Abstract
Background
Invasive candidiasis is an important nosocomial infection associated with high mortality among immunosuppressive or critically ill patients. We described the incidence of invasive candidiasis in our hospital over 6 years and showed the antifungal susceptibility and genotypes of the isolated yeast.
Method
The yeast species were isolated on CHROMagar Candida medium and identified using an yeast identification card, followed by analysis of the D1/D2 domain of 26S rDNA. The susceptibilities of the isolates to flucytosine, amphotericin B, fluconazole, itraconazole, and voriconazole were tested using the ATB FUNGUS 3 system, and that to caspofungin was tested using E-test strips. C. albicans was genotyped using single-strand conformation polymorphism of CAI (Candida albicans I) microsatellite DNA combined with GeneScan data.
Results
From January 2006 to December 2011, a total of 259 isolates of invasive Candida spp. were obtained from 253 patients, among them 6 patients had multiple positive samples. Ninety-one stains were from blood and 168 from sterile fluids, accounting for 6.07% of all pathogens isolated in our hospital. Most of these strains were C. albicans (41.29% in blood/59.06% in sterile body fluids), followed by C. tropicalis (18.06%/25.72%), C. parapsilosis (17.42%/5.43%), C. glabrata (11.61%/3.99%) and other Candida spp. (11.61%/5.80%). Most Candida spp. were isolated from the ICU. The new species-specific CLSI candida MIC breakpoints were applied to these date. Resistance to fluconazole occurred in 6.6% of C. albicans isolates, 10.6% of C. tropicalis isolates and 15.0% of C. glabrata isolates. For the 136 C. albicans isolates, 54 CAI patterns were recognized. The C. albicans strains from blood or sterile body fluids showed no predominant CAI genotypes. C. albicans isolates from different samples from the same patient had the same genotype.
Conclusions
Invasive candidiasis has been commonly encountered in our hospital in the past 6 years, with increasing frequency of non-C. albicans. Resistance to fluconazole was highly predictive of resistance to voriconazole. CAI SSCP genotyping showed that all C. albicans strains were polymorphic. Invasive candidiasis were commonly endogenous infection.
Keywords: Invasive Candida, Epidemiology, Antifungal susceptibility, Genotyping
Background
In the early 1980s, fungi have emerged as major causes of human disease, especially in immunocompromised and hospitalized patients [1]. Candida spp. are the fourth most common pathogens in nosocomial bloodstream infections (BSIs), and they account for 12% of all hospital-acquired BSIs in the United States [2]. The mortality of candidemia in adults and neonatal children was estimated to be 15%–25% and 10%–15% in the United States, respectively [3]. Further, studies have shown that Candida infection can extend the duration of hospital stay [4], increase the cost of medical care [5], and lead to a high mortality (30%–40%) [6]. In recent years, non-C. albicans spp. have been detected more commonly in candidemia [7,8]. Both C. albicans and non-C. albicans cause invasive candidiasis, and the antifungal resistance of invasive Candida aggravates the situation. Some reports on Candida resistance to antifungals showed that resistance could make therapy more difficult [9,10]. The species and antifungal resistance of Candida differ among geographies, and it is necessary to survey the prevalence and drug resistance of Candida spp. in different locations in order to ensure effective antifungal therapy against invasive Candida.
Few studies on the long-term monitoring of the incidence and antifungal susceptibilities of invasive Candida spp. have been reported from China. This study aims to describe the trend of invasive candidiasis in a teaching hospital during the last 6 years.
Methods
Study population and data collection
The study was conducted in Beijing Chaoyang Hospital, a teaching facility of the Capital Medical University in Beijing, China. According to the diagnostic criteria, all patients with invasive candidiasis were enrolled from different wards from 2006 to 2011. The following information was collected including age, sex, underlying diseases, et al. This investigation had the approval of the Ethics Committee of Beijing Chaoyang Hospital and informed consent was obtained from all study subjects or their next of kin.
Candida strain collection
A total of 259 isolates of invasive Candida spp. were obtained from 253 patients, among them 6 patients had multiple positive samples. Invasive Candida strains were isolated from blood (91) and sterile samples (168) (including ascitic fluid, bile,central venous catheter, pleural fluid); the total of 259 samples accounted for 6.07% of all pathogens (5.75% of 1583 from blood and 6.27% of 2684 pathogens from sterile fluids) isolated in Beijing Chao-yang Hospital during the study period. Patients come from 13 provinces in china, mainly in northern china (including Beijing, Shanxi, Inner Mongolia, Heilongjiang et al.) Most of the Candida spp. were isolated from patients in the intensive care unit (ICUs). The Candida strains were primarily cultured on CHROMAgar Candida medium (JinZhang, Tianjing China) and identified using an yeast identification card (BioMérieux, France) or the API 20C AUX system (BioMérieux, France), followed by sequence analysis of the D1/D2 domain in the 26S rDNA [11]. Quality control isolates included C. albicans ATCC90028, C. parapsilosis ATCC22019, and C. krusei ATCC6258.
Antifungal drug susceptibility test
A total of 240 Candida strains were tested for antifungal susceptibility. The susceptibility of these strains to 6 antifungal drugs was tested in vitro. Flucytosine, amphotericin B, fluconazole, voriconazole, and itraconazole were tested using the ATB FUNGUS 3 system (1000723260; BioMérieux, France), and caspofungin, using the E-test (BJ3625 AB Biodisk; Solna, Sweden). For fluconazole, voriconazole, and itraconazole, the minimal inhibitory concentration (MIC) was determined by determining the concentration at which a prominent reduction in the yeast cell count was observed after 24 h of treatment. For the caspofungin, the MIC was determined by detection of a significant reduction in growth after 24 h of incubation. The MIC for amphotericin B and flucytosine were defined as the lowest concentration at which no visible growth was detected. Recently approved CLSI 24 h resistance breakpoints for fluconazole,voriconazole and echinocandins were used [12]: isolates of C. albicans, C. tropicalis and C. parapsilosis with an MIC ≥ 8 μg/ml and isolates of C. glabrata with an MIC ≥ 64 μg/ml were considered resistant to fluconazole; isolates of C. albicans, C. tropicalis and C. parapsilosis with an MIC ≥ 1 μg/ml were considered resistant to voriconazole; isolates of C. albicans and C. tropicalis with an MIC ≥ 1 μg/ml were considered resistant to caspofungin, isolates of C. parapsilosis with an MIC ≥ 8 μg/ml were considered resistant to caspofungin, isolates of C. glabrata with an MIC ≥ 0.5 μg/ml were considered resistant to caspofungin. Data were reported as the MIC ranges; the MIC50 and MIC90 values; and the number of resistant (R) isolates.
DNA extraction and PCR
DNA extraction and PCR amplification of CAI microsatellite DNA were performed according to the methods of [13]. The microsatellite locus CAI was amplified by PCR (forward primer, 5′-ATG CCA TTG AGT GGA ATT GG-3′; reverse, 5′-AGT GGC TTG TGT TGG GTT TT-3′). For GeneScan analysis, the forward primer was fluorescently labeled with 6-carboxyfluorescein at the 5′ end. PCR amplification was performed in a thermocycler (ICycler; Bio-Rad, Hercules, CA) with the following program: initial denaturing step at 95°C for 4 min; 33 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min; and a final extension step at 72°C for 7 min.
Genotype analysis
Genotype analysis of the amplified CAI fragments was carried out using the methods described by [14]. The sizes of the PCR products were automatically determined using the ABI 370 genetic analyzer (Applied Biosystem, Foster City, CA). Because of the diploid nature of C. albicans, the CAI genotypes of the strains were designated by the number of trinucleotide repeat units in both alleles of the microsatellite locus [14].
Statistical analysis
Drug susceptibility testing was analyzed using WHONET 5.6. Data were analyzed with SPSS 17.0 (SPSS Inc., Chicago, IL). Categorical variables were compared using the chi-square or Fisher’s exact test. A P value of 0.05 was considered significant.
Results
Patient characteristics
According to the diagnostic criteria of invasive candidiasis [15], a total of 253 patients were enrolled: male (170/253) 67.19%, female (83/253) 32.81%. The mean age was 65.08 ± 13.87 ys. The incidence rate of invasive candidiasis was 0.25 cases per 1,000 admissions from 2006–2011. The mortality rate at 6 weeks in ICUs (39.73%) was higher than that in other wards (17.39%), P < 0.05. The types of infection were bloodstream infection (approximately 35%), abdominal infection (approximately 35%), biliary tract infection (approximately 18%), catheter related blood stream infection (approximately 7%), and chest infection (approximately 6%), respectively.
Distribution of Candida
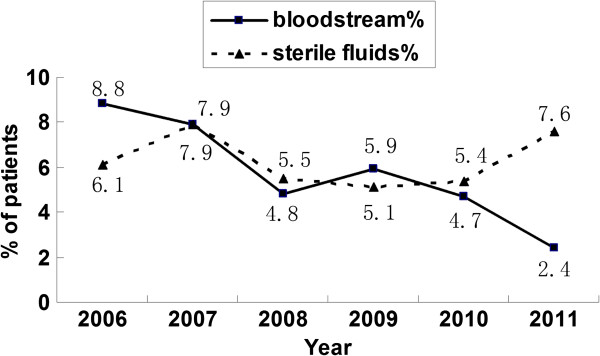
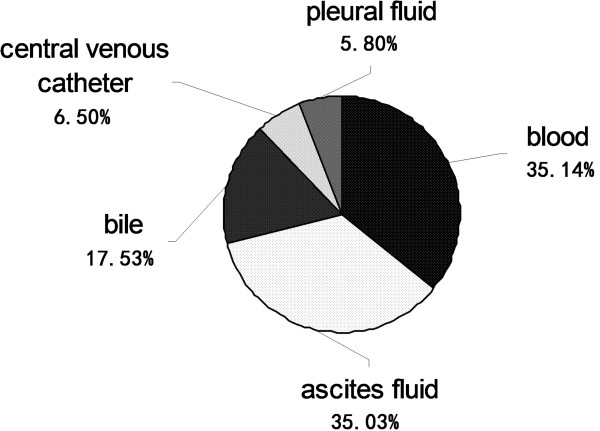
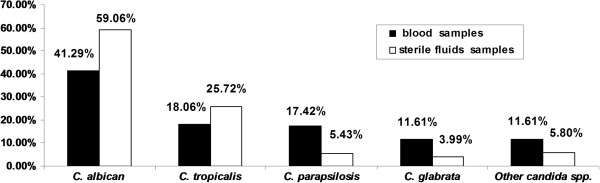
A total of 91 Candida strains were isolated from blood, which account for 5.75% of the 1583 pathogens isolated from blood from January 2006 to December 2011. The annual rates of candidemia over the 6 years of the study were as follows: 8.8%, 7.9%, 4.8%, 5.9%, 4.7%, and 2.4%. Further, 168 Candida strains were collected from sterile fluids (ascitic fluid, bile, central venous catheter and pleural fluid), which accounted for 6.27% of the 2684 pathogens isolated from sterile fluids (Figure 1). The distribution of these 259 invasive candidiasis specimens was as follows: blood 35.14%, ascitic fluid 35.03%, bile 17.53%, central venous catheter fluid 6.50%, and pleural fluids 5.80% (Figure 2). Most of these strains were C. albicans (41.29% in blood/59.06% in sterile body fluids), followed by C. tropicalis (18.06%/25.72%), C. parapsilosis (17.42%/5.43%), C. glabrata (11.61%/3.99%) and other Candida spp. (11.61%/5.80%) (Figure 3).
Figure 1.
Changes in the isolation rate of Candida spp. in the last 6 years. The annual rates of invasive candidiasis from 2006 to 2011 were showed in the figure, according to the identification of Candida strains from bloodstream and sterile fluids (ascitic fluid, bile, central venous catheter and pleural fluid).
Figure 2.
Distribution of Candida spp. in different samples (n = 259). The distribution of the 259 invasive candidiasis specimens was as followed: blood 35.14%, ascitic fluid 35.03%, bile 17.53%, central venous catheter fluid 6.50%, and pleural fluids 5.80%.
Figure 3.
Distribution of Candida spp. in blood and sterile fluids samples. Most of these strains were C. albicans (41.29% in blood/59.06% in sterile fluids), followed by C. tropicalis (18.06%/25.72%), C. parapsilosis (17.42%/5.43%), C. glabrata (11.61%/3.99%) and other Candida spp. (11.61%/5.80%).
The top 3 wards in which candidemia occurred were the ICUs (43.23%) and the hematology (16.13%) and the urology (10.32%). The wards in which Candida spp. were most frequently isolated from sterile fluids were the ICUs (41.91%) and the interventional radiology (18.15%) and the hepatobiliary (10.56%). Thus, our data showed that most patients with nosocomial candidasis were from the ICUs.
Susceptibility of antifungal drugs
The susceptibilities of the 240 yeast strains to the 6 antifungal drugs are shown in Table 1. Most Candida strains were sensitive to all the antifungal drugs, although C. glabrata and C. tropicalis showed decreased sensitivity to triazole drugs (less than 90%). The percentages of isolates in the R categories were 7.5% for fluconazole and 5.4% for voriconazole, respectively. Caspofungin and amphotericin B exhibited excellent antifungal activity against all Candida species. The MIC of fluconazole was ≥8 μg/ml for C. albicans isolates (6.6%) and that of voriconazole was ≥1 μg/ml (2.9%). Further, the MIC of fluconazole was ≥8 μg/ml for C. tropicalis isolates (10.6%) and that of voriconazole was ≥1 μg/ml (10.6%). The MIC of fluconazole was ≥64 μg/ml for C. glabrata isolates (15.0%). Three of 136 C. albicans isolates (approximately 2%) were resistant to both FLC (MIC ≥ 8 μg/ml) and VRC (MIC ≥ 1 μg/ml). Six of 59 C. tropicalis isolates (approximately 10%) had FLC MIC ≥ 8 μg/ml and VRC ≥ 1 μg/ml. Three of 20 C. glabrata isolates (approximately 15%) had FLC MIC ≥ 64 μg/ml and VRC ≥ 8 μg/ml. No isolate was resistant to all existing classes. Cross-resistance to triazole antifungal drugs was found in 2.9% C. albicans and 10.6% C. tropicalis isolates. Resistance to fluconazole was highly predictive of voriconazole resistance. No statistically significant differences in azole resistance (P > 0.05) were observed for C. albicans, C. tropicalis, and C. glabrata isolated from either blood or sterile fluids.
Table 1.
In vitro antifungal susceptibility testing of 240 clinical isolates to 6 antifungal agents
|
Species (No of isolates) |
Antifungal agent |
MIC (μg/ml) |
% Resistant |
||
|---|---|---|---|---|---|
| Range | MIC50a | MIC90a | |||
| All isolates (240) |
Fluconazole |
1– > 128 |
1 |
4 |
7.5 |
| Voriconazole(except C. glabrata) |
0.06– > 8 |
0.064 |
0.25 |
5.4 |
|
| Itraconazole(limit C.albicans only) |
0.125– > 4 |
0.125 |
0.5 |
4.9 |
|
| Caspofunginb |
0.032–2 |
0.125 |
0.5 |
0 |
|
| Flucytosine |
< 4–16 |
4 |
4 |
|
|
| Amphotericin B |
< 0.5–0.5 |
0.5 |
0.5 |
|
|
|
C. albicans (136) |
Fluconazole |
1– > 128 |
1 |
2 |
6.6 |
| Voriconazole |
0.06– > 8 |
0.064 |
0.064 |
2.9 |
|
| Itraconazole |
0.125– > 4 |
0.125 |
0.125 |
4.9 |
|
| Caspofunginb |
0.032–0.38 |
0.094 |
0.19 |
0 |
|
| Flucytosine |
< 4–16 |
4 |
4 |
|
|
| Amphotericin B |
< 0.5–0.5 |
0.5 |
0.5 |
|
|
|
C.tropicalis (59) |
Fluconazole |
1– > 128 |
1 |
128 |
10.6 |
| Voriconazole |
0.06– > 8 |
0.064 |
8 |
10.6 |
|
| Itraconazole |
0.125– > 4 |
0.125 |
4 |
|
|
| Caspofunginb |
0.032–0.38 |
0.094 |
0.19 |
0 |
|
| Flucytosine |
< 4–4 |
4 |
4 |
|
|
| Amphotericin B |
< 0.5–0.5 |
0.5 |
0.5 |
|
|
|
C. paraps-ilosis (25) |
Fluconazole |
< 1–2 |
1 |
1 |
0 |
| Voriconazole |
< 0.06–0.25 |
0.064 |
0.064 |
0 |
|
| Itraconazole |
< 0.125–0.25 |
0.125 |
0.125 |
|
|
| Caspofunginb |
0.38–2 |
1 |
1.5 |
0 |
|
| Flucytosine |
< 4– > 16 |
4 |
16 |
|
|
| Amphotericin B |
< 0.5–0.5 |
0.5 |
0.5 |
|
|
|
C.glabrata (20) |
Fluconazole |
1–64 |
2 |
64 |
15.0 |
| Voriconazole |
0.06–8 |
0.125 |
8 |
|
|
| Itraconazole |
0.125– > 4 |
0.5 |
4 |
|
|
| Caspofunginb |
0.047–0.38 |
0.19 |
0.25 |
0 |
|
| Flucytosine |
< 4–4 |
4 |
4 |
|
|
| Amphotericin B | < 0.5–0.5 | 0.5 | 0.5 | ||
Footnote: aMIC50, 50% minimal inhibitory concentration; MIC90: 90% minimal inhibitory concentration. bCaspofungin tested using E-test strips.
CAI genotypes of C. albicans
Genotype analysis was conducted for all 136 C. albicans, 54 CAI patterns were recognized. The overall frequencies of the dominant genotypes were as follows: 18–26, 7.40%; 16–21, 6.48%; 21–21, 5.55%; 11–11, 5.55%; 26–26, 4.63%; 25–33, 4.63%; 11–20, 4.46%; 11–18, 3.70%; and 18–18, 3.70%. In 9 cases, the genotypes of C. albicans isolates from different samples (blood, catheter fluid, ascitic fluid, and bile) from the same patient were identical.
Discussion
This study described the distribution, in vitro antifungal susceptibilities, and genotyping results of invasive Candida spp. isolated between 2006 and 2011 from Beijing Chaoyang hospital in China.
The results showed that invasive candidiasis has frequently occurred in our hospital in the past 6 years. The average isolation rate of Candida spp. was 6.07% among all pathogens isolated from blood and sterile fluid samples. In most cases, the invasive candidiasis occurred in the ICUs, indicating that more attention must be paid to ICU patients in order to prevent them from contracting this disease. Although C. albicans was the most predominant species, the frequency at which non-albicans Candida were detected increased obviously over the study period. In our hospital, in more than half the recent cases of candidemia, the causative organisms were non-albicans Candida strains, C. tropicalis, C. parapsilosis, and C. glabrata were the most common among the non-albicans Candida spp. detected, in accordance with reports by Pereira GH, et al. [16,17]. In the case of the blood samples, C. albicans no longer comprised the majority of isolated but remained the most frequently isolated species (41.29%), followed by C. tropicalis (18.06%), C. parapsilosis (17.42%), and C. glabrata (11.61%). In the case of the sterile samples, C. albicans was the most commonly isolated organism (59.06%), followed by C. tropicalis (25.72%), C. parapsilosis (5.43%), and C. glabrata (3.99%). It is reported that C. glabrata is No.4 among the most isolated species, which are similar to our findings, but different from those reported from Europe and the US [1,18,19].
C. abicans has traditionally been the leading cause of candidemia worldwide, with the proportion of C.abicans infections decreasing and proportion of non-C.abicans infections increasing, especially C. tropicalis, C. parapsilosis and C. glabrata. The trend has also been observed in this study, the proportion of C. abicans candidemia cases decreased from 54% to 41%. At the same time, the proportion of C. parapsilosis candidemia, C. glabrata candidemia and C. tropicalis candidemia cases rose from 12% to 17%, 7% to 12% and 15% to 18%, respectively. In the ARTEMIS DISK global antifungal surveillance project undertaken between 1997 and 2007, 90% of infections were found to be caused by 4 Candida species, i.e., C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis[20]. This distribution is similar to our study, C. abicans, C.tropicalis,C. parapsilosis and C. glabrata still comprise 92.67% of all isolates, that has not substantially changed in 20 years in the US [21]. During the last decade, the frequency of detection of the latter 3 species has been increasing in many countries, including China [22-24]. The widespread use of fluconazole may change the epidemiology of infections by non-albicans Candida, especially azole-resistant Candida spp. [25,26]. The shift to non-albicans Candida species is also significant because of the newest class of antifungal agents, so the caspofungin, are not currently recommended as primary therapy against C. parapsilosis, the third most common species.
Few studies have conducted long-term research on invasive Candida in China. The antifungal resistance rate in our study was higher than that reported by Guiyang in 2009 [27] and Hong Kong in 2011 [28]. However, none of the isolates in this study were resistant to caspofungin and amphotericin B. More than 90% of the C. albicans isolates were susceptible to all antifungal drugs between 2006 and 2011. We found that fluconazole resistance in C. albicans increased from 2.2% to 6.6% and in C.tropicalis, increased from 5.1% to 10.6%. There was also,a trend toward increased resistance to azoles in an Italian study [29]. We also found cross-resistance in our study, in agreement with previous reports from other countries [9,30]. A surveillance from 41 institutions participated study had shown that resistance to fluconazole was highly predictive of resistance to voriconazole [22]. In our study, all of the C. tropicalis isolates resistant to FLC (MIC ≥ 8 μg/ml) were also resistant to VRC (MIC ≥ 1 μg/ml). The same result in C. glabrata isolates which had FLC MIC ≥ 64 μg/ml and VRC ≥ 8 μg/ml was also observed. Therefore, we inferred that resistance to fluconazole highly tend to predictive of voriconazole resistance for C. glabrata and C. tropicalis.
Fifty-four CAI patterns were recognized from the 136 C. albicans isolates. CAI SSCP genotyping showed that the C. albicans strains were polymorphic. C. albicans isolates in different samples from the same patient had the same genotype. Compared to Li’s study [13], we found more patterns. These results implied that the genotypic distribution of C. albicans strains may be various in different samples. Isolates in different samples from the same patient showed exactly the same CAI SSCP pattern (e.g. isolates from patients CY61, CY65, CY66, CY69, CY141, CY119, CY123, CY68 and CY154 in Table 2), indicating that the infection was mainly endogenous [14,31].
Table 2.
Different specimens, isolation times, and genotypes of clinical samples
| No. patient | Isolation time | Specimen | CAI* genotype |
|---|---|---|---|
| CY61A |
2010-08-20 |
blood |
25-35 |
| CY61B |
2010-08-20 |
ascitic fluid |
25-35 |
| CY65A |
2010-02-25 |
blood |
11-20 |
| CY65B |
2010-02-25 |
catheter fluid |
11-20 |
| CY66A |
2010-03-30 |
blood |
21-21 |
| CY66B |
2010-03-29 |
catheter fluid |
21-21 |
| CY69A |
2010-08-20 |
blood |
16-21 |
| CY69B |
2010-08-23 |
catheter fluid |
16-21 |
| CY141A |
2011-01-02 |
blood |
32-46 |
| CY141B |
2011-01-07 |
catheter fluid |
32-46 |
| CY119A |
2010-01-10 |
blood |
16-18 |
| CY119B |
2010-01-07 |
ascitic fluid |
16-18 |
| CY123A |
2010-04-14 |
ascitic fluid |
17-28 |
| CY123B |
2010-04-14 |
catheter fluid |
17-28 |
| CY68A |
2010-03-23 |
blood |
25-33 |
| CY68B |
2010-03-20 |
catheter fluid |
25-33 |
| CY68C |
2010-03-22 |
ascitic fluid |
25-33 |
| CY154A |
2011-09-26 |
blood |
16-21 |
| CY154B |
2011-09-26 |
catheter fluid |
16-21 |
| CY154C |
2011-10-01 |
bile |
16-21 |
| CY154D | 2011-10-04 | ascitic fluid | 16-21 |
*CAI CA is the abbreviation of Candida albicans, followed by the roman numeral I.
Conclusions
In summary, this study showed the epidemiology and antifungal resistance of invasive Candida spp. isolated from Beijing Chaoyang hospital between 2006 and 2011. C. albicans was the most frequently isolated species, compared to the non-albicans Candida spp. Although the overall rates of resistance to antifungal agents remain low, species-specific raise some concerns, especially C. tropicalis and C. glabrata were the most frequent detected among organisms resistant to fluconazole. Currently, Caspofungin and amphotericin B are highly effective in vitro against all Candida species. Most of the invasive candidiasis cases occurred in the ICU, so careful attention should be paid to ICU patients to prevent them from contracting this disease. Genotyping of CAI SSCP showed that the C. albicans strains were polymorphic from blood and sterile samples and isolates from different samples of the same patient had the same genotype.
Abbreviations
CAI SSCP: Single-strand conformation polymorphism; ICU: Intensive care unit; MIC: Minimal inhibitory concentration; R: Resistant.
Competing interests
The authors declare that they have no competing interests.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Fang Li, Email: lifang280@sohu.com.
Lin Wu, Email: ptwm_80@yahoo.com.cn.
Bin Cao, Email: caobin1999@gmail.com.
Yuyu Zhang, Email: rainrain0828@sina.com.
Xiaoli Li, Email: 5162202@163.com.
Yingmei Liu, Email: lym13601063223@126.com.
Acknowledgements
This study was supported by a grant from the National Major S & T Research Projects for the Control and Prevention of Major Infectious Diseases in China (2012ZX10004–206), a grant WS931592 from the CHINA_ANTIF_2010 Pfizer China Antifungal Research Fund and the 2010 Janssen Research Council China (JRCC) Fund of financial support. We thank the Institute of Microbiology, Chinese Academy of Sciences. We also thank Fengyan Bai and Jing Xie for their help in the molecular biology experiments.
References
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenigün Koçak B, Kuloğlu F, Doğan Çelik A, Akata F. Evaluation of epidemiological characteristics and risk factors of candidemia in adult patients in a tertiary-care hospital. Mikrobiyol Bul. 2011;45:489–503. [PubMed] [Google Scholar]
- Arnold HM, Micek ST, Shorr AF, Zilberberg MD, Labelle AJ, Kothari S. Hospital resource utilization and costs of inappropriate treatment of candidemia. Pharmacotherapy. 2010;30:361–368. doi: 10.1592/phco.30.4.361. [DOI] [PubMed] [Google Scholar]
- Chen LY, Liao SY, Kuo SC, Chen SJ, Chen YY, Wang FD. Changes in the incidence of candidaemia during 2000–2008 in a tertiary medical centre in northern Taiwan. J Hosp Infect. 2011;78:50–53. doi: 10.1016/j.jhin.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Chow JK, Golan Y, Ruthazer R, Karchmer AW, Carmeli Y, Lichtenberg D. Factors associated with candidemia caused by non-albicans Candida species versus Candida albicans in the intensive care unit. Clin Infect Dis. 2008;46:1206–1213. doi: 10.1086/529435. [DOI] [PubMed] [Google Scholar]
- Sipsas NV, Kontoyiannis DP. Invasive fungal infections in patients with cancer in the Intensive Care Unit. Int J Antimicrob Agents. 2012;39:464–471. doi: 10.1016/j.ijantimicag.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panackal AA, Gribskov JL, Staab JF, Kirby KA, Rinaldi M, Marr KA. Clinical significance of azole antifungal drug cross-resistance in Candida glabrata. J Clin Microbiol. 2006;44:1740–1743. doi: 10.1128/JCM.44.5.1740-1743.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreghini E, Orlando F, Sanguinetti M, Posteraro B, Giannini D, Manso E. Comparative Effects of Micafungin, Caspofungin, and Anidulafungin against a Difficult-To-Treat Fungal Opportunistic Pathogen, Candida glabrata. Antimicrob Agents Chemother. 2012;56:1215–1222. doi: 10.1128/AAC.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of Candida spp. by use of clinical and laboratory standards institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50:2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bai FY. Single-strand conformation polymorphism of microsatellite for rapid strain typing of Candida albicans. Med Mycol. 2007;45(7):629–635. doi: 10.1080/13693780701530950. [DOI] [PubMed] [Google Scholar]
- Sampaio P, Gusmao L, Correia A, Alves C, Rodrigues AG, Pina-Vaz C. New microsatellite multiplex PCR for Candida albicans strain typing reveals microevolutionary changes. J Clin Microbiol. 2005;43:3869–3876. doi: 10.1128/JCM.43.8.3869-3876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner JS, Jarvis WR, Emori TG. CDC definations for nosocomial infections. Am J Infect Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- Pereira GH, Muller PR, Szeszs MW, Levin AS, Melhem MS. Five-year evaluation of bloodstream yeast infections in a tertiary hospital: the predominance of non-C. albicans Candida species. Med Mycol. 2010;48:839–842. doi: 10.3109/13693780903580121. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- Lockhart SR, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Bolden CB. Species identification and antifungal susceptibility of candida bloodstream isolates from population-based surveillance in two US cities from 2008–2011. J Clin Microbiol. 2012;50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xiao M, Chen SC, Kong F, Sun ZY, Liao K, Lu J, Shao HF, Yan Y, Fan H, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Xu YC. In Vitro Susceptibilities of Yeast Species to Fluconazole and Voriconazole as Determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J Clin Microbiol. 2012;50:3952–3959. doi: 10.1128/JCM.01130-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema D, Gibbs D, Newell V, Ellis D, Tullio V. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010;48:1366–1377. doi: 10.1128/JCM.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao AS, Brandt ME, Pruitt WR, Conn LA, Perkins BA, Stephens DS. The epidemiology of candidemia in two United States cities:results of a population-based active surveillance. Clin Infect Dis. 1999;29:1164–1170. doi: 10.1086/313450. [DOI] [PubMed] [Google Scholar]
- Lyon GM, Karatela S, Sunay S, Adiri Y. Antifungal susceptibility testing of Candida isolates from the Candida surveillance study. J Clin Microbiol. 2010;48:1270–1275. doi: 10.1128/JCM.02363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mean M, Marchetti O, Calandra T. Bench-to-bedside review: Candida infections in the intensive care unit. Crit Care. 2008;12:204. doi: 10.1186/cc6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Zhu LP, Ou XT, Xu B, Hu XP, Wang X, Weng XH. Epidemiology and risk factors for non-Candida albicans candidemia in non-neutropenic patients at a Chinese teaching hospital. Med Mycol. 2011;49:552–555. doi: 10.3109/13693786.2010.541948. [DOI] [PubMed] [Google Scholar]
- Cuenca-Estrella M, Gomez-Lopez A, Mellado E, Monzon A, Buitrago MJ, Rodriguez-Tudela JL. Activity profile in vitro of micafungin against Spanish clinical isolates of common and emerging species of yeasts and molds. Antimicrob Agents Chemother. 2009;53:2192–2195. doi: 10.1128/AAC.01543-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni P, Leonessa M, Galletto P, Latino MA, Arisio R, Maule M. Routine use of fluconazole prophylaxis in a neonatal intensive care unit does not select natively fluconazole-resistant Candida subspecies. Pediatr Infect Dis J. 2008;27:731–737. doi: 10.1097/INF.0b013e318170bb0c. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kang Y, Yokoyama K, Gonoi T, Mikami Y. Molecular differentiation and antifungal susceptibility of Candida albicans isolated from patients with respiratory infections in Guiyang Medical College Hospital, China. Nihon Ishinkin Gakkai Zasshi. 2009;50:175–178. doi: 10.3314/jjmm.50.175. [DOI] [PubMed] [Google Scholar]
- Seneviratne CJ, Wong SS, Yuen KY, Meurman JH, Parnanen P, Vaara M. Antifungal susceptibility and virulence attributes of bloodstream isolates of Candida from Hong Kong and Finland. Mycopathologia. 2011;172:389–395. doi: 10.1007/s11046-011-9444-4. [DOI] [PubMed] [Google Scholar]
- Tortorano AM, Prigitano A, Dho G, Grancini A, Passera M. Antifungal susceptibility profiles of Candida isolates from a prospective survey of invasive fungal infections in Italian intensive care units. J Med Microbiol. 2012;61:389–393. doi: 10.1099/jmm.0.037895-0. [DOI] [PubMed] [Google Scholar]
- Chen TC, Chen YH, Chen YC, Lu PL. Fluconazole exposure rather than clonal spreading is correlated with the emergence of Candida glabrata with cross-resistance to triazole antifungal agents. Kaohsiung J Med Sci. 2012;28:306–315. doi: 10.1016/j.kjms.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Hattori H, Iwata T, Nakagawa Y, Kawamoto F, Tomita Y, Kikuchi A. Genotype analysis of Candida albicans isolates obtained from different body locations of patients with superficial candidiasis using PCRs targeting 25S rDNA and ALT repeat sequences of the RPS. J Dermatol Sci. 2006;42:31–46. doi: 10.1016/j.jdermsci.2005.12.003. [DOI] [PubMed] [Google Scholar]