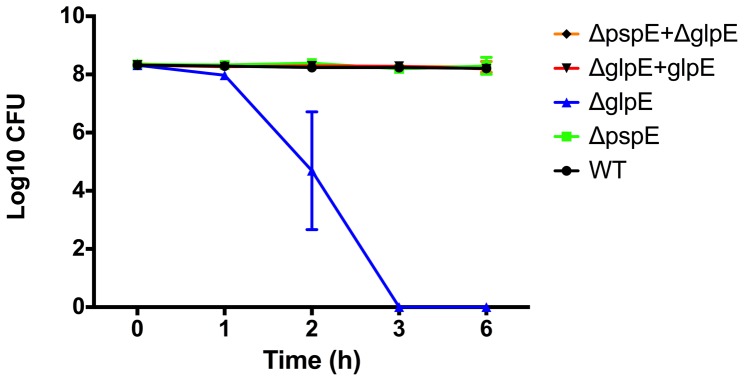
Wild type (black), ΔglpE (blue), ΔpspE (green), ΔglpE/ΔpspE (red) and ΔglpE+glpE strains were inoculated to an OD600 value of 0.05, H2O2 was added and growth was followed for 16 hours. Results shown are representative of two biological repeats.
pspE and glpE are not Important for S. Typhimurium Resistance Towards Cyanide
Another possible physiological role of sulfur transferases is the detoxification of cyanide as shown for the RhdA rhodanese in Pseudomonas aeruginosa [57]. In a standard biochemical classification S. enterica serovars are classified as non-detoxifying bacteria of cyanide at a concentration of 75 mg/l KCN [58]. However, we speculated that glpE and pspE might contribute to cyanide resistance at concentrations below this threshold. To test this, S. Typhimurium wild type, glpE and pspE single and double mutant strains were grown in the presence of KCN at concentrations ranging from 0.3 mg/l to 75 mg/l. The wild type and the ΔglpE, ΔpspE and ΔglpE/ΔpspE strains showed similar sensitivity towards KCN with the expected growth inhibition at 75 mg/l, poor growth at 15 mg/l and normal growth at 3 mg/l, 0.6 mg/l and 0.3 mg/l (data not shown), indicating that GlpE and PspE proteins are not involved in cyanide tolerance in S. Typhimurium.
Intracellular Survival and Cytotoxicity Towards Macrophages is Independent of GlpE and PspE
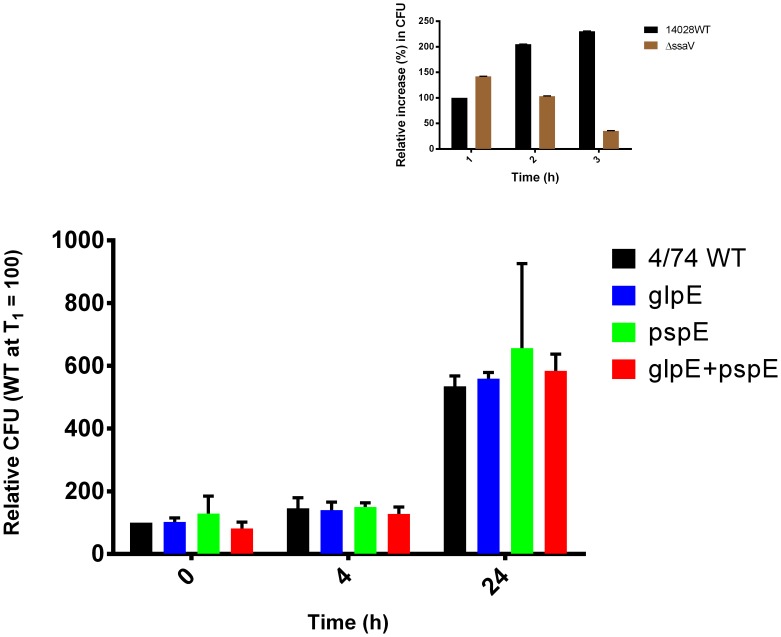
Virulence of S. Typhimurium in the mouse model of systemic disease was decreased in the absence of glpE and pspE (Table 3). In order to determine what might have caused this reduction, we tested the ability of glpE and pspE deficient and complemented strains to infect and survive inside J774 macrophages. This was considered relevant as these features play a role in the development of systemic disease of S. Typhimurium [1]–[2] and as both genes are expressed during infection of cultured macrophages [31]. Survival/replication inside J774 macrophages 1h p.i., 4h p.i. and 24 h p.i. was found to be similar to the wild type strain (Figure 5). A mutant in the SPI-2 gene ssaV which is attenuated for macrophage survival [49] was included as control, and showed the expected phenotype, as it was taken up to the same extend as the wild type strain, but showed reduced intracellular propagation. Theoretically, since cytotoxic effects results in exposure to gentamicin in such cell culture experiments, a strain with increased multiplication ability could be masked by an increase in cytotoxicity. However, the mutated strains did not differ significantly from the wild type in cytotoxicity 24 hours post infection (data not shown). Thus, S. Typhimurium intracellular survival and replication in macrophages was independent of the presence of the two putative TSTs GlpE and PspE. A possible explanation for the discrepancies in the in vivo and in vitro virulence data could be the limitations in use of cell culture experiments to study the complex interaction of S. Typhimurium with host cells and tissues [59]–[60].
Figure 5. Infection of J774 macrophages with wild type 4/74 and ΔpspE, ΔglpE and ΔpspE/glpE strains.
J774A.1 macrophages were infected with complemented-opsonized bacteria in a MOI of 10 and incubated for 25 min. Extracellular bacteria were killed by treatment with 100 µg ml−1 gentamicin. Bacteria were released from the macrophages 1 h p.i., 4 h p.i. and 24 h p.i. and the number of intracellular bacteria was determined by CFU ml−1 calculations and was expressed relatively to CFU at T1. The results show mean values ± SEM of at least three independent replicates. The small inserts shows results of an internal control experiment conducted with a SPI-2 (ΔssaV) deficient strain showing the expected reduced intracellular survival/replication of the mutated strain.
GlpE and PspE are also Dispensable for Invasion of Epithelial Cells
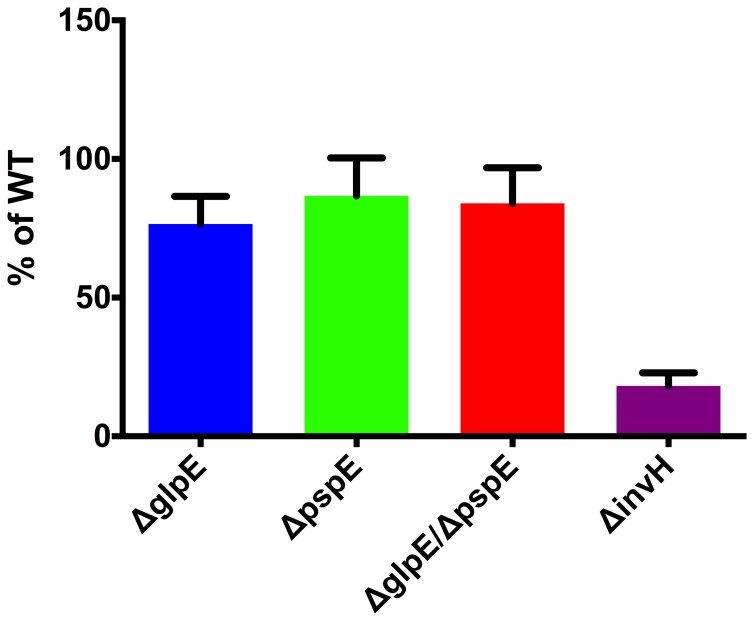
Infection of the intestinal epithelial layer is the first critical step in Salmonella virulence. This aspect of infection was bypassed in our mice experiments, since we used intra peritoneal challenge. The ability of S. Typhimurium to infect epithelial cells largely depends on expression of the SPI-1 encoded T3SS (T3SS1) and release of effector molecules through this system [1]. During infection of epithelial cells, glpE is constitutive and pspE is highly expressed [32]. We therefore tested the role of GlpE and PspE in invasion in an epithelial cell infection model. Moreover, we tested the ability of glpE and pspE mutant strains to express genes encoding regulatory, structural and effector molecules of the T3SS1. Single and double deletion of glpE and pspE genes did not change the ability of S. Typhimurium to invade epithelial INT-407 cells compared to the wild type, whereas the control strain ΔinvH, an S. Typhimurium strain with a deficiency in T3SS1 [37], was decreased in this phenotype (p<0.01) (Figure 6). In line with these findings, single or double deletion of glpE and pspE genes in S. Typhimurium 4/74 did not change expression of hilA, invG, prgH and sopB genes compared to the wild type as determined from qPCR experiments (data not shown). The growth conditions we used to demonstrate this were not optimal for induction of SPI-1, but the genes have previously been demonstrated to be expressed under this condition [42]. Overall, glpE and pspE are dispensable for in vitro invasion of S. Typhimurium and expression of genes that are associated with the T3SS1.
Figure 6. Infection of epithelial INT-407 cells wild type 4/74 and ΔpspE, ΔglpE and ΔpspE/glpE strains.
Monolayers of INT-407 cells were infected with bacteria grown to logarithmic phase in a MOI of 100 and incubated for 15 min. Extracellular bacteria were killed by treatment with 100 µg ml−1 gentamicin. Number of intracellular bacteria was determined by CFU ml−1 calculations and values were adjusted against values for the wild type. The results represent mean values of at least three independent experiments ± SEM. Significant difference (**p<0.01) was determined by one-sample t-test analysis.
Conclusion
This worked revealed that parallel deletion of glpE and pspE genes decreased virulence of S. Typhimurium in the mouse model of typhoid fever, suggesting a role of TST activity in systemic infection. Deletion of glpE but not pspE significantly affected H2O2 resistance, but since the double mutant was not affected in this assay we found it unlikely that reduced oxidative stress was the reason for the virulence phenotype. Thus the mechanism by which GlpE and PspE contribute to virulence in S. Typhimurium remains to be characterized in future research.
Acknowledgments
We greatly appreciate Tatjana Kristensen and Tony Bønnelycke for skillful technical assistance.
Funding Statement
This work was funded by the Danish Research Council for Technology and Production through grant no. 274-07-0328. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haraga A, Ohlson MB, Miller SI (2008) Salmonellae interplay with host cells. Nat Rev Microbiol 6: 53–66. [DOI] [PubMed] [Google Scholar]
- 2. Portillo FGD (2001) Salmonella intracellular proliferation: where, when and how? Microb Infect 3: 1305–1311. [DOI] [PubMed] [Google Scholar]
- 3. Ibarra JA, Steele-Mortimer O (2009) Salmonella- the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol 11: 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foster JW, Spector MP (1995) How Salmonella survive against the odds. Ann Rev Microbiol 49: 145–174. [DOI] [PubMed] [Google Scholar]
- 5. Rychlik I, Barrow PA (2005) Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev 29: 1021–1040. [DOI] [PubMed] [Google Scholar]
- 6. Fang FC (1997) Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest 99: 2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherayil BJ, Antos D (2001) Inducible nitric oxide synthase and Salmonella infection. Microb Infect 3: 771–776. [DOI] [PubMed] [Google Scholar]
- 8. Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC (2007) Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. . Proc Natl Acad Sci, USA 104: 8484–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jarboe LR, Hyduke DR, Tran LM, Chou KJ, Liao JC (2008) Determination of the Escherichia coli S-nitrosoglutathione response network using integrated biochemical and systems analysis. J Biol Chem 283: 5148–5157. [DOI] [PubMed] [Google Scholar]
- 10. Henard CA, Vazquez-Torres A (2011) Nitric oxide and salmonella pathogenesis. Front Microbiol 2: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bang IS, Liu L, Vazquez-Torres A, Crouch ML, Stamler JS, Fang FC (2006) Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J Biol Chem 281: 28039–28047. [DOI] [PubMed] [Google Scholar]
- 12. Bordo D, Bork P (2002) The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep 3: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cipollone R, Ascenzi P, Visca P (2007) Common themes and variations in the rhodanese superfamily. IUBMB Life 59: 51–59. [DOI] [PubMed] [Google Scholar]
- 14. Westley J (1981) Thiosulfate: cyanide sulfurtransferase (rhodanese). Methods Enzymol 77: 285–291. [DOI] [PubMed] [Google Scholar]
- 15. Donadio S, Shafiee A, Hutchinson CR (1990) Disruption of a rhodaneselike gene results in cysteine auxotrophy in Saccharopolyspora erythraea . J Bacteriol 172: 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cipollone R, Frangipani E, Tiburzi F, Imperi F, Ascenzi P, et al. (2007) Involvement of Pseudomonas aeruginosa rhodanese in protection from cyanide toxicity. Appl Environ Microbiol 73: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vennesland B, Castric PA, Conn EE, Solomonson LP, Volini M, et al. (1982) Cyanide metabolism. Fed Proc 41: 2639–2648. [PubMed] [Google Scholar]
- 18. Bonomi F, Pagani S, Cerletti P, Cannella C (1977) Rhodanese-mediated sulfur transfer to succinate dehydrogenase. Eur J Biochem 72: 17–24. [DOI] [PubMed] [Google Scholar]
- 19. Pagani S, Galante YM (1983) Interaction of rhodanese with mitochondrial NADH dehydrogenase. Biochim Biophys Acta 742: 278–284. [DOI] [PubMed] [Google Scholar]
- 20. Pagani S, Bonomi F, Cerletti P (1984) Enzymic synthesis of the iron-sulfur cluster of spinach ferredoxin. Eur J Biochem 142: 361–366. [DOI] [PubMed] [Google Scholar]
- 21. Bonomi F, Pagani S, Kurtz DM Jr (1985) Enzymic synthesis of the 4Fe-4S clusters of Clostridium pasteurianum ferredoxin. Eur J Biochem 148: 67–73. [DOI] [PubMed] [Google Scholar]
- 22. Ray WK, Zeng G, Potters MB, Mansuri AM, Larson TJ (2000) Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J Bacteriol 182: 2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams H, Teertstra W, Koster M, Tommassen J (2002) PspE (phage-shock protein E) of Escherichia coli is a rhodanese. FEBS Lett 518: 173–176. [DOI] [PubMed] [Google Scholar]
- 24. Cheng H, Donahue JL, Battle SE, Ray WK, Larson TJ (2008) Biochemical and genetic characterization of PspE and GlpE, two single-domain sulfurtransferases of Escherichia coli . Open Microbiol J 2: 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chng SS, Dutton RJ, Denoncin K, Vertommen D, Collet JF, et al. (2012) Overexpression of the rhodanese PspE, a single cysteine-containing protein, restores disulphide bond formation to an Escherichia coli strain lacking DsbA. Mol Microbiol 85: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Darwin AJ (2005) The phage-shock-protein response. Mol Microbiol 57: 621–628.30. [DOI] [PubMed] [Google Scholar]
- 27. Joly N, Engl C, Jovanovic G, Huvet M, Toni T, et al. (2010) Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. Fems Microbiol Rev 34: 797–827. [DOI] [PubMed] [Google Scholar]
- 28. Lloyd LJ, Jones SE, Jovanovic G, Gyaneshwar P, Rolfe MD, et al. (2004) Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein g (PspG). J Biolog Chem 279: 55707–55714. [DOI] [PubMed] [Google Scholar]
- 29. Huvet M, Toni T, Sheng X, Thorne T, Jovanovic G, et al. (2011) The evolution of the phage shock protein response system: interplay between protein function, genomic organization, and system function. Mol Biol Evol 28: 1141–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brissette JL, Weiner L, Ripmaster TL, Model P (1991) Characterization and sequence of the Escherichia coli stress-induced psp operon. J Mol Biol 220: 35–48. [DOI] [PubMed] [Google Scholar]
- 31. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica . Mol Microbiol 47: 103–118. [DOI] [PubMed] [Google Scholar]
- 32. Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, et al. (2008) During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10: 958–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang B, Larson TJ (1998) Multiple promoters are responsible for transcription of the glpEGR operon of Escherichia coli K-12. Biochem Biophys Acta 1396: 114–126. [DOI] [PubMed] [Google Scholar]
- 34. Choi YL, Kawase S, Kawamukai M, Sakai H, Komano T (1991) Regulation of glpD and glpE gene expression by a cyclic AMP-cAMP receptor protein (cAMP-CRP) complex in Escherichia coli. . Biochem Biophys Acta 1088: 31–35. [DOI] [PubMed] [Google Scholar]
- 35. Zeng G, Ye S, Larson TJ (1996) Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J Bacteriol 178: 7080–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wallis TS, Paulin SM, Plested JS, Watson PR, Jones PW (1995) The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun 63: 2755–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watson PR, Paulin SM, Bland AP, Jones PW, Wallis TS (1995) Characterization of intestinal invasion by Salmonella Typhimurium and Salmonella Dublin and effect of a mutation in the invH gene. Infect Immun 63: 2743–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Nat Academ Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang AC, Cohen SN (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134: 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Enomoto M, Stocker BA (1974) Transduction by phage P1kc in Salmonella typhimurium . Virology 60: 503–514. [DOI] [PubMed] [Google Scholar]
- 41. Thomsen LE, Chadfield MS, Bispham J, Wallis TS, Olsen JE, et al. (2003) Reduced amounts of LPS affect both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol Lett 228: 225–231. [DOI] [PubMed] [Google Scholar]
- 42. Jelsbak L, Thomsen LE, Wallrodt I, Jensen PR, Olsen JE (2012) Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS One 7: e36149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kundinger MM, Zabala-Diaz IB, Chalova VI, Moore RW, Ricke SC (2008) Real-time polymerase chain reaction quantification of Salmonella Typhimurium hilA expression during agitation and static incubation. J Rapid Method Automat Microbiol 16: 273–283. [Google Scholar]
- 44. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 45. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nuc Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frees D, Qazi SNA, Hill PJ, Ingmer H (2003) Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol 48: 1565–1578. [DOI] [PubMed] [Google Scholar]
- 47. Galan JE, Curtiss R (1989) Cloning and molecular characterization of genes whose products allow Salmonella Typhimurium to penetrate tissue-culture cells. Proc Nat Acad Sci USA 86: 6383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eriksson S, Bjorkman J, Borg S, Syk A, Pettersson S, et al. (2000) Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell Microbiol 2: 239–250. [DOI] [PubMed] [Google Scholar]
- 49. Beuzon CR, Salcedo SP, Holden DW (2002) Growth and killing of a Salmonella enteric serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiol 148: 2705–2715. [DOI] [PubMed] [Google Scholar]
- 50. Beuzón CR, Holden DW (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo . Microb Infect 3: 1345–42. [DOI] [PubMed] [Google Scholar]
- 51. Model P, Jovanovic G, Dworkin J (1997) The Escherichia coli phage-shock-protein (psp) operon. Mol Microbiol 24: 255–261. [DOI] [PubMed] [Google Scholar]
- 52. Karlinsey JE, Maguire ME, Becker LA, Crouch MLV, Fang FC (2010) The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol Microbiol 78: 669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adamowicz M, Kelley PM, Nickerson KW (1991) Detergent (sodium dodecyl sulfate) shock proteins in Escherichia coli . J Bacteriol 173: 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Troxell B, Fink RC, Porvollic S, McClelland M, Hassan HM (2011) The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol 11: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Velayudhan J, Castor M, Rickardson A, Main-Hester KL, Fanc FC (2007) The role of ferritins in physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron sulfur cluster repair and viurlence. Mol Microbiol 63: 1495–1507. [DOI] [PubMed] [Google Scholar]
- 56. Djaman O, Outten FW, Imlay JA (2004) Repair of oxidized iron-sulfur clusters in Escherichia coli. . J Biol Chem 279: 44590–44599. [DOI] [PubMed] [Google Scholar]
- 57. Cipollone R, Frankipani B, Tiburzi F, Imperi F, Visca P (2007) Involvement of Pseudomonas aeruginosa rhodanase from cyanide toxicity. Appl Environ Microbiol 73: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Edwards RR, Fife MA (1988) Cyanide media in the differentiation of enteric bacteria. Appl Microbiol 4: 46–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Finlay BB, Brumell JH (2000) Salmonella interactions with host cells: in vitro to in vivo . Philos Trans R Soc Lond B Biol Sci 355: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Holzer SU, Hensel M (2012) Divergent roles of Salmonella pathogenicity island 2 and metabolic traits during interaction of S. enterica serovar Typhimurium with host cells. PLoS One 7: e33220. [DOI] [PMC free article] [PubMed] [Google Scholar]