Abstract
Repeated sequence signatures are characteristic features of all genomic DNA. We have made a rigorous search for repeat genomic sequences in the human pathogens Neisseria meningitidis, Neisseria gonorrhoeae and Haemophilus influenzae and found that by far the most frequent 9–10mers residing within coding regions are the DNA uptake sequences (DUS) required for natural genetic transformation. More importantly, we found a significantly higher density of DUS within genes involved in DNA repair, recombination, restriction-modification and replication than in any other annotated gene group in these organisms. Pasteurella multocida also displayed high frequencies of a putative DUS identical to that previously identified in H.influenzae and with a skewed distribution towards genome maintenance genes, indicating that this bacterium might be transformation competent under certain conditions. These results imply that the high frequency of DUS in genome maintenance genes is conserved among phylogenetically divergent species and thus are of significant biological importance. Increased DUS density is expected to enhance DNA uptake and the over-representation of DUS in genome maintenance genes might reflect facilitated recovery of genome preserving functions. For example, transient and beneficial increase in genome instability can be allowed during pathogenesis simply through loss of antimutator genes, since these DUS-containing sequences will be preferentially recovered. Furthermore, uptake of such genes could provide a mechanism for facilitated recovery from DNA damage after genotoxic stress.
INTRODUCTION
A prominent feature of many eubacterial genomes is the abundance and diversity of DNA sequence repeats (1,2). For example, efficient transformation of the naturally competent bacterial species Neisseria gonorrhoeae, Neisseria meningitidis and Haemophilus influenzae requires the presence in the transforming DNA of 9- or 10mer sequence elements that occur genome-wide in very high numbers (3,4). These are referred to as DNA uptake sequences (DUS) and the neisserial genome sequences contain ∼1900 DUS (5,6), whereas the H.influenzae genome harbors 1471 copies of such repeat elements (7,8). When exposed to a mixture of homologous and foreign DNAs, these human pathogens show preferential uptake of DUS-containing DNA (4,9). The family Neisseriaceae comprise the genera Neisseria, Kingella and Eikenella, which all contain naturally competent species (10), although neisserial DUS-requirement has only been demonstrated for N.gonorrhoeae and N.meningitidis (4,11). The H.influenzae DUS, which often is referred to as an uptake signal sequence (USS) (8), represents the most frequent oligomeric sequence found in this genome (1,8). The family Pasteurellaceae contains the genera Haemophilus, Actinobacillus and Pasteurella. Actinobacillus actinomycetemcomitans is also naturally competent (12), and competence in this bacterium is dependent on the presence of the same DUS as in H.influenzae (13).
Transposon-mediated DNA rearrangements involving repetitive elements are often regarded as a major evolutionary driving force (14). However, repetitive DNA sequences may have other important functions, such as the regulation of gene expression by control of mRNA stability (15) and, when present in the protein coding sequence (CDS), in the generation of antigenic variation by phase variation (16) or in recombination of modular genes (17). Rocha et al. (18) used a computational approach to compare stress response genes in Escherichia coli with the rest of the genome with regard to presence of various kinds of repeat elements known to promote genetic variability. Long repeats engaged in homologous recombination were found to be rare in stress response genes. However, high numbers of short close repeats were observed, capable of inducing phenotypic variability by strand slippage during DNA, RNA or protein synthesis. An increase in phenotypic heterogeneity via sequence repeat rearrangements may provide increased fitness to subpopulations of cells under stress conditions.
There is a limited understanding to why sequence-specific DNA uptake has evolved, and how DUS became disseminated and over-represented within their resident genomes. Many hypotheses addressing these questions have been proposed (1,19,20). Similarly, the mechanisms by which other repeated sequence elements have evolved in microbial genomes are also poorly understood. One step towards understanding the evolutionary role and biological significance of repeated elements has been to examine the genomic distribution of DUS, which has indicated that these sequences are randomly distributed throughout the genomes (1,19). Previous reports have emphasized the potential role of DUS as transcriptional terminators located downstream of open reading frames, often in the form of inverted repeats (8,21).
During our studies on neisserial genome maintenance functions, we observed that several genes engaged in DNA repair and recombination harboured DUS within their CDS. In contrast, we noted a reduced tendency for genes encoding surface components and factors involved in protein secretion to carry DUS. To analyse this notion in a more systematic fashion, we have performed comprehensive analysis of the most frequently occurring 9- and 10mer sequences in the CDS of the Neisseria species and the family Pasteurellaceae. The repeat sequences already identified as DUS were found to be the most frequent 9- or 10mers in the CDS of the genomes studied. Most importantly, analyses of DUS occurring inside coding sequences revealed a clear bias of DUS towards gene functions involved in genome metabolism and maintenance.
MATERIALS AND METHODS
Genome sequences and data sets
Complete CDS files and gene annotation files for the genomes of the bacterial species listed in Table 1 except N.gonorrhoeae and A.actinomycetemcomitans were downloaded from the Clusters of Orthologous Groups (COG) system at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The sequence data for the N.gonorrhoeae genome were kindly provided by Thomas S. Brettin at the Los Alamos National Laboratory. For this genome, COG annotation was far from complete. The genome sequence of A.actinomycetemcomitans HK1651 was kindly provided by David Dyer at the Genome Sequence Project at the University of Oklahoma. The A.actinomycetemcomitans genome has not been COG classified.
Table 1. Bacterial strains included in the study: genome characteristics.
| Bacterial species, strains and reference | Accession no. | Genome size (bp) | G+C (%) |
|---|---|---|---|
| Naturally competent bacteria | |||
| With DUS: 5′-GCCGTCTGAA-3′ | |||
| N.meningitidis Z2491 (5) |
NC_003116 |
2 184 406 |
51.8 |
| N.meningitidis MC58 (6) |
NC_003112 |
2 272 351 |
51.5 |
| N.gonorrhoeae FA1090 (43) |
NC_002946 |
2 153 944 |
51.4 |
| With DUS: 5′-AAGTGCGGT-3′ | |||
| H.influenzae Rd/KW20 (7) |
NC_000907 |
1 830 138 |
39.1 |
| A.actinomycetemcomitans HK1651 (43) |
NC_002924 |
2 105 329 |
42.7 |
| Not known to be competent | |||
| E.coli K12 (44) |
NC_000913 |
4 639 221 |
50.8 |
| P.multocida PM70 (45) | NC_002663 | 2 257 487 | 41.0 |
Gene classification system
The CDS of the genomes were grouped into genome maintenance genes (GMGs) and non-GMGs. GMGs were prospectively defined as genes involved in DNA replication, recombination, repair and restriction/modification. To avoid any bias from our side in the selection of GMGs versus non-GMGs, we employed the COGs of proteins (22) (http://www.ncbi.nlm.nih.gov/COG/). This is a commonly used gene classification system, and one by which nearly all the genomes listed in Table 1 are classified. GMGs are generally contained in COG group L. However, COG group L also contains genes that are not strictly GMGs; these are mainly transposases. COG group L was therefore split into two subgroups: GMGs and the remainder, COG subgroup L-. Furthermore, two COG entries not in group L were included among GMGs as they obviously belong to this group: these were COG1066 (formerly in COG group O) and COG0553 (formerly in COG group K) encoding a RadA-homolog and a helicase, respectively. The definition of GMGs is in concordance with the gene classification systems used by the Sanger Centre and TIGR. The complete list of the COG numbers defining GMGs can be found in the Supplementary Material.
Coding sequences that were not COG-classified were excluded from analyses comparing GMGs with non-GMGs, since they may include incorrectly predicted genes (23).
Statistical analyses
Frequencies of oligomeric sequences in CDS. Occurrences of DUS on either strand within CDS were identified and counted. Similar counts were made of other kmers, i.e. oligomers of length k, where k is the length of the DUS, and the most frequent kmers were identified. DUS counts were made for each COG group and for GMGs. The frequency of a given kmer is the number of occurrences divided by the total number of kmers, where the number of kmers in a gene is the gene length minus k – 1.
The relative abundance of a kmer is the number of occurrences divided by the expected number of occurrences. The expected number of occurrences of the kmer w1…wk is found using a fourth order Markov model taking into account the position relative to the reading frame as presented in Reinert et al. (24): i.e. the expected number is calculated from the number of 4- and 5mers as:
![]()
where ![]() denotes the number of words w1w2 … wl relative to the reading frame
denotes the number of words w1w2 … wl relative to the reading frame ![]() as counted modulo 3. Increasing the order to 5, the DUS would contain up to half of the 6mers, which would strongly influence the estimates by substantially elevating the 6mer counts. Lower order, on the other hand, might reduce the quality of the model. Hence, the order 4 was found to be most appropriate. As an abundance of DUS will increase the estimate of the expected number, this may lead to an underestimation of the relative abundance.
as counted modulo 3. Increasing the order to 5, the DUS would contain up to half of the 6mers, which would strongly influence the estimates by substantially elevating the 6mer counts. Lower order, on the other hand, might reduce the quality of the model. Hence, the order 4 was found to be most appropriate. As an abundance of DUS will increase the estimate of the expected number, this may lead to an underestimation of the relative abundance.
Distribution of DUS. Confidence intervals for the number of kmers, e.g. DUS, were calculated assuming a Poisson distribution. DUS in intergenic regions frequently occur in pairs as inverted repeats, whereas in CDS they generally occur singly; they are generally sufficiently far apart to consider them independent occurrences. Hence, the Poisson assumption seems appropriate for DUS in CDS. The number of CDS with multiple DUS was assessed to see if this was in compliance with the Poisson assumption (Supplementary Material).
The number of DUS and the corresponding 95% confidence intervals are divided by the expected number and presented as relative abundance. The relative abundances of DUS per COG group of N.meningitidis Z2491 is presented. Similar tables for the other genomes are found in the Supplementary Material.
The frequencies of DUS in GMGs and non-GMGs were calculated, and the bias towards GMGs is given by the DUS frequencies in GMG divided by non-GMG frequencies. Confidence intervals and P values were determined by assuming a binomial distribution of DUS between GMGs and non-GMGs.
Control analyses. The number of DUS is assumed to be Poisson distributed, and is compared with the expected number of DUS. The deviance residual is used to assess the statistical significance of DUS abundances (see Supplementary Material for further details). If the Markov model is assumed to give the true expectancies of all oligomers, this should be close to normally distributed with mean 0 and standard deviation 1. In reality, the standard deviation may be larger. Significance testing is performed assuming only that the deviance residuals are normally distributed; means and standard deviations of these distributions are found weighing kmers by their expected numbers.
The analyses might be influenced by inherent differences in the composition of different gene groups. The expected number of DUS was calculated separately for GMGs and non-GMG COG groups using the Markov model. Because groups were much smaller, providing fewer data per group, the order of the Markov model was reduced to 3 in all groups. Also, to prevent DUS abundances from influencing the Markov model, DUS were masked prior to counting 3- and 4mers. This will cause the expected number of DUS to be under-estimated, and relative abundances to be over-estimated; but this bias will be consistent across the groups, allowing a comparison of expected frequencies between them.
A final control analysis was performed on a set of control sequences to check if the distribution of these were generally skewed towards GMG. To imitate DUS and avoid repetitive homo-polymeric oligomers, we used the 99 most frequent oligomers containing all four nucleotides other than the DUS itself. We then counted the occurrences of control sequences that did not overlap a DUS; this was done by masking the DUS and doing a recount (Fig. 1). The control sequences generally did not appear in sufficient numbers to allow for analyses of individual sequences without a large degree of uncertainty. Instead, all 99 control sequences were grouped to allow for a more powerful analysis. In order to reduce the problem of overlapping control sequences, an occurrence was not counted if the kmer one base ahead also was a control sequence (Fig. 1). The bias towards GMGs was defined for these controls as for the DUS. Fisher’s exact one-sided test was used to assess if DUS were more strongly biased towards GMGs than the control sequences. As kmers similar to and potentially related to DUS might be included as control sequences, this control analysis may be somewhat conservative.
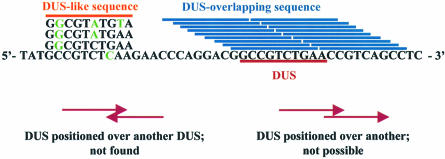
Figure 1.
Designation of DUS overlapping and DUS-associated sequences, as exemplified by the Neisseria DUS. The DNA uptake sequence 5′-GGCCGTCTGAA-3′ is marked in red. Oligomers that physically overlap with the DUS are termed DUS-overlapping sequences and are marked in blue. Oligomers that are mutated in one or more of the 10 bp DUS (green denotes the mutation in the examples given) are called DUS-like sequences and are marked in orange. DUS overlapping each other with one DUS located on each strand were not detected (the Neisseria sp.) or could not occur (H.influenzae). DUS-overlapping sequences were excluded from the analysis, while the DUS-like sequences were included among the control sequences.
The control analyses, both methods and results, are extensively covered in the Supplementary Material.
Search for DUS-like oligomers in CDS. DUS-like oligomers, i.e. kmers that are equal to DUS except at one base (Fig. 1), were counted, and their expected numbers calculated using the Markov model. If DUS degenerate over time by point mutations, there should be an abundance of DUS-like kmers: i.e. more than expected.
RESULTS
Search for frequently occurring oligomer sequences
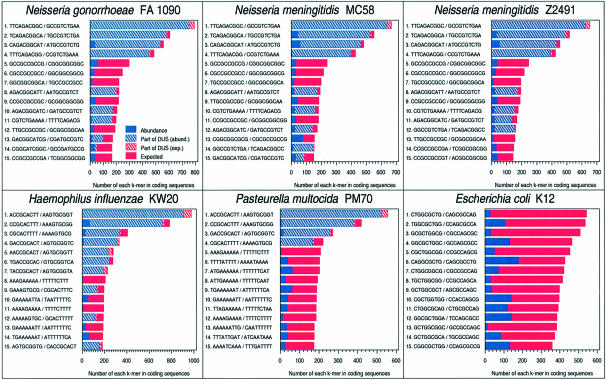
The DUS previously identified for the pathogenic neisseriae (5′-GCCGTCTGAA-3′) and H.influenzae (5′-AAGTGCGGT-3′) were clearly the most frequently occurring 9- and 10mers in the CDS of their respective genomes (Fig. 2) and many times as frequent as expected from the 5mer composition of CDS. In all cases, the deviance residuals exceeds 30 standard deviations; and this makes both the P-value and the E-value (P times the number of different kmers) <10–100, which is highly significant from a statistical point of view. Other abundant kmers appear, but are mostly part of a DUS as shown in Figure 2: for example, the neisseriae DUS often comes as ATGCCGTCTGAAA which would contribute significantly to the count of ATGCCGTCTG.
Figure 2.
The most frequent oligonucleotides (9- and 10mers) containing all four nucleotides in the CDS of N.meningitidis Z2491, N.meningitidis MC58, N.gonorrhoeae FA1090, H.influenzae KW20, P.multocida PM70 and E.coli K12 were counted; E.coli K12 is included to illustrate the distribution in a negative control. DUS denotes either the neisserial 10 bp DUS or the H.influenzae 9 bp DUS. The length of the bars represent the number of DUS found in CDS. The bar is split into the expected number (red) and the abundance (blue), which is the actual number found minus the expected number. DUS and DUS-overlapping sequences (see Fig. 1) were counted separately and illustrated as the shaded areas; the solid areas illustrate oligonucleotide counts if DUS were masked before counting.
The H.influenzae DUS signature was the most frequent oligomer present in the CDS of Pasteurella multocida, and found to be as abundant as in H.influenzae. This sequence was therefore assessed as a DUS candidate in P.multocida.
For comparison, Figure 2 also includes a 9mer count for E.coli K12. It may be noted that the most frequent 9mers are not much more frequent than expected, at least not to the extent found for DUS. It should be noted that a fourth order Markov model might not handle highly repetitive regions particularly well, and that most of the more abundant kmers (the solid blue peaks in Fig. 2) apparently belong to homo-oligomeric elements.
The H.influenzae DUS signature was also the most frequent oligomer in the complete genome of A.actinomycetemcomitans (1760 copies) (Table 2). In A.actinomycetemcomitans the second most frequent kmer not overlapping the H.influenzae DUS was the 9mer 5′-AATAAAAAA-3′, which was found in 260 copies.
Table 2. Number of DUS (kmers) in complete bacterial genomes, CDS and GMGs.
| |
|
No. of selected kmer in | No. of CDS | Average CDS length (bp) | ||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Frequent k-mer | Genome | CDS | GMGs | All | GMGs | All | GMGs |
|
N.meningitidis Z2491 |
Neisseria DUSa |
1892 |
653 |
97 |
2065 |
95 |
854 |
1369 |
|
N.meningitidis MC58 |
Neisseria DUSa |
1935 |
669 |
88 |
2079 |
95 |
868 |
1336 |
|
N.gonorrhoeae FA1090 |
Neisseria DUSa |
1965 |
792 |
92 |
2185 |
93 |
827 |
1451 |
|
H.influenzae Rd/KW20 |
H.influenzae DUSa |
1471 |
977 |
112 |
1714 |
93 |
941 |
1403 |
|
P.multocida PM70b |
H.influenzae DUSa |
927 |
557 |
58 |
2015 |
92 |
998 |
1468 |
| A.actinomycetemcomitans HK1651c | H.influenzae DUSa | 1760 | nad | nad | nad | nad | nad | nad |
aThe neisserial and H.influenzae DUS are related to the natural competence of these bacteria.
bPasteurella multocida is not know to be competent; the H.influenzae DUS was selected because of its high frequency.
cSince the A.actinomycetemcomitans HK1651 genome is not yet classified according to the COG system, an analysis of the DUS distribution in CDS was not performed.
dNot applicable.
Occurrence and direction of DUS in CDS
In the genomes of N.meningitidis and H.influenzae respectively, 35 and 65% of DUS are located inside CDS (CDS cover ∼80% of these genomes), with 22 and 38% of the CDS containing at least one DUS element (19) (Table 2). DUS were generally found in both orientations. However, the neisserial DUS appears to have a bias towards the reverse orientation of open reading frames, with 293 forward oriented and 360 reversely oriented DUS in the N.meningitidis Z2491 genome (Supplementary Material). The Markov model does not explain the prevalence towards the reverse orientation, though it may be due to a stop codon occurring in one of the reading frames in the forward direction.
Distribution of DUS between GMGs and non-GMGs
Table 3 shows the distribution of DUS between GMGs and the non-GMG COG groups for N.meningitidis Z2491. GMGs were prospectively defined to contain genes involved in DNA replication, recombination, repair and restriction/modification mostly as defined by the COG L classification (Materials and Methods).
Table 3. Distribution of the neisserial DUS in CDS of N.meningitidis Z2491 by COG groupa.
| COG groups | No. of DUS | No. of CDS | Average CDS length | Relative abundance (95% CI) | |
|---|---|---|---|---|---|
| All |
|
653 |
2065 |
854 |
22.0 (20.4-23.8) |
| C |
Energy production, conversion |
23 |
104 |
1111 |
11.8 (7.5-17.7) |
| D |
Cell division, chromosome partitioning |
14 |
24 |
1129 |
30.7 (16.8-51.5) |
| E |
Amino acid transport, metabolism |
69 |
137 |
1141 |
26.2 (20.4-33.1) |
| F |
Nucleotide transport, metabolism |
13 |
48 |
1005 |
16.0 (8.5-27.4) |
| G |
Carbohydrate transport, metabolism |
14 |
56 |
1093 |
13.6 (7.4-22.8) |
| H |
Coenzyme metabolism |
39 |
74 |
939 |
33.4 (23.7-45.6) |
| I |
Lipid metabolism |
10 |
40 |
930 |
16.0 (7.7-29.4) |
| J |
Translation, ribosomal structure and biogenesis |
53 |
142 |
830 |
26.8 (20.1-35.0) |
| K |
Transcription |
15 |
61 |
875 |
16.7 (9.4-27.6) |
| L |
DNA replication, recombination, repair |
98 |
163 |
1087 |
32.8 (26.7-40.0) |
| GMG |
Genome maintenance genes |
97 |
95 |
1369 |
44.2 (35.9-53.9) |
| L- |
Others: mostly transposases |
1 |
68 |
693 |
1.3 (0.03-7.1) |
| M |
Cell envelope biogenesis, outer membrane |
61 |
123 |
1178 |
25.0 (19.1-32.1) |
| N |
Cell motility and secretion |
6 |
49 |
985 |
7.4 (2.7-16.1) |
| O |
Posttranslational modification, protein turnover, chaperones |
17 |
66 |
1031 |
14.8 (8.7-23.8) |
| P |
Inorganic ion transport, metabolism |
22 |
74 |
1200 |
14.7 (9.2-22.3) |
| Q |
Secondary metabolites biosynthesis, transport and catabolism |
8 |
27 |
1133 |
15.5 (6.7-30.6) |
| R |
General function prediction only |
60 |
148 |
906 |
26.6 (20.3-34.3) |
| S |
Function unknown |
37 |
147 |
684 |
22.0 (15.5-30.3) |
| T |
Signal transduction mechanisms |
7 |
27 |
966 |
16.0 (6.4-32.9) |
| nab | Unclassified | 87 | 555 | 467 | 20.1 (16.1-24.8) |
aGMG were defined as genes involved in DNA replication, recombination, repair and restriction/modification and made into a separate group; otherwise, the COG classification was used. The number of DUS per group was counted. DUS frequencies are reported by their relative abundance: the number of DUS found divided by the expected number of DUS as estimated by a fourth order Markov model. We find 95% confidence intervals for the number of DUS assuming a Poisson distribution, and divide by the expected number of DUS as for the relative abundance.
bNot applicable.
In the CDS of N.meningitidis Z2491, the DUS is overall 22.0 times as frequent as expected (relative abundance as given in Table 3). In GMGs, however, the relative abundance is 44.2; in non-GMGs it is 20.2. Comparing the DUS frequency in GMGs to that in non-GMGs, this makes DUS 2.19 times more frequent in GMGs than in non-GMGs, which is statistically highly significant (P < 0.0001) (Table 4).
Table 4. Distribution of DUS between GMGs and non-GMGs in selected genomes.
| Species |
Number in | Bias towards GMGa ratio (95% CI) |
Test of even distribution (one-sided) |
|
|---|---|---|---|---|
| GMGs | non-GMGs | |||
|
N.meningitidis Z2491 |
97 |
469 |
2.19 (1.74-2.73) |
P < 0.0001 |
|
N.meningitidis MC58 |
88 |
487 |
2.00 (1.58-2.52) |
P < 0.0001 |
|
N.gonorrhoeae FA1090 |
91 |
436 |
1.69 (1.33–2.12) |
P < 0.0001 |
|
H.influenzae Rd KW20 |
112 |
820 |
1.45 (1.18–1.77) |
P = 0.0002 |
| P.multocida PM70 | 58 | 469 | 1.56 (1.17–2.05) | P = 0.0015 |
aBias towards GMGs is the ratio of the abundance of DUS in GMGs versus the abundance of DUS non-GMGs; these are given with 95% confidence intervals. The significance test is a one-sided binomial test of ratio = 1 against ratio > 1: i.e. if the portion of DUS in GMGs is higher than the portions of coding sequences made up of GMGs in terms of length.
When this analysis was extended to other genomes with known or putative DUS, we consistently found an overrepresentation of DUS within GMGs, with strong statistical significance (Table 4). The bias is particularly pronounced for the neisseriae genomes (P < 0.0001). One may argue GMG were analysed because they were observed to harbour DUS, and that this implies an implicit selection of GMG from amongst other groups of genes. However, even if the choice of GMG might have explained some degree of abundance in one genome, this cannot account for the strength of the bias nor the consistency across all the genomes.
GMGs and each COG group was analysed separately to see if differences in composition might explain the bias of DUS towards GMGs. In most of the genomes, the codon usage in different gene groups indicates that DUS should be slightly less frequent in GMGs than in non-GMGs, rather than more frequent. Only in N.meningitidis MC58 was a slight bias of DUS towards GMG expected, but this was only 8% whereas the actual bias found was 100%: i.e. twice as frequent in GMG as in non-GMG.
In the analyses of the control sequences, we found a slight bias of the control sequence group towards GMGs in N.meningitidis and H.influenzae. However, this bias was generally much weaker than that of DUS. This bias is strongest in H.influenzae (13% more in GMG), which also had the weakest bias of DUS (45% more in GMG), which is still statistically significant (P = 0.015). Again, due to the strength and consistency of DUS bias towards GMGs in diverse genomes with different DUS signatures, we conclude that this bias is specific to the biological function of DUS rather than a consequence of some kind of compositional difference between GMGs and non-GMGs. Also, the control analyses did not indicate alternative explanations.
In addition to the bias of DUS towards GMGs in the genomes of N.meningitidis, N.gonorrhoeae, H.influenzae and P.multocida, patterns of uneven DUS distribution were observed between other non-GMG COGs. DUS were generally more frequent in COG groups H and R, which encode components involved in coenzyme metabolism and those with a general function prediction only. DUS were generally less frequent in COGs C (energy production), N (cell motility and secretion) and P (inorganic ion transport); possibly also in groups F (nucleotide transport), G (carbohydrate transport) and K (transcription), although the lower DUS density in these COGs were less evident than in the former groups. It should also be noted that COG L- (non-GMGs, mainly transposases) had a very low DUS density in the N.meningitidis genomes.
Specificity of DUS
DUS-like signatures, i.e. kmers that are equal to DUS except for one base mutation, were also counted. We found that DUS-like kmers were generally no more frequent than expected from the Markov model, with only a few exceptions. In particular, in H.influenzae we also find the DUS-like oligomer AAGAGCGGT and sometimes with a change of the last base; and in the Neisseria genomes, GTCGTCTGAA and GCCGTCCGAA. Whereas DUS occur in several hundreds, DUS-like kmers generally count no more than 20–30. More details of these analyses are included in the Supplementary Material.
DISCUSSION
The identification of unique genomic signatures can provide important clues to the understanding of essential functional and evolutionary processes. Using frequent word analyses, we assessed the occurrence of the previously established DUS within the CDS of naturally competent bacterial species. Specifically, we found that the DUS of the genus Neisseria and the family Pasteurellaceae were much more frequent than any other oligomer. We have demonstrated that this abundance is far greater than can be explained from evolutionary considerations other than postulating an important biological significance of DUS over time. For H.influenzae, this agrees with previous analysis showing that the 9 bp DUS is the most frequent oligomeric sequence present in the Haemophilus genome (1,8). In contrast, the E.coli genome did not exhibit particularly frequent kmers within its coding sequences. The genome of P.multocida also displayed a putative DUS signature identical to that of H.influenzae and A.actinomycetemcomitans, but with lower frequencies (Table 2). However, natural competence for transformation has so far not been described in P.multocida or any other Pasteurella species. Nevertheless, these results indicate that P.multocida might be naturally transformable under certain environmental conditions, similar to H.influenzae. It could be argued that P.multocida has lost its competence in the recent past but one would then expect a higher number of degenerative DUS than what was found.
More strikingly, analysis of DUS representation in the annotated coding sequences of both N.meningitidis, N.gonorrhoeae and H.influenzae showed a higher density of DUS in genes involved in DNA repair, recombination, restriction-modification and replication than in other annotated gene groups. Among the 20 genes in the N.meningitidis MC58 genome harbouring the highest numbers of DUS, 9 were GMGs. The genome of P.multocida PM70 also exhibited a clear bias of DUS towards GMGs. Since the skewed DUS distribution is identified in bacterial species that are only distantly related and contain completely different DNA uptake signal sequences, we must conclude that this is a phenomenon with highly significant biological implications.
Reports have claimed that most DUS are located as inverted repeats downstream of open reading frames throughout the genome, serving as putative transcriptional terminators (21,25). However, 82% of the H.influenzae DUS occur singly (7) and a high proportion of H.influenzae DUS (66%) and neisserial DUS (35%) are located within the coding sequences which make up ∼80% of these genomes (5–8) (Table 2), thus arguing against a primary role of DUS in the regulation of gene expression. The fact that most DUS occur singly probably reflects difficulties in embedding dual repeats within functional proteins. We have also considered the possibility that DUS are arbitrarily inserted into CDS due to its abundance in intergenic regions. In this event one would expect a high selection against DUS in CDS. However, DUS inside CDS are remarkably well preserved, and there is no clear abundance of point-mutated copies of DUS (1). This strongly indicates a selective pressure for preserving DUS in CDS. The question still remains: why do the DUS so frequently form inverted repeats in the intergenic regions, while DUS inside CDS occur singly and seldom as inverted repeats? Collectively, these findings might indicate that DUS either have dual functions or that its function takes on dual forms.
Bacteria are the only organisms known to actively take up DNA from the environment and recombine it into their genomes. A variety of transformation systems are found; both the structural components involved and their functional dynamics differ among the divergent bacterial entities (26,27). The pathogenic Neisseria are reported to be naturally competent throughout their entire life cycle (28,29), while in H.influenzae competence is induced under certain environmental stimuli including starvation and nucleotide acquisition (25,30,31). Streptococcus pneumoniae and Bacillus subtilis have well-characterized pheromone- and density-regulated competence for transformation (32), while transformation in Helicobacter pylori is mediated by the basic components of a type IV secretion system (33). Although representing a completely different strategy than the quorum-sensing system of S.pneumoniae and B.subtilis, the DUS dependent transformation mechanism also ensures that competent cells preferentially take up relevant homologous DNA. It is known that not all transformation systems, even in Gram-negative bacterial species, depend on the presence of DUS for DNA uptake (27). For example, the naturally competent bacterium Acinetobacter calcoaceticus apparently lack homo-specific DUS and other markers of DNA specificity (25,34).
Considering the dissimilarities of N.meningitidis and H.influenzae DUS and their respective transformation systems, DUS in other naturally competent bacterial species are not likely to be similar. The reported absence of DUS in the H.pylori genome (35) may therefore not be definite. The two DUS-signifying properties of being overly frequent in the genome and exhibiting a biased distribution towards genome maintenance genes might be used as tools to identify new DNA uptake sequences.
The role of DUS in the long-term history of naturally competent bacterial species is unclear. It has been suggested that bacterial surface components yet to be identified facilitate selective binding and uptake of DUS-containing exogenous DNA in the initial phase of the transformation process. Since DNA taken up in nature most often originate from a mixture of dead lysed bacteria, a selective pressure on DNA uptake would then be ensured already at the bacterial surface mediating uptake of DNA from the same or a closely related bacterial species. Once DNA is inside the cell, however, DUS may serve one or more as yet unidentified functions. An even spacing of DUS around the H.influenzae genome has previously been demonstrated (1), which might indicate a role for DUS in even packaging of the chromosome in the nucleoid. Repeat elements have previously been shown to function as a signal or tool for DNA repair and/or recombination (36). Single-stranded DUS might function in a way similar to neisserial Correia elements inducing recombination events (37) or to Chi elements in E.coli promoting the production of recombinogenic strands in RecBCD-mediated recombination (38). A clustering of DUS-containing genes around the origin of replication might be an indication of a Chi-like function. This is, however, not the case. Competence for transformation is not found to be regulated by DNA damage in the naturally transformable bacteria B.subtilis and H.influenzae (39), so no clear evidence for a role for DUS in recombinational repair has yet been presented. Nevertheless, our findings strongly indicate a link between DUS and DNA metabolism. Redfield and co-workers have pointed out that nutrient acquisition may potentially be an even stronger selective force than either DNA repair or recombination, as nucleotides of DNA always are in great demand, as are other basic nutrient components (20,36). This hypothesis is not easily reconciled with a high DUS occurrence inside CDS and a skewed distribution of DUS towards GMGs.
The bias of DUS towards genome maintenance genes in the phylogenetically divergent N.meningitidis and H.influenzae clearly indicates an evolutionary conservation of this phenomenon and strongly suggests functional significance. However, an alternative interpretation could be that GMGs represent ancient genes, which have had a higher probability of acquiring DUS than other gene groups. To answer this, we have analyzed the dinucleotide sequence (DNS) frequencies in different gene groups of N.meningitidis and H.influenzae. DNS varies between organisms but is characteristically conserved within the entire genome of a given organisms and particularly so within ancient genes (40). We find as expected that GMGs have average DNS composition, but that other gene groups with average DNS do not have an overrepresentation of DUS, thus arguing against this interpretation.
Transforming DNA has traditionally been perceived as generating genetic variability and conferring change such as antibiotic resistance (18). However, transforming DNA may also mediate conservation and act to restore damaged genes. Assuming that increased DUS density enhances the propensity for uptake of particular DNA fragments, the overrepresentation of DUS in DNA repair genes may reveal the benefits of maintaining or restoring the integrity of the repair machinery through preferential uptake of ‘advantageous’ DNA. An obvious interpretation of this phenomenon is that genome maintenance genes are particularly important and must be replaced by new copies if irreparably damaged or lost, as evidenced by typical mutator genes (Table 5). In a population of dying bacteria, after for instance exposure to oxygen radicals or other DNA damaging agents, easy access by survivors to undamaged gene copies from lysed cells will contribute to the preservation of the genome stability of the population as a whole. Such a form of mutation avoidance might be highly significant during the temporary elimination described for genes with high mutation loads (41) and may contribute to alleviate the mutation load after stress-induced mutagenesis in bacteria (42). The need for this pathway is evident by the deterioration of MMR genes (41,42). This might be a reflection of a potential mechanism whereby pathogens can temporarily accept loss of genome stability in order to rapidly adapt to new conditions (41), after which genome maintenance can be re-established by uptake of the genes in demand, by transformation of DNA from the environment. Furthermore, uptake of such genes could also provide a mechanism for facilitated recovery from DNA damage after genotoxic stress. Other important questions arising relate to how such elements accumulate in bacterial genomes and what factors contribute to their distribution, density, and conservation.
Table 5. The occurrence of Neisseria and Haemophilus DUS in mutator genes.
| Strain | mutS | mutL | MutH | COG0494 (mutT)d |
|---|---|---|---|---|
|
N.meningitidis MC58a |
4 |
4 |
–c |
2/1/0/0/0 |
|
N.meningitidis Z2491a |
4 |
4 |
–c |
2/0/0/0/0 |
|
N.gonorrhoea FA1090a |
2 |
4 |
–c |
2/1/0/0 |
|
H.influenzae Rd/KW20b |
2 |
3 |
0 |
2/0/0/0 |
| P.multocida PM70b | 2 | 1 | 0 | 0/0/0/0 |
aNeisseria DUS.
bHaemophilus influenzae DUS.
cNot present in genome.
dThe Nudix NTP pyrophosphohydrolases including oxidative damage repair enzymes, occur as four or five gene homologs.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful for preliminary sequence data obtained from the Institute for Genomic Research, the Sanger Centre, the Los Alamos National Laboratory and David Dyer at the Genome Sequencing Project at the University of Oklahoma. We highly appreciate the contribution of S. A. Frye to the kmer analysis. We thank M. Koomey for stimulating discussions and O. H. Ambur for critical reading of the manuscript. Support from the Research Council of Norway, the Norwegian Cancer Society, the University of Oslo and Rikshospitalet is greatly acknowledged.
REFERENCES
- 1.Karlin S., Mrazek,J. and Campbell,A.M. (1996) Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res., 24, 4263–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocha E.P., Danchin,A. and Viari,A. (1999) Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol., 16, 1219–1230. [DOI] [PubMed] [Google Scholar]
- 3.Danner D.B., Deich,R.A., Sisco,K.L. and Smith,H.O. (1980) An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene, 11, 311–318. [DOI] [PubMed] [Google Scholar]
- 4.Elkins C., Thomas,C.E., Seifert,H.S. and Sparling,P.F. (1991) Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol., 173, 3911–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkhill J., Achtman,M., James,K.D., Bentley,S.D., Churcher,C., Klee,S.R., Morelli,G., Basham,D., Brown,D., Chillingworth,T. et al. (2000) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature, 404, 502–506. [DOI] [PubMed] [Google Scholar]
- 6.Tettelin H., Saunders,N.J., Heidelberg,J., Jeffries,A.C., Nelson,K.E., Eisen,J.A., Ketchum,K.A., Hood,D.W., Peden,J.F., Dodson,R.J. et al. (2000) Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science, 287, 1809–1815. [DOI] [PubMed] [Google Scholar]
- 7.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.F., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 8.Smith H.O., Tomb,J.F., Dougherty,B.A., Fleischmann,R.D. and Venter,J.C. (1995) Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science, 269, 538–540. [DOI] [PubMed] [Google Scholar]
- 9.Scocca J.J., Poland,R.L. and Zoon,K.C. (1974) Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J. Bacteriol., 118, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tønjum T. (2004) In Garrity,G. (ed.), Bergey’s Manual of Systematic Bacteriology. Springer Co., pp. 299–367. [Google Scholar]
- 11.Graves J.F., Biswas,G.D. and Sparling,P.F. (1982) Sequence-specific DNA uptake in transformation of Neisseria gonorrhoeae. J. Bacteriol., 152, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tønjum T., Bukholm,G. and Bøvre,K. (1990) Identification of Haemophilus aphrophilus and Actinobacillus actinomycetemcomitans by DNA-DNA hybridization and genetic transformation. J. Clin. Microbiol., 28, 1994–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Goodman,S.D., Redfield,R.J. and Chen,C. (2002) Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol., 184, 3442–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedoroff N.V. (1999) Transposable elements as a molecular evolutionary force. Ann. NY Acad. Sci., 870, 251–264. [DOI] [PubMed] [Google Scholar]
- 15.Newbury S.F., Smith,N.H., Robinson,E.C., Hiles,I.D. and Higgins,C.F. (1987) Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell, 48, 297–310. [DOI] [PubMed] [Google Scholar]
- 16.Snyder L.A., Butcher,S.A. and Saunders,N.J. (2001) Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology, 147, 2321–2332. [DOI] [PubMed] [Google Scholar]
- 17.Feil E.J., Enright,M.C. and Spratt,B.G. (2000) Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol., 151, 465–469. [DOI] [PubMed] [Google Scholar]
- 18.Rocha E.P., Matic,I. and Taddei,F. (2002) Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions? Nucleic Acids Res., 30, 1886–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith H.O., Gwinn,M.L. and Salzberg,S.L. (1999) DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol., 150, 603–616. [DOI] [PubMed] [Google Scholar]
- 20.Redfield R.J., Schrag,M.R. and Dean,A.M. (1997) The evolution of bacterial transformation: sex with poor relations. Genetics, 146, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman S.D. and Scocca,J.J. (1988) Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl Acad. Sci. USA, 85, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skovgaard M., Jensen,L.J., Brunak,S., Ussery,D. and Krogh,A. (2001) On the total number of genes and their length distribution in complete microbial genomes. Trends Genet., 17, 425–428. [DOI] [PubMed] [Google Scholar]
- 24.Reinert G., Schbath,S. and Waterman,M.S. (2000) Probabilistic and statistical properties of words: an overview. J. Comput. Biol., 7, 1–46. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz M.G., Reipschlager,K. and Wackernagel,W. (1992) Plasmid transformation of naturally competent Acinetobacter calcoaceticus in non-sterile soil extract and groundwater. Arch. Microbiol., 157, 355–360. [DOI] [PubMed] [Google Scholar]
- 26.Dubnau D. (1999) DNA uptake in bacteria. Annu. Rev. Microbiol., 53, 217–244. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz M.G. and Wackernagel,W. (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev., 58, 563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sparling P.F. (1966) Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol., 92, 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonjum T. and Koomey,M. (1997) The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships—a review. Gene, 192, 155–163. [DOI] [PubMed] [Google Scholar]
- 30.Chandler M.S. (1992) The gene encoding cAMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc. Natl Acad. Sci. USA, 89, 1626–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacFadyen L.P., Chen,D., Vo,H.C., Liao,D., Sinotte,R. and Redfield,R.J. (2001) Competence development by Haemophilus influenzae is regulated by the availability of nucleic acid precursors. Mol. Microbiol., 40, 700–707. [DOI] [PubMed] [Google Scholar]
- 32.Havarstein L.S., Coomaraswamy,G. and Morrison,D.A. (1995) An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl Acad. Sci. USA, 92, 11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofreuter D., Odenbreit,S. and Haas,R. (2001) Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol., 41, 379–391. [DOI] [PubMed] [Google Scholar]
- 34.Palmen R., Vosman,B., Buijsman,P., Breek,C.K. and Hellingwerf,K.J. (1993) Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol., 139, 295–305. [DOI] [PubMed] [Google Scholar]
- 35.Saunders N.J., Peden,J.F. and Moxon,E.R. (1999) Absence in Helicobacter pylori of an uptake sequence for enhancing uptake of homospecific DNA during transformation. Microbiology, 145, 3523–3528. [DOI] [PubMed] [Google Scholar]
- 36.Redfield R.J. (2001) Do bacteria have sex? Nature Rev. Genet., 2, 634–639. [DOI] [PubMed] [Google Scholar]
- 37.Correia F.F., Inouye,S. and Inouye,M. (1988) A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem., 263, 12194–12198. [PubMed] [Google Scholar]
- 38.Myers R.S. and Stahl,F.W. (1994) Chi and the RecBC D enzyme of Escherichia coli. Annu. Rev. Genet., 28, 49–70. [DOI] [PubMed] [Google Scholar]
- 39.Redfield R.J. (1993) Evolution of natural transformation: testing the DNA repair hypothesis in Bacillus subtilis and Haemophilus influenzae. Genetics, 133, 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlin S. (1998) Global dinucleotide signatures and analysis of genomic heterogeneity. Curr. Opin. Microbiol., 1, 598–610. [DOI] [PubMed] [Google Scholar]
- 41.Denamur E., Lecointre,G., Darlu,P., Tenaillon,O., Acquaviva,C., Sayada,C., Sunjevaric,I., Rothstein,R., Elion,J., Taddei,F. et al. (2000) Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell, 103, 711–721. [DOI] [PubMed] [Google Scholar]
- 42.Bjedov I., Tenaillon,O., Gerard,B., Souza,V., Denamur,E., Radman,M., Taddei,F. and Matic,I. (2003) Stress-induced mutagenesis in bacteria. Science, 300, 1404–1409. [DOI] [PubMed] [Google Scholar]
- 43.http://www.genome.ou.edu. 2003.
- 44.Blattner F.R., Plunkett,G., III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 45.May B.J., Zhang,Q., Li,L.L., Paustian,M.L., Whittam,T.S. and Kapur,V. (2001) Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl Acad. Sci. USA, 98, 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]