Abstract
In human patients, a wide range of mutations in keratin (K) 5 or K14 lead to the blistering skin disorder epidermolysis bullosa simplex. Given that K14 deficiency does not lead to the ablation of a basal cell cytoskeleton because of a compensatory role of K15, we have investigated the requirement for the keratin cytoskeleton in basal cells by inactivating the K5 gene in mice. We report that the K5−/− mice die shortly after birth, lack keratin filaments in the basal epidermis, and are more severely affected than K14−/− mice. In contrast to the K14−/− mice, we detected a strong induction of the wound-healing keratin K6 in the suprabasal epidermis of cytolyzed areas of postnatal K5−/− mice. In addition, K5 and K14 mice differed with respect to tongue lesions. Moreover, we show that in the absence of K5 and other type II keratins, residual K14 and K15 aggregated along hemidesmosomes, demonstrating that individual keratins without a partner are stable in vivo. Our data indicate that K5 may be the natural partner of K15 and K17. We suggest that K5 null mutations may be lethal in human epidermolysis bullosa simplex patients.
INTRODUCTION
Keratin (K) intermediate filaments (IFs) belong to a gene family that is organized into two subfamilies, coding for type I (K9–20) and type II keratins (K1–8). At least one member of each family is necessary to form heterodimeric IFs. In epidermis, keratins display a complex expression pattern that is widely assumed to reflect the structural requirements of distinct epidermal compartments (Fuchs and Cleveland, 1998).
The major keratins in the basal layer of stratified epithelia are K5, K14, and K15 (Fuchs and Green, 1978, 1980; Moll et al., 1982; Leube et al., 1988; Lloyd et al., 1995). In wound healing or in hyperproliferative disorders, the keratin pair K6 and K16 is transiently expressed in the suprabasal layers instead of K1, K2e, and K10 (for review, see McGowan and Coulombe, 1998). The functional significance of the highly patterned expression profile of keratins is still poorly understood, but evidence is accumulating that they have distinct functions in epidermis (Paladini and Coulombe, 1999).
The structural function of keratins was elucidated by the discovery of point mutations in human epidermal keratin genes (Corden and McLean, 1996), preceded by studies on transgenic mice expressing mutant keratin subunits (Vassar et al., 1991). These mutations cause a number of inherited human skin disorders such as epidermolysis bullosa simplex (EBS) (Bonifas et al., 1991; Coulombe et al., 1991; Lane et al., 1992; Rothnagel et al., 1995; Corden and McLean, 1996). The characteristic feature of EBS is epidermal blistering resulting from cytolysis of basal keratinocytes. On the basis of these observations, it was suggested that the overall function of epidermal keratins was to provide the reinforcement of the epidermis and the maintenance of cellular integrity under mechanical and thermal stress. Several mice carrying null or dominant keratin mutations that affect epidermal integrity have added further support to this notion (Fuchs et al., 1992; Lloyd et al., 1995; Porter et al., 1996; Ness et al., 1998; Wojcik et al., 2000; Wong et al., 2000).
So far, however, none of these mouse models have addressed the question of whether the deletion of the corresponding partner keratin in the same tissue leads to the same phenotype. This is an important issue, because it was suggested that types I and II keratins may associate with distinct sets of proteins. Transient transfection experiments and yeast 2 hybrid assays demonstrated that the amino terminal head domain of type II keratins (e.g., K5) binds to the C-terminal tail of desmoplakin I, whereas no association was seen with type I keratins (Kouklis et al., 1994; Meng et al., 1997). More recently, it was shown that the major hemidesmosomal plaque protein plectin interacts with K14 but not with K5 (Geerts et al., 1999). On the basis of these findings it is conceivable that the ablation of a type II keratin might affect a different subset of partner proteins than that of a type I keratin, leading to different phenotypes in mice.
The reported phenotype of K14−/− mice that exhibited a generalized blistering of the skin accompanied by an increased mortality was compatible with that of severe cases of Dowling–Meara EBS; however, some mice survived the first 3 mo of life, possibly because of a partial compensation of the loss of K14 by the endogenous K15, which was shown to form poorly organized IFs with K5 (Lloyd et al., 1995). With the use of K14−/− keratinocytes, even filaments between the unusual K5 and K17 were reported (Troy and Turksen, 1999).
These findings were complemented by the rare occurrence of human EBS patients who lack K14 because of a point mutation leading to a premature stop codon (Chan et al., 1994; Rugg et al., 1994; Jonkman et al., 1996; Batta et al., 2000). As in K14−/− mice, basal keratinocytes of these patients were found to contain “wispy” filaments consisting of K5 and K15 that did not form higher order keratin bundles. Taken together, the analysis of K14 deficiency in humans suggested that a minimal residual keratin cytoskeleton was sufficient to maintain the integrity of basal keratinocytes and allow the formation of a functional epidermis.
To investigate the role of type II keratins in stratified epithelia and to address the question of whether keratins fulfill distinct functions in the same cell type, we generated K5−/− mice by homologous recombination. Here we report that the deletion of K5 renders the basal epidermis deficient of keratin IFs and leads to perinatal lethality with full penetrance. Moreover, we demonstrate that K5−/− and K14−/− mice display distinct defects: whereas the former showed extensive lesions along the ventral tongue and the induction of the wound-healing keratin K6 next to cytolyzing cells, the latter displayed dorsal tongue lesions and were not reported to induce K6. Finally, on the basis of the severity of the phenotype of K5−/− mice, we suggest that a K5 deficiency in humans may be lethal.
MATERIALS AND METHODS
Construction of the Targeting Vector
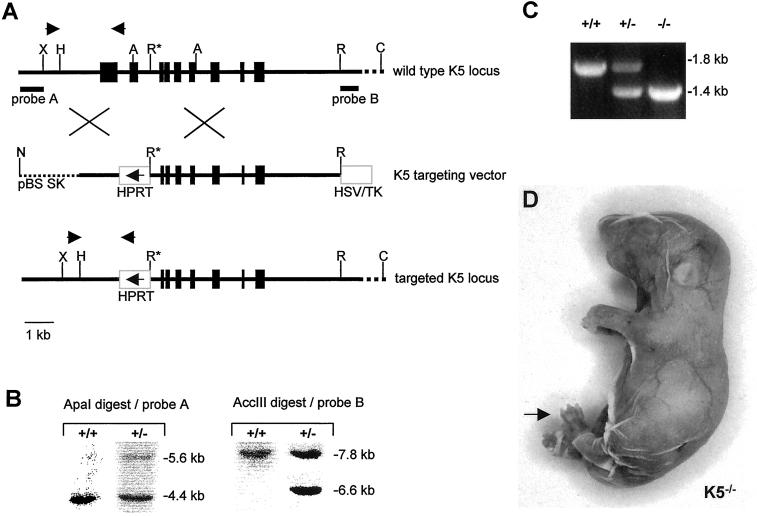
The targeting vector was constructed from a 129/ola mouse genomic library and designed to replace the first two exons of the K5 gene, including the promoter region by the mouse HPRT minigene (Porter et al., 1996). The hypoxanthin-phosphorisosyl-transferase (HPRT) minigene was flanked by sequences homologous to the K5 locus (short arm of homology: 1.4 kb; long arm of homology: 6.3 kb) and by an HSV-TK (herpes simplex virus thymidin kinase) cassette (Figure 1A). The construct was cloned into a Bluescript II SK+ vector (Stratagene, Heidelberg, Germany) and electroporated into HM-1 embryonal stem (ES) cells (Magin et al., 1998). Positive clones were detected by PCR analysis and verified by Southern blotting (Figure 1B). The position of 5′ and 3′ probes is indicated in Figure 1A. Correctly targeted ES cells were injected into blastocysts of BALB/c mice and returned to CBA recipients. Male chimeras showing germ-line transmission were used to generate K5−/− mice in the BALB/c background. The different genotypes were identified by PCR (Figure 1C). Sequence data for the mouse keratin 5 genomic sequences are available from European Molecular Biology Laboratory/GenBank/DDBJ under accession number AF306785.
Figure 1.
Targeting strategy and confirmation of gene targeting event. (A) Map of the K5 gene locus, the targeting vector, and the recombinant K5 locus. The core promoter and the first two exons of the K5 gene up to the 5′ EcoRI (R*) site were replaced by the HPRT minigene. The arrow in the HPRT minigene indicates the direction of transcription. In addition, an HSV/TK minigene was inserted as a negative selectable marker. The NotI restriction site was used to linearize the vector for transfection. Probes A and B mark the position of the 5′ and 3′ probe used in Southern blotting. Arrows above the K5 gene locus and the recombinant allele indicate primer positions for PCR-based genotyping. Letters indicate restriction sites: A, ApaI; C, AccIII; H, HindIII; N, NotI; R, EcoRI; X, XbaI. (B) Example of Southern blot analysis of ES cells. To confirm the correct targeting event, genomic DNA was digested with ApaI for detection with the 5′ probe, which led to a 5.6-kb band for the targeted allele and a 4.4-kb band for the wild-type allele. For detection with the 3′ probe, an AccIII digest was performed resulting in a 6.6-kb fragment for the targeted allele and a 7.8-kb fragment for the wild-type allele. (C) Identification of genotypes by PCR. Primers designed to identify wild-type and mutant alleles (A) were used for genotyping of the litters. The wild-type allele resulted in a product of ∼1.8-kb size, the targeted allele in a 1.4-kb product. (D) Neonatal homozygous K5−/− mouse. The fragile epidermis almost completely lost contact with the dermis after the mechanical stress of birth. Paws were sometimes denuded (arrow). K5−/− animals died within 1 h after birth.
Analysis of DNA Sequence Data
K5 genomic sequence data were obtained by automated DNA sequencing (DKFZ, Heidelberg, Germany) and analyzed with the use of the Heidelberg Unix Sequence Analysis Resources (HUSAR) program based on the Genetics Computer Group (GCG) programs.
Preparation and Analysis of Genomic DNA
For Southern blot analysis, 10 μg of genomic DNA were digested with either ApaI or AccIII and separated on a 0.8% agarose gel. Gels and blots were processed as described previously (Porter et al., 1996). The 5′ hybridization probe was a SalI–XbaI fragment deduced upstream of the K5 promoter, and the 3′ probe was an EcoRI–PstI fragment downstream of the stop codon (Figure 1A). Both were random prime labeled (Roche Diagnostics, Mannheim, Germany) with the use of [32P]-dCTP and processed as described (Magin et al., 1998).
PCR Conditions for Identification of Transgenic Mice
Genotyping of mice was performed by PCR based on three primers: primer 1 (K5 forward): 5′-tgggacaggaagagaggtgatc-3′; primer 2 (K5 reverse): 5′-accaaaaccaaatccactgccg-3′; and primer 3 (HPRT reverse): 5′-gcagtgttggctgtattttccc-3′. The PCR with the use of Taq polymerase (MBI Fermentas, St.Leon-Rot, Germany) was performed as follows: 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 1 min. The wild-type allele produces a 1.8-kb product; the targeted allele produces a 1.4-kb product (Figure 1C).
Electron Microscopy
Because the K5−/− skin was very fragile, pregnant mice were killed on embryonic day 18.5 (E18.5) of pregnancy, and pups were delivered by Caesarean section. Decapitated pups were transferred to ice-cold Karnovsky fixative for a short time. Then, back skin was dissected from the animals and incubated briefly in Karnovsky fixative to minimize splitting of epidermis from dermis. Finally, the edges of each skin sample were cut with a razor blade to yield small pieces of 3 mm2. Further processing of skin samples was performed as described earlier (Bussow, 1978).
Antibodies
The following antibodies were used: AF138 (α-K5) and AF109 (α-K1) (BAbCO, Richmond, CA); 693–1 (α-K6), α-K14 (used in Western blotting) (M. Blessing, Mainz, Germany); α-K15 (E. Fuchs, Chicago, IL); LL001 (α-K14, used in immunofluorescence analysis), LH2 (α-K10) (I.M. Leigh, London, UK); α-K17, α-K6 (used in immunofluorescence analysis) (P. Coulombe, Baltimore, MD); and RPmK161 (α-K16) (R. Porter, Dundee, UK). Alexa-conjugated secondary antibodies were purchased from Molecular Probes (Leiden, The Netherlands), and HRP-conjugated anti-mouse and anti-rabbit antisera were purchased from Dianova (Hamburg, Germany) or Roche Diagnostics.
Immunofluorescence Analysis
Back skin was taken from newborn mice, frozen in liquid nitrogen, and stored at −80°C. Cryosectioning was performed as described previously (Magin et al., 1998). The primary antibody dilutions were as follows: α-K5 1:5000; α-K14 1:5; α-K15 1:200; α-K1 1:500; α-K6 1:600; α-K16 1:500; and α-K17 1:100. Sections were viewed with a fluorescence photomicroscope (Axiophot2, Zeiss, Oberkochen, Germany).
RNA Analysis
Skin was removed from neonatal mice and immediately frozen in liquid nitrogen. RNA was extracted with TRIzol reagent (Life Technologies-BRL, Karlsruhe, Germany) according to the protocol of the supplier. Twenty micrograms of RNA were separated on a 1.2% formaldehyde/agarose gel, transferred to a nylon membrane (Genescreen, New Life Science Products, Boston, MA), and hybridized to [32P]-dCTP random prime-labeled probes (Roche). The probe for K5 was derived from the 3′ noncoding region (position 8207–8695). Probes for mouse K1, K10, and K14 were derived from the 3′ noncoding region. Final washes were done at 68°C in 0.1× SSC/0.1% SDS; then the blot was subjected to autoradiography at −80°C. For loading control, the blots were reprobed with a mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (Ambion, Austin, TX). For the analysis of K6 protein expression, we performed semiquantitative RT-PCR. Total RNA from mouse skin was prepared as described above. K6 primers were deduced downstream of the 3′ stop codon of K6b but were found to amplify both isoforms of K6, K6a as well as K6b. K6 primers were as follows: primer 1 (K6 forward): 5′-ggaggctgtgtcctctcg-3′; primer 2 (K6 reverse): 5′-tagaaaaagttactttttataaatctg-3′; K8 primers: primer 1 (K8 forward): 5′-tgcagaacatgagcattc-3′, primer 2 (K8 reverse): 5′-cagaggattagggctgat-3′; and GAPDH primers: primer 1 (forward): 5′-accacagtccatgcca-tcac-3′, primer 2 (reverse): 5′-tccaccaccatgttgctgta-3′.
In the reverse transcription reaction, 1 μg of total RNA and 20 pmol of oligonucleotide primers (Roche) were incubated at 65°C for 10 min. After a short incubation on ice, 10 mM dithiothreitol, 1 mM dNTPs (sodium salts), 20 U of RNAsin (Promega, Mannheim, Gemany), and 50 U of Expand reverse transcriptase (Roche) were added to reach a total volume of 20 μl. The reaction was incubated at 42°C for 1 h and then stopped on ice. RT-PCR for K6 and GAPDH was performed in a 50-μl reaction mixture containing 5 μl of template cDNA. Amplification was performed with Platinum Taq polymerase (Life Technologies) according to the protocol of the supplier. After 15 cycles, we took 5 μl of each reaction and performed a second PCR with 40 cycles. K8 RT-PCR was performed as follows: 5 μl of the same cDNA as for K6 were mixed with 5 μl of 10× PCR buffer, 3 μl of 25 mM MgCl2, 2 μl of 5 mM dNTPs, 25 pmol of each primer, and 1.5 U of Taq polymerase (MBI Fermentas) and filled up to 50 μl total volume. Thirty-five cycles of 94°C for 25 s, 60°C for 25 s, and 72°C for 1 min were performed.
Protein Analysis of Skin
Total proteins from skin of newborn mice were extracted by homogenization in 1× SDS sample buffer (Schroder et al., 1999). Gel electrophoresis was performed by standard procedures. The proteins were electrotransferred to polyvinylidene difluoride membranes as described above. Membranes were blocked in 0.1% Tween-20 including 5% milk powder. Primary antibodies were used in the following dilutions: α-K5 1:50,000; α-K14, 1:50,000; α-K15 1:5,000; α-K17 1:10,000; α-K1 1:200,000; α-K10 1:10,000; and α-K6 1:10,000. HRP-conjugated goat anti-mouse (Dianova) or goat anti-rabbit (Roche) antiserum was used as a secondary antibody in a 1:30,000 dilution. Bound antibodies were detected with the use of the Pierce SuperSignal Chemiluminescence Detection System (Pierce, Rockford, IL).
Two-dimensional Gel Electrophoresis
For two-dimensional gel electrophoresis of keratin complexes, cytoskeletal protein extracts were prepared from neonatal mouse skin (Hatzfeld and Franke, 1985).The cytoskeletal proteins were resuspended in isoelectric focussing sample buffer containing 4 M urea. Dialysis to urea concentrations up to 9.5 M was performed in colloidion bags (Sartorius, Göttingen, Germany). The samples were first separated by isoelectric focusing at the respective urea concentration and subsequently by SDS-PAGE (Porter et al., 1996). Ampholine composition was as follows (Pharmacia, Freiburg, Germany): 0.8% 4–6, 0.8% 5–7, and 0.4% 3–10. For Western blotting and antibodies, see above.
Analysis of Skin Barrier Formation
Skin barrier formation was analyzed according to the protocol of Hardman et al. (1998). In short, embryos were dehydrated by taking up and down a methanol series for 1 min per step. After equilibration in PBS for 1 min each, the pups were stained in 0.2% toluidine blue in water. For destaining, pups were washed in water and rinsed in 90% ethanol until the skin color differentiated. Embryos were photographed with the use of a Leica MS 5 binocular microscope (Leica, Solms, Germany).
RESULTS
K5−/− Mice Have an Extremely Fragile Epidermis and Die Shortly after Birth
To generate K5−/− mice, we deleted the promoter and the first two exons of the K5 gene (Figure 1A). Correctly targeted ES cell clones were received at high frequency (one of eight) and produced germ-line chimeras. These were used to establish heterozygous lines of BALB/c mice. Heterozygous pups appeared inconspicuous at birth and displayed no overt phenotype. Homozygotes were easily identified at birth because they displayed an extremely delicate, loose, and fragile epidermis that lost contact with the dermis (Figure 1D). Paw areas were denuded in some pups. The pups appeared to have breathing problems because sometimes the torn off epidermis had moved to close their nostrils and mouth. Stomachs of homozygous pups contained no milk. All K5−/− animals died within the first hour after birth. Obviously, the structural weakening of the epidermis caused by the loss of K5 rendered the epidermis extremely fragile and prone to rupturing during the physical trauma of birth. The analysis of >300 mice showed that all three genotypes were born at the expected Mendelian ratio excluding an embryonic phenotype.
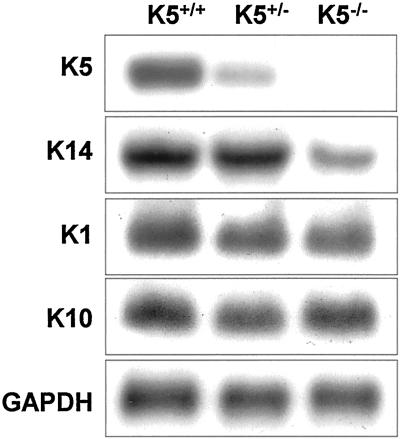
To investigate whether we had generated a true null allele, Northern and Western blot analyses were performed. As expected, the K5 mRNA was reduced in the K5+/− and completely absent in the K5−/− mice (Figure 2). Consequently, Western blot analysis of skin protein extracts of K5−/− mice confirmed the absence of K5 protein; in heterozygous K5-deficient animals the amount of K5 appeared closely similar to the wild type (Figure 3).
Figure 2.
Analysis of RNA expression by Northern blotting. K5 mRNA expression was reduced in K5+/− animals and absent in K5−/− mice. The expression of K14 was diminished in K5−/− animals, whereas that of suprabasal keratins K1 and K10 remained unaltered. Probes for the detection of K5, K14, K1, and K10 were derived from the 3′ untranslated regions. For loading controls, the RNAs were reprobed with a GAPDH probe.
Figure 3.
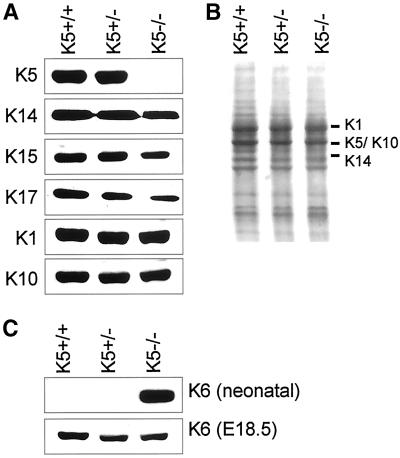
Keratin protein expression in neonatal skin. (A) No K5 protein could be detected in K5−/− mice. K14 and K15 were only slightly reduced. In addition, we found a reduction in K17 expression. K1 and K10 protein expression was unaltered in K5−/− mice. (B) Coomassie blue staining of Western blot and skin proteins for loading control. Marker sizes are denoted on the left side of the lanes. (C) In agreement with the RT-PCR data, the K6 expression was strongly induced in neonatal K5−/− mice. One day before birth (E18.5), no K6 induction was seen.
Single Type I Keratins Persist in K5−/− Mice
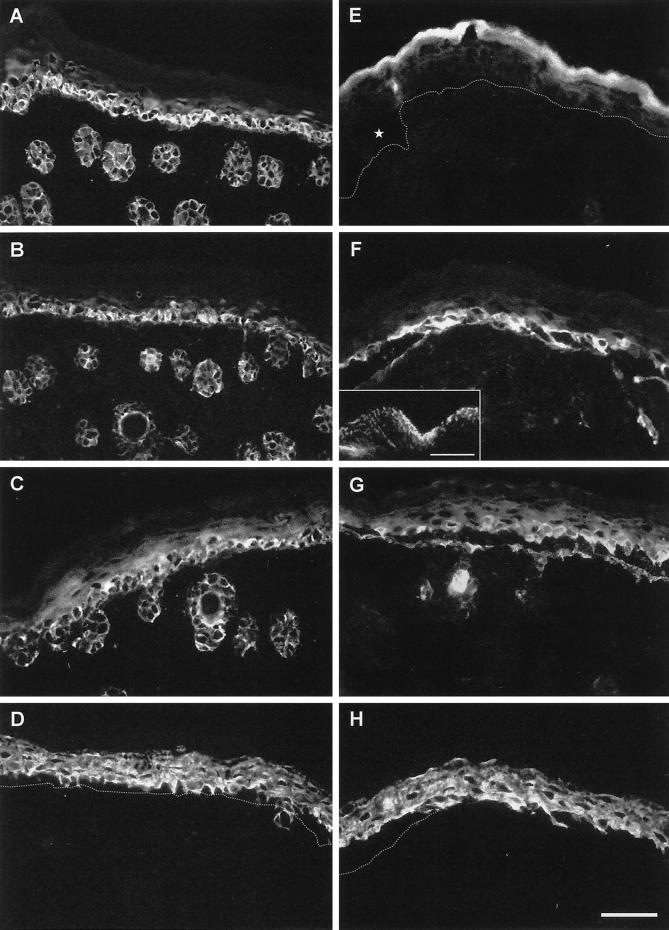
To investigate the basis for the skin fragility after the absence of K5, we performed immunofluorescence analysis of neonatal K5−/− and wild-type mice. Staining of frozen skin sections confirmed the absence of K5 in homozygous mice (Figure 4E), whereas it was strongly expressed in the basal and spinous layers of wild-type animals (Figure 4A). With the use of antibodies for K14 on adjacent sections, the basal layer of the epidermis and the outer root sheath of the hair follicles were stained for K14 in wild-type mice (Figure 4B). Surprisingly, the expression of K14 and K15 persisted in the absence of K5 (Figure 4, F and G). In K5−/− mice, a punctate K14 staining was detected along residual hemidesmosomes and in the blister roof of cytolyzed basal keratinocytes of interfollicular epidermis (Figure 4F, inset). At the same time, sparse K14 staining filaments remained in hair follicles. Because we were not able to show a colocalization with any epithelial type II keratin, we suspected that these filaments might result from the up-regulation of a yet unspecified hair keratin. The proximity of the two type I keratins to hemidesmosomes was in agreement with a recent report identifying K14 but not K5 as a binding partner for the hemidesmosomal plaque protein plectin (Geerts et al., 1999). In fact, staining for plectin revealed an almost complete colocalization with K14. The persistence of K14 is at odds with previous data showing that the stability of a keratin in cell culture is dependent on the expression of its partner keratin (Kulesh and Oshima, 1988; Kulesh et al., 1989; Lersch et al., 1989; Magin et al., 1990; Bader et al., 1991).
Figure 4.
Immunofluorescence analysis of keratin expression in neonatal K5 wild-type (A–D) and K5−/− (E–H) mice. Note the complete absence of K5 in the K5−/− mouse (E) compared with the wild type (A), whereas its partners K14 (F) and K15 (G) still persisted at levels similar to the wild type. Along the basement membrane, a punctuate staining of K14 and K15 was observed (F, inset; bar, 4 μm), whereas filaments of K14 and K15 were seen in the spinous layer. The K1 and K10 expression were unaltered (D, H). In addition to the intraepidermal disruption, we noticed that hair follicles were frequently torn out of the dermis by the mechanical stress of birth and that sometimes nuclei of cytolyzed cells had accumulated in the epidermal cleft. The dotted line denotes the position of the basement membrane. Bar, 50 μm.
To ascertain the expression of K14, we performed Northern and Western blots. Northern blot analysis of total RNA revealed that the K14 mRNA expression was unaffected in the absence of one wild-type allele of K5 but decreased in K5-deficient mice (Figure 2). This decrease is possibly a result of cytolyzed basal cells where K14 is predominantly expressed. In line with the immunofluorescence analysis, K14 protein was detected in skin protein extracts of K5+/+, K5+/−, and K5−/− mice (Figure 3). K14 was not reduced in heterozygous animals, in line with our Northern blotting results. In K5−/− mice, only a minor reduction of K14, K15, and K17 proteins was noted. The analysis of keratins 1 and 10, regarded as markers of terminal differentiation, revealed that their expression remained unaltered (Figure 4H). Most significantly, we observed an intense filamentous K14 staining in the spinous layer of K5−/− mice. Double immunofluorescence with the use of K14 and K1 antibodies revealed a colocalization. This corresponded to a small number of basal keratinocytes in wild-type skin that coexpressed K1 and K14 (Figure 4, compare B and D, F and H [see also Bickenbach et al., 1996]) and extended even into rare intact cells of the basal layer of K5−/− mice where both intact filaments and aggregates along the basement membrane stained positive for K1 and K14. Although we have not yet performed immunogold electron microscopy, this colocalization was reminiscent of that reported for K14 and K1 in K10-deficient mice (Reichelt et al.).
Because K5 is part of all stratifying epithelia, we extended our immunofluorescence analysis to other K5-expressing tissues. Besides epidermis, we studied the keratin expression in tongue, esophagus, palate, fore stomach, foot, and tail (our unpublished results). In general, the intraepidermal cleft could be observed in every stratifying epithelium that we examined, emphasizing the extreme fragility of the basal cells as a consequence of the loss of K5.
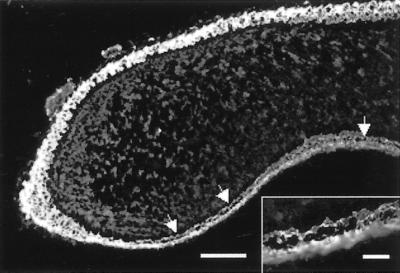
In contrast to K14−/− and K6−/− mice (Lloyd et al., 1995; Wong et al., 2000), the basal cells on the dorsal side of the tongue were largely unaffected (Figure 5). Here, suprabasal expression of K6 was recently shown to be essential for the maintenance of epidermal integrity (Wong et al., 2000). Along the ventral tongue, we observed cytolysis even before the first milk uptake. This observation supports different roles for K5 and K6. Although the former is required along the ventral epithelium, the latter seems to be essential in the filiform papillae of the dorsal tongue (Wong et al., 2000). In all tissues analyzed, K14 expression in basal keratinocytes persisted in the absence of K5 and showed a punctuate distribution in cytolyzed areas that colocalized with plectin. On the other hand, no colocalization with a type II keratin could be detected. Finally, staining for alpha-6 integrin and for constituent proteins of desmosomes did not reveal any changes, indicating that the absence of keratin filaments in basal epidermis caused a severe cytolysis but no overall change of the epidermal differentiation program.
Figure 5.
Immunofluorescence analysis of K14 expression in the tongue of neonatal K5−/− mice. In the absence of K5, increased tissue fragility was observed on the ventral side of the tongue. Bar, 140 μm; inset, 5 μm.
K5−/− and K14−/− Mice Differ with Respect to the Induction of the Wound-Healing Keratin K6
Keratins K6 and K16 are being regarded as the most sensitive indicators of disturbances in epidermal homeostasis (McGowan and Coulombe, 1998). In unaffected animals, they are restricted to hair follicles and foot sole epidermis and are absent in interfollicular epidermis (Takahashi et al., 1998; Rothnagel et al., 1999). After mechanical stress or tissue injury or as a result of hyperproliferation or hypoproliferation (Weiss et al., 1984; Stoler et al., 1988; Sellheyer et al., 1993; Wilson et al., 1994; Porter et al., 1998), particularly K6 is strongly induced in interfollicular epidermis. Therefore, we investigated the relationship between the extent of tissue damage in E18.5 and neonatal K5−/− mice and the induction of K6. In neonatal homozygous K5-deficient mice, K6 was strongly induced in most cells of the spinous and lower granular layers in cytolyzing areas (Figure 6D). With the use of a K6 antiserum that recognizes both K6a and K6b, a weak K6 expression was also noticed in the basal layer along the blister roof of cytolyzing cells. Basal keratinocytes in unaffected regions of epidermis were clearly negative for K6 expression. When K5−/− mice were analyzed at E18.5, the expression of K6 was similar to that of wild-type mice and remained restricted to the outer root sheath of hair follicles. This suggests that the induction of K6 occurs between E18.5 and the day of birth, most likely after the mechanical trauma on cytolyzing keratinocytes (Takahashi et al., 1998).
Figure 6.
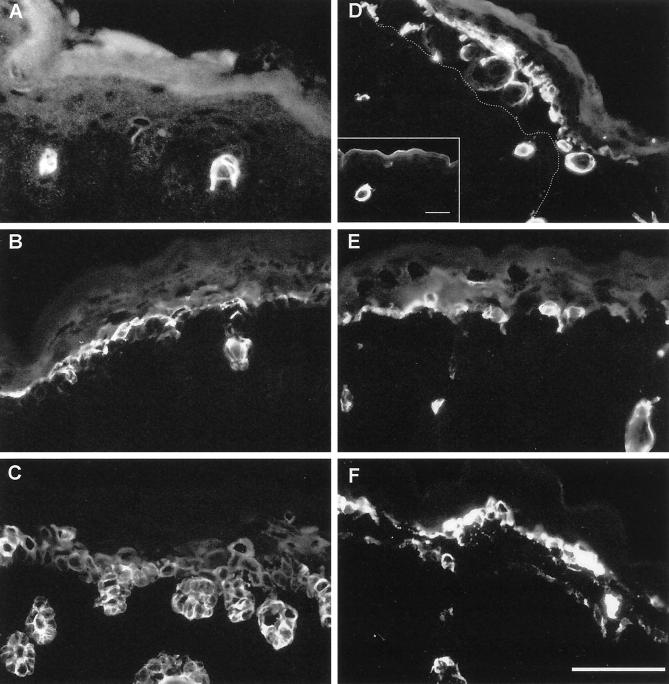
Immunofluorescence analysis of wound-healing keratin expression in skin of wild-type (A–C) and K5−/− mice (D–F). In wild-type mice, K6 expression was restricted to hair follicles and single suprabasal cells (A), whereas it was strongly increased in blisters of K5−/− mice (D). In the blister, hair follicles torn out of the dermis were noted. In unaffected areas, K6 induction was comparable to the wild type Bar, inset, 60 μm. The dotted line denotes the position of the blister base. The expression of K16 (E) was not changed remarkably when compared with the wild type (B). Note that the K17 expression in the wild-type skin (C) was almost comparable to K14 staining. In K5−/− mice, K17 (F) did not show a significant increase in expression. We noted, however, the same punctuate staining as observed before for K14 and K15. Bar, 50 μm.
In wild-type mice, K16 was found in hair follicles and in most cells of the spinous layer (Figure 6B), whereas K17 was present not only in hair follicles (Figure 6C) but also in the basal and suprabasal layers of the interfollicular epidermis, comparable to the distribution of K14 and K15. The expression of K16 (Figure 6E) and K17 (Figure 6F) remained almost unchanged in K5−/− animals as judged by immunofluorescence, except that K17 was distributed in a punctuate manner in the remains of basal epidermis of K5−/− back skin, reminiscent of the K14 and K15 pattern. These data are compatible with the notion that K5 can serve as a keratin partner not only for K14 and K15 but also for K17.
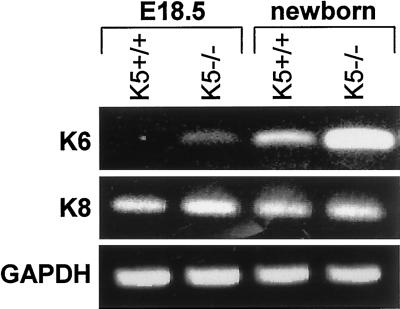
To corroborate our immunofluorescence findings on the induction of K6, we performed semiquantitative RT-PCR with the use of skin RNA from E18.5 and neonatal K5+/+ and K5−/− animals. This demonstrated a strong induction of K6 in the neonatal K5−/− mice (Figure 7). On E18.5, the expression of K6 mRNA was weakly induced compared with wild-type mice, indicating that the induction of K6 mRNA was already initiated, possibly close to cytolyzing basal keratinocytes. Subsequent Western blotting demonstrated that the strong increase in K6 mRNA was also translated into the corresponding protein in neonatal mice, whereas we were unable to visualize K6 on E18.5 by this technique.
Figure 7.
Semiquantitative RT-PCR of K6 in skin of E18.5 and neonatal mice. Note the strong induction of K6 expression in neonatal K5−/− mice. K8 and GAPDH expression were used as internal controls.
The induction of K6 in K5−/− mice seems to be in contrast to the K14−/− mice (Lloyd et al., 1995). Whether this difference is due to the extent of tissue damage in both mice or whether there are other reasons remains to be clarified.
Biochemical Analysis of Keratin Complexes
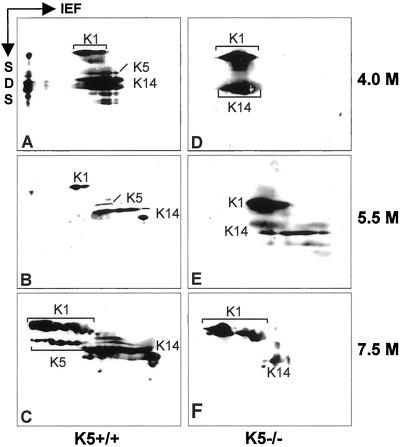
Previous data had shown that the stability of a keratin depended on the expression of its partner keratin (Kulesh et al., 1989; Lersch et al., 1989). Yet, the immunofluorescence analysis had revealed the persistence of K14 expression in K5−/− mice. Moreover, we noticed a colocalization of K14 and K1 in a few spinous cells and in addition even in a few basal cells. Therefore, we examined whether K14 might become stabilized in suprabasal epidermis by complex formation with K1. To that end, cytoskeletal keratin preparations were isolated from both genotypes of mice and resuspended in 4 M urea, a condition known to maintain oligomeric building blocks of keratins (Franke et al., 1983). Dialysis of keratin complexes against increasing concentrations of urea leads to the dissociation of individual keratin complexes. At appropriate urea concentrations, keratin complexes can be visualized by isoelectric focusing followed by SDS gel electrophoresis (Franke et al., 1983; Hatzfeld and Franke, 1985). In wild-type skin, type I and type II keratins migrated in their authentic complexes up to a concentration of 5.5 M urea. At higher urea concentrations, complexes dissociated and keratins migrated to their individual isoelectric points (Figure 8, A–C). In extracts from K5−/− mouse skin, a significant portion of K14 has already moved to its isoelectric point at 5.5 M urea, indicating the absence of its type II keratin partner K5 (Figure 8E). The remaining K14, however, resided in a complex position together with K1 (Figure 8E). This complex was dissociated at 7.5 M urea (Figure 8F). Together, these data indicate that in the absence of its natural partner, K14 can form novel heteromeric keratin complexes (see also Reichelt et al.).
Figure 8.
Analysis of keratin complexes in epidermis. Cytoskeletal extracts from neonatal wild-type (A–C) and K5−/− mice (D–F) were dialyzed against increasing urea concentrations and separated by two-dimensional gel electrophoresis (isoelectric focussing in the first dimension; SDS-PAGE in the second dimension) to analyze in vivo keratin complexes. In the wild type, keratins migrated in a complex at 4 M (A) and 5.5 M urea (B). At 7.5 M urea, complexes had dissolved and the keratins migrated according to their individual isoelectric points (C). In K5−/− mice, a small fraction of K14 was found in a complex with K1 at 4 and 5.5 M urea (E).
Ultrastructural Analysis of the Epidermis of K5−/− Mice
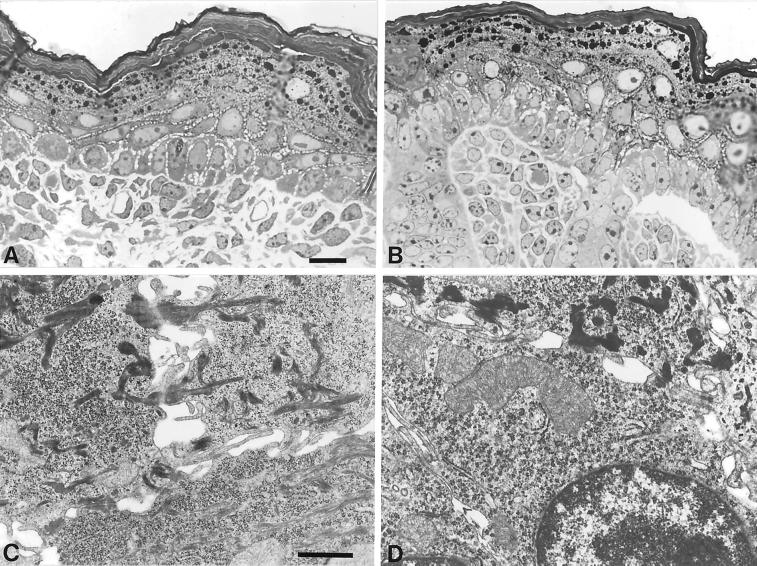
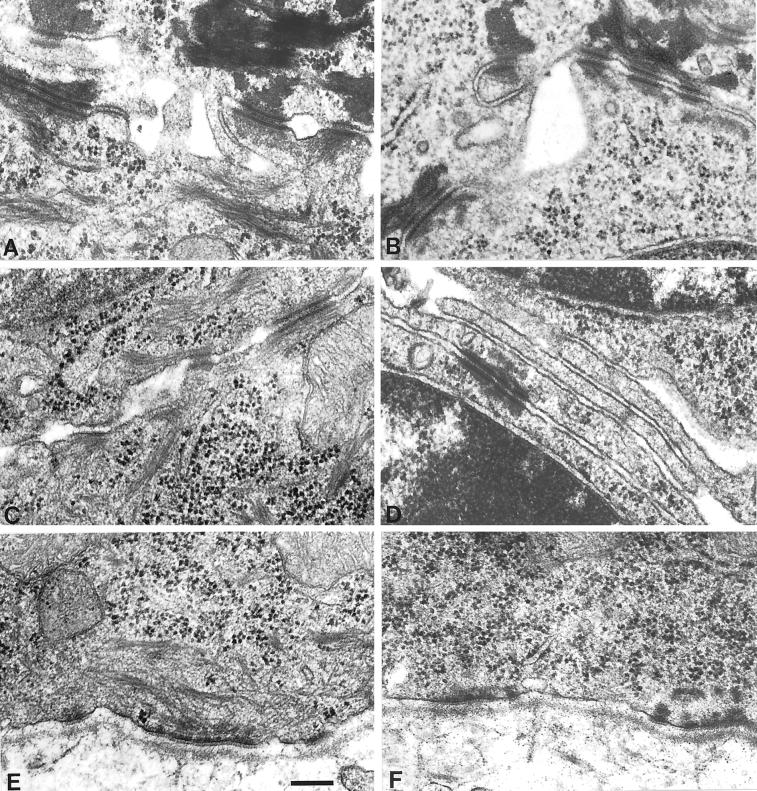
To gain insight into the ultrastructure of K5−/− epidermis, we decided to prepare the tissue specimen on E18.5 by cesarean section, because several attempts to analyze epidermis from newborn mice were unsuccessful because of the extreme fragility of K5 pups delivered by natural birth. Although this reduced the extent of cytolysis in K5−/− epidermis, considerable cell damage prevailed. In agreement with the observations made in transmission electron microscopy of EBS in humans, semithin sections of the K5−/− back skin revealed an intraepidermal cytolysis in those areas in which cytolysis occurred. The cleavage in the epidermis took place in the subnuclear cytoplasm just superficial to the hemidesmosomes, with remnants of the cytoplasm still attached to the floor of the blister (Figure 9B). Suprabasal cells on the other hand appeared unaltered compared with the wild type and showed the presence of late differentiation markers, including keratohyalin granules and a cornified envelope (Figure 9A). Although back skin of K5 heterozygous animals was indistinguishable from wild-type skin and rich in keratin filaments attached to hemidesmosomes and desmosomes (Figure 9C), we found the complete absence of keratin filaments in the basal layer of K5−/− mice (Figures 9D and 10, B, D, and F). This is in strong contrast to the K14−/− mice (Lloyd et al., 1995) and the human K14-deficient patients (Chan et al., 1994; Rugg et al., 1994; Jonkman et al., 1996; Batta et al., 2000), which still contain few and differently organized keratin IFs formed between K5 and K15 or K17. Despite the considerable strain on basal keratinocytes, neither hemidesmosomes (Figure 10F) nor desmosomes between two basal (Figure 9D) or between a basal and a suprabasal cell (Figure 10B) appeared altered. This is in agreement with the observations made in K14−/− patients or in K18−/− mice (Jonkman et al., 1996; Magin et al., 1998). The absence of keratin IFs did not seem to alter the position of basal cell nuclei or organelles.
Figure 9.
Semithin sections and electron microscopy of E18.5 back skin. (A, B) Semithin sections of back skin revealed an unaltered appearance of unaffected areas of the basal layer and the suprabasal layers of epidermis of the K5−/− skin (A) compared with the wild type (B). In K5−/− mice, mechanical stress led to the cytolysis of basal cells. The split occurred in the subnuclear cytoplasm, above the hemidesmosomes. Bar, 10 μm. (C, D) Ultrastructural analysis of the basal–suprabasal transition in epidermis of wild-type (C) and K5−/− (D) mice revealed the absence of keratin filaments in the basal layer of K5−/− epidermis. In the suprabasal layer, the keratin filaments appeared unaltered. Bar, 1 μm.
Figure 10.
Ultrastructural analysis of the epidermis in E18.5 wild-type (A, C, E) and K5−/− (B, D, F) back skin. In the basal layer of K5−/− mice, no keratin filaments were visible (F). The structure of the hemidesmosomes appeared unaltered when compared with the wild type (E). (C, D) Desmosomes between two cells in the basal layer. Despite the complete absence of the keratin filaments, no difference could be seen between the desmosomes of the wild-type (C) and K5−/− (D) mice. (A, B) Keratin filaments in the suprabasal layers appeared unchanged. Bar, 0.25 μm.
Cytoskeletal density and organization of suprabasal cells were normal when compared with those in the wild type (Figures 9D and 10B). Interestingly, with the transition from basal IFs to suprabasal layers, keratin filaments were present and showed a distribution similar to that in wild-type littermates. In agreement with Chan et al. (1994) and Lloyd et al. (1995), we concluded that K1 and K10 do form a proper suprabasal IF cytoskeleton without K5/K14 acting as a scaffold (Kartasova et al., 1993). Taken together, our data show that the presence of keratin IFs, even if organized in a different way as seen in the various K14−/− settings, is essential to maintain the integrity of the basal epidermis.
Barrier Formation in K5−/− Mice Is Not Disturbed
Earlier investigations had shown that the time course of skin barrier formation allowed the analysis of correlative changes in epidermal morphology (Aszterbaum et al., 1992). To validate our observations of an unaltered suprabasal development in the absence of K5, we performed a dye permeability assay (Hardman et al., 1998). Mice were taken each day starting at day 16.5 of embryonic development including day 19.5 by cesarean section to prevent the epidermis of the K5−/− mice from rupturing. The assay did not reveal a difference between barrier formation of the wild-type and homozygous deficient mice. At places where the epidermis was disrupted by dissection, the extreme fragility of the skin led to an intense intraepidermal staining of K5−/− mice (our unpublished results). Small lesions in the skin were stained in the K5−/− mice but not in the wild-type mouse. As a result of the fragility of the epidermis, a small lesion resulted in many cytolyzed cells, which enhanced the distribution of the stain.
DISCUSSION
Severity of K5 versus K14 Phenotype
In view of the impressive number of keratin disorders and knockout mice analyzed up to now, it is surprising that the genotype/phenotype correlation is still unclear. This is exemplified by the range of defects resulting from K14 deficiencies reported in rare human “knockouts” (Chan et al. 1994; Rugg et al. 1994; Jonkman et al. 1996; Batta et al., 2000). K14−/− mice presented with fragile stratified epithelia, causing death in the majority of homozygous litters a few days after birth (Lloyd et al., 1995). Although the notion that in those few surviving mice K15 was able to compensate for K14, this could not explain why the majority of pups died prematurely. In fact, studies with the use of keratinocytes from K14−/− mice strongly suggested that K17, another type I keratin, can form filaments with K5 (Troy and Turksen, 1999). Collectively, these data raise the issue of whether the presence of missense mutations, the formation of atypical keratin filaments, e.g., from K5/15 or K5/17, or the absence of keratins altogether leads to the most severe cell and tissue pathology. The generation of K5−/− mice has enabled us to resolve this issue.
Here, we have shown that the targeted deletion of the K5 gene resulted in the loss of a keratin cytoskeleton in the basal layer of K5−/− epidermis, accompanied by extensive cytolysis and resulting in embryonic lethality immediately after birth. Given that K5−/− mice are more severely affected than K14−/− mice and K14-deficient patients (Chan et al., 1994; Rugg et al., 1994; Jonkman et al., 1996; Batta et al., 2000), our data argue that the lack of keratin IFs in basal epidermal keratinocytes is far more detrimental than the presence of mutant or atypical keratin filaments.
Troy and Turksen (1999) were able to show that in keratinocytes cultured from K14-deficient mice, K17 forms filaments with K5 that compensate for the loss of K14. In addition, Paladini and Coulombe (1999) demonstrated a rescue of K14−/− mice, despite remaining abnormalities in the organization of the keratin filament network, by expressing either human K16 or a K14/K16 hybrid under the control of the K14 promoter. Supporting the idea of divergent and specialized functions for keratins, K18, a major keratin of simple epithelia, could not fully rescue the K14 phenotype and left mice with a fragile paw epidermis, whereas back skin was less affected. Under conditions of mechanical stress, K5/K18 filaments were unable to maintain tissue integrity (Hutton et al., 1998). In addition, the analysis of K14−/− mice had shown that the residual sparse K5/K15 filament network in the basal layer of epidermis enabled some mice to survive for up to 3 month after birth. Therefore, the absence of a keratin cytoskeleton in the basal layer of K5−/− mice strongly suggests that K5 is the only type II keratin in basal interfollicular keratinocytes. We cannot exclude the possibility, however, that individual basal cells in specialized locations, e.g., in the hair follicle, express an additional type II keratin.
An additional explanation for the phenotypic differences between K5−/− and K14−/− mice might be related to the observation that types I and II keratins interact with distinct subsets of associated proteins. The hemidesmosomal proteins plectin and BPAG1, for example, were found to associate with type I keratins, providing the stability of the intermediate filament system in the cell (Geerts et al., 1999). In K5−/− mice, K14 colocalized with hemidesmosomes. Although this association might have provided stability to K14, it did not contribute sufficiently to the epidermal integrity of K5−/− mice. We therefore interpret their phenotype as a result of the lack of a cytoskeleton in the basal cells of epidermis. In light of these data, we suggest that K5−/− mutations in humans might be lethal.
K5 Null Mice Indicate an Additional Role for K6
As outlined below, K6 together with K16 or K17 has long been considered to be a keratin that is essential in wound healing or alternative routes of differentiation (McGowan and Coulombe, 1998a). Therefore, it was unexpected that the targeted deletion of K6a resulted in only a minor wound-healing defect (Wojcik et al., 2000). In line with these findings, targeting both K6a and K6b generated mice with severe tissue degeneration confined to the posterior region of the dorsal tongue and upper palate (Wong et al., 2000). On the tongue, the filiform papillae, which may be particularly sensitive to trauma, showed the most severe defects in K6−/− animals (Wong et al., 2000). Although wound healing could not be examined because of the early death of K6 null mice, these data support a primary role for K6 as a structural keratin in the dorsal tongue, similar to K5 and K14. In support of these findings, we noted that the dorsal tongue of K5−/− mice, where K6 is constitutively expressed, remained largely unaffected, whereas the ventral aspect showed extensive tissue fragility even in pups delivered by cesarean section. It follows that along the ventral tongue, it is the basal epidermis that is predominantly susceptible to the absence of IFs as seen in K5−/− mice, whereas on the dorsal side, it is both the basal and upper strata as seen in K6−/− and K14−/− mice (Lloyd et al., 1995; Wong et al., 2000). Collectively, these data support the idea that one important function of K6 is that of a constitutive cytoskeletal keratin similar to K5 or other type II keratins. In the light of accumulating data, the presumed role of K6 in wound healing merits reevaluation. Finally, it will be a future challenge to understand the nature of those signals responsible for K6 expression at constitutive and inducible sites.
Mechanism of K6 Induction
In line with the idea of different roles for types I and II keratins, K5−/− mice show a strong induction of K6 in suprabasal interfollicular epidermis. This seems to be in contrast to K14−/− mice (Lloyd et al., 1995). K6 is unique among keratins because it is regulated in two different ways. First, it is constitutively expressed in several stratified epithelia (McGowan and Coulombe, 1998a). Second, K6, sometimes together with K16 and K17, is regarded as an indicator of epidermal disturbance and becomes strongly induced after wounding or in hyperproliferative disorders. The induction of these keratins occurs within 6–12 h in suprabasal cells after epidermis full-thickness injury (McGowan and Coulombe, 1998a) and takes place at the mRNA level (Coulombe, 1997; Takahashi et al., 1998). With the accumulation of K6, K16, and K17, the protein levels of K1 and K10 are decreased (Mansbridge and Knapp, 1987; Paladini et al., 1996). First insights into the regulation of the K6 induction in wound healing came from in vitro studies with the use of human skin keratinocytes, which revealed AP-1 and epidermal growth factor-responsive elements in the human and bovine K6 promoter region (Jiang et al., 1993; Navarro et al., 1995). Epidermal growth factor receptor signaling was found to be rapidly induced after injury (Martin and Nobes, 1992; Martin, 1997). Most recently, tumor necrosis factor-α was suggested to induce K6 through NFκB and C/EBPβ (Komine et al., 2000).
In K5−/− mice, the marked increase in the suprabasal expression of K6 was observed only in the cytolyzed areas of the epidermis. This induction was confirmed on the mRNA level and could already be seen 1 d before birth, although K6 was not detectable at the protein level. The restricted induction of K6 would be compatible with the hypothesis that a local release of tumor necrosis factor-α is triggered by keratinocyte cytolysis. Given that similar cell damage was found in K14−/− mice that survived longer than our animals (Lloyd et al., 1995), one might have expected an induction of K6 as well. The major difference between the two animal models is the absence of keratin filaments in K5−/− mice. Possibly, the larger extent of cell fragility in these mice provides a trigger sufficient for K6 induction. Therefore, our mice provide an excellent model for studying the mechanism involved in K6 induction. Understanding this pathway might allow regulated gene expression under the control of the K6 regulatory region (Takahashi and Coulombe, 1996), which would be useful for the development of gene therapy protocols for skin diseases.
Although the in vivo expression of K6 has not been analyzed in K14−/− humans, the expression of its partner K16, however, was unaltered in humans (Jonkman et al., 1996), except for the expression of K16 in some suprabasal cells noticed by Rugg et al. (1994). Although K16 and K17 are assumed to be induced in wound healing together with K6, we observed no difference in K16 and K17 expression in wild-type and K5−/− mice, despite the fact that IL-6, an additional mediator of host response to tissue injury, was found to be an inducer of K17 (Komine et al., 1996). Therefore, our in vivo data provide strong evidence that K6, K16, and K17 can be regulated individually (Porter et al., 1998). In addition, we found no alterations in K1 and K10 expression or in the number of desmosomes. Moreover, the K17 expression observed in immunofluorescence analysis of wild-type and K5−/− mice epidermis was found to resemble the expression of K14. The K17 expression reported here is in contrast to the one described by McGowan and Coulombe (1998b) and most likely is a result of mouse strain differences. On the basis of this observation, we suggest that K5 could be the natural partner of K15 and K17, which is in line with other findings (Troy and Turksen, 1999).
Expression and Accumulation of Type I Keratins
In vitro data have supported the notion that single keratins are unable to form IFs (Steinert et al., 1976; Hatzfeld and Franke, 1985) and quickly become proteolyzed in the absence of their partner (Domenjoud et al., 1988; Kulesh and Oshima, 1988; Kulesh et al., 1989; Lersch et al., 1989; Magin et al., 1990; Bader et al., 1991). In addition to a consistent suprabasal expression of type I keratins, a punctuate basal staining was observed in epidermal blisters of K5−/− mice. Surprisingly, the amount of type I keratin proteins was not reduced in the absence of the type II partner. Whether the increased stability of K14, K15, and K17 in the absence of the type II partner keratin might be mediated by other intermediate filament-associated proteins such as hsp27 (Perng et al., 1999) will be analyzed in future experiments. The ongoing expression of K1 and K10 in the absence of basal keratin IFs demonstrates that the formation of suprabasal K1/K10 keratin filaments does not rely on the preceding expression of K5/K14 as suggested by Kartasova et al. (1993). Clearly, the formation of IFs is an intrinsic property of all in vivo keratin pairs.
K5−/− Mouse as a Model for EBS and a Tool for Therapeutic Approaches
Until now, a number of transgenic mouse models have been developed for EBS. The first model was introduced by Vassar et al. (1991), who expressed a dominant negative mutant form of the human K14 gene on top of the wild-type alleles in mice. The dominant negative mutation created a truncated form of the K14 protein without the C terminus and ∼30% of the rod domain. Although the phenotype exhibited by those transgenic mice provided the first evidence that mutations in the K14 gene are the underlying cause of EBS, this model was not identical to the human disease. Because the integration of the mutant keratin was at random sites, effects of surrounding sequences could not be excluded, and finally the transgene expression levels varied as did the phenotypic severity. With the use of the same approach, Coulombe et al. (1991) were able to show that different mutations in one gene can be responsible for different subclasses of EBS by analyzing transgenic mice expressing human K14 mutants. In addition, they provided evidence that a major function of keratin filaments is to impart mechanical integrity to cells. The third model was provided by K14 knockout mice (Lloyd et al., 1995). These animals had either a recessive or a null phenotype. In patients, so far only a few K14 null cases have been reported. These patients showed a Koebner form of EBS, which is less severe than the Dowling–Meara form exhibited by K14-deficient mice. The absence of K14 lead to the formation of K5/K15 filaments, which enabled some mice to survive for months after birth.
Our K5 null mouse represents a new model for EBS, and the postnatal lethality of these mice might offer an explanation of why K5 “knockout” EBS patients have not been reported so far. The results of the analysis of K5−/− mice have an important implication for therapeutic approaches of skin diseases resulting from keratin mutations: subtle differences in IF density appear to be less important than the existence of an IF network, and a network less perfect than the K5/K14 network seems sufficient to rescue the phenotype. To test this hypothesis, we are currently investigating whether the expression of the type III IF protein desmin can compensate for the lack of K5 in the basal layer of epidermis of K5−/− mice.
In summary, we have shown that the targeted deletion of K5 differs from that of K14 not only in the extent of tissue damage but also with respect to the induction of K6. Our findings would suggest that the EBS patients carrying K5 mutations may not be viable. Moreover, our data provide evidence that K5 may be the natural partner of K15 and K17.
ACKNOWLEDGMENTS
We thank T. Schwaluk for her excellent technical assistance with blastocyst injection and embryo transfer, M. Lindemann for her technical assistance on the electron microscope analysis, and L. Casanova for her help in the characterization of the murine K5 locus. A special thank you goes to all collaborators who provided us with antibodies. M.V. acknowledges financial support from the Fundación Ramón Areces. B.P. is supported by the Graduiertenkolleg Funktionelle Proteindomaenen. J.K. is supported by a DEBRA grant to T.M.M.
Abbreviations used:
- IF
intermediate filament
- K
keratin
REFERENCES
- Aszterbaum M, Menon GK, Feingold KR, Williams ML. Ontogeny of the epidermal barrier to water loss in the rat: correlation of function with stratum corneum structure and lipid content. Pediatr Res. 1992;31:308–317. doi: 10.1203/00006450-199204000-00002. [DOI] [PubMed] [Google Scholar]
- Bader BL, Magin TM, Freudenmann M, Stumpp S, Franke WW. Intermediate filaments formed de novo from tail-less cytokeratins in the cytoplasm and in the nucleus. J Cell Biol. 1991;115:1293–1307. doi: 10.1083/jcb.115.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Rugg EL, Wilson NJ, West N, Goodyear H, Lane EB, Gratian M, Dopping-Hepenstal P, Moss C, Eady RA. A keratin 14 “knockout” mutation in recessive epidermolysis bullosa simplex resulting in less severe disease. Br J Dermatol. 2000;143:621–627. doi: 10.1111/j.1365-2133.2000.03722.x. [DOI] [PubMed] [Google Scholar]
- Bickenbach JR, Longley MA, Bundman DS, Dominey AM, Bowden PE, Rothnagel JA, Roop DR. A transgenic mouse model that recapitulates the clinical features of both neonatal and adult forms of the skin disease epidermolytic hyperkeratosis. Differentiation. 1996;61:129–139. doi: 10.1046/j.1432-0436.1996.6120129.x. [DOI] [PubMed] [Google Scholar]
- Bonifas JM, Rothman AL, Epstein EH., Jr Epidermolysis bullosa simplex: evidence in two families for keratin gene abnormalities. Science. 1991;254:1202–1205. doi: 10.1126/science.1720261. [DOI] [PubMed] [Google Scholar]
- Bussow H. Schwann cell myelin ensheathing C.N.S. axons in the nerve fiber layer of the cat retina. J Neurocytol. 1978;7:207–214. doi: 10.1007/BF01217919. [DOI] [PubMed] [Google Scholar]
- Chan Y, Anton LI, Yu QC, Jackel A, Zabel B, Ernst JP, Fuchs E. A human keratin 14 “knockout”: the absence of K14 leads to severe epidermolysis bullosa simplex and a function for an intermediate filament protein. Genes Dev. 1994;8:2574–2587. doi: 10.1101/gad.8.21.2574. [DOI] [PubMed] [Google Scholar]
- Corden LD, McLean WH. Human keratin diseases: hereditary fragility of specific epithelial tissues. Exp Dermatol. 1996;5:297–307. doi: 10.1111/j.1600-0625.1996.tb00133.x. [DOI] [PubMed] [Google Scholar]
- Coulombe PA. Towards a molecular definition of keratinocyte activation after acute injury to stratified epithelia. Biochem Biophys Res Commun. 1997;236:231–238. doi: 10.1006/bbrc.1997.6945. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Hutton ME, Vassar R, Fuchs E. A function for keratins and a common thread among different types of epidermolysis bullosa simplex diseases. J Cell Biol. 1991;115:1661–1674. doi: 10.1083/jcb.115.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenjoud L, Jorcano JL, Breuer B, Alonso A. Synthesis and fate of keratins 8 and 18 in nonepithelial cells transfected with cDNA. Exp Cell Res. 1988;179:352–361. doi: 10.1016/0014-4827(88)90274-1. [DOI] [PubMed] [Google Scholar]
- Franke WW, Kapprell HP, Mueller H. Isolation and symmetrical splitting of desmosomal structures in 9 M urea. Eur J Cell Biol. 1983;32:117–130. [PubMed] [Google Scholar]
- Fuchs E, Cleveland DW. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Esteves RA, Coulombe PA. Transgenic mice expressing a mutant keratin 10 gene reveal the likely genetic basis for epidermolytic hyperkeratosis. Proc Natl Acad Sci USA. 1992;89:6906–6910. doi: 10.1073/pnas.89.15.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Green H. The expression of keratin genes in epidermis and cultured epidermal cells. Cell. 1978;15:887–897. doi: 10.1016/0092-8674(78)90273-8. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980;19:1033–1042. doi: 10.1016/0092-8674(80)90094-x. [DOI] [PubMed] [Google Scholar]
- Geerts D, Fontao L, Nievers MG, Schaapveld RQ, Purkis PE, Wheeler GN, Lane EB, Leigh IM, Sonnenberg A. Binding of integrin alpha6beta4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J Cell Biol. 1999;147:417–434. doi: 10.1083/jcb.147.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M, Franke WW. Pair formation and promiscuity of cytokeratins: formation in vitro of heterotypic complexes and intermediate-sized filaments by homologous and heterologous recombinations of purified polypeptides. J Cell Biol. 1985;101:1826–1841. doi: 10.1083/jcb.101.5.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton E, Paladini RD, Yu QC, Yen M, Coulombe PA, Fuchs E. Functional differences between keratins of stratified and simple epithelia. J Cell Biol. 1998;143:487–499. doi: 10.1083/jcb.143.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CK, Magnaldo T, Ohtsuki M, Freedberg IM, Bernerd F, Blumenberg M. Epidermal growth factor and transforming growth factor alpha specifically induce the activation- and hyperproliferation-associated keratins 6 and 16. Proc Natl Acad Sci USA. 1993;90:6786–6790. doi: 10.1073/pnas.90.14.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, et al. Effects of keratin 14 ablation on the clinical and cellular phenotype in a kindred with recessive epidermolysis bullosa simplex. J Invest Dermatol. 1996;107:764–769. doi: 10.1111/1523-1747.ep12365805. [DOI] [PubMed] [Google Scholar]
- Kartasova T, Roop DR, Holbrook KA, Yuspa SH. Mouse differentiation-specific keratins 1 and 10 require a preexisting keratin scaffold to form a filament network. J Cell Biol. 1993;120:1251–1261. doi: 10.1083/jcb.120.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine M, Freedberg IM, Blumenberg M. Regulation of epidermal expression of keratin K17 in inflammatory skin diseases. J Invest Dermatol. 1996;107:569–575. doi: 10.1111/1523-1747.ep12582820. [DOI] [PubMed] [Google Scholar]
- Komine M, Rao LS, Kaneko T, Tomic-Canic M, Tamaki K, Freedberg IM, Blumenberg M. Inflammatory vs. proliferative processes in epidermis: TNF{alpha} induces K6b keratin synthesis through transcriptional factors NF{kappa}B and C/EBP{beta} J Biol Chem. 2000;275:32077–32088. doi: 10.1074/jbc.M001253200. [DOI] [PubMed] [Google Scholar]
- Kouklis PD, Hutton E, Fuchs E. Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J Cell Biol. 1994;127:1049–1060. doi: 10.1083/jcb.127.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesh DA, Cecena G, Darmon YM, Vasseur M, Oshima RG. Posttranslational regulation of keratins: degradation of mouse and human keratins 18 and 8. Mol Cell Biol. 1989;9:1553–1565. doi: 10.1128/mcb.9.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesh DA, Oshima RG. Cloning of the human keratin 18 gene and its expression in nonepithelial mouse cells. Mol Cell Biol. 1988;8:1540–1550. doi: 10.1128/mcb.8.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane EB, Rugg EL, Navsaria H, Leigh IM, Heagerty AH, Ishida YA, Eady RA. A mutation in the conserved helix termination peptide of keratin 5 in hereditary skin blistering. Nature. 1992;356:244–246. doi: 10.1038/356244a0. [DOI] [PubMed] [Google Scholar]
- Lersch R, Stellmach V, Stocks C, Giudice G, Fuchs E. Isolation, sequence, and expression of a human keratin K5 gene: transcriptional regulation of keratins and insights into pairwise control. Mol Cell Biol. 1989;9:3685–3697. doi: 10.1128/mcb.9.9.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube RE, Bader BL, Bosch FX, Zimbelmann R, Achtstaetter T, Franke WW. Molecular characterization and expression of the stratification-related cytokeratins 4 and 15. J Cell Biol. 1988;106:1249–1261. doi: 10.1083/jcb.106.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, Yu QC, Cheng J, Turksen K, Degenstein L, Hutton E, Fuchs E. The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J Cell Biol. 1995;129:1329–1344. doi: 10.1083/jcb.129.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin TM, Bader BL, Freudenmann M, Franke WW. De novo formation of cytokeratin filaments in calf lens cells and cytoplasts after transfection with cDNAs or microinjection with mRNAs encoding human cytokeratins. Eur J Cell Biol. 1990;53:333–348. [PubMed] [Google Scholar]
- Magin TM, Schroder R, Leitgeb S, Wanninger F, Zatloukal K, Grund C, Melton DW. Lessons from keratin 18 knockout mice: formation of novel keratin filaments, secondary loss of keratin 7 and accumulation of liver-specific keratin 8-positive aggregates. J Cell Biol. 1998;140:1441–1451. doi: 10.1083/jcb.140.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbridge JN, Knapp AM. Changes in keratinocyte maturation during wound healing. J Invest Dermatol. 1987;89:253–263. doi: 10.1111/1523-1747.ep12471216. [DOI] [PubMed] [Google Scholar]
- Martin P. Wound healing-aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- Martin P, Nobes CD. An early molecular component of the wound healing response in rat embryos—induction of c-fos protein in cells at the epidermal wound margin. Mech Dev. 1992;38:209–215. doi: 10.1016/0925-4773(92)90054-n. [DOI] [PubMed] [Google Scholar]
- McGowan K, Coulombe PA. The wound repair-associated keratins 6, 16, and 17. Insights into the role of intermediate filaments in specifying keratinocyte cytoarchitecture. Subcell Biochem. 1998a;31:173–204. [PubMed] [Google Scholar]
- McGowan K, Coulombe PA. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 1998b;143:469–86. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng JJ, Bornslaeger EA, Green KJ, Steinert PM, Ip W. Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J Biol Chem. 1997;272:21495–21503. doi: 10.1074/jbc.272.34.21495. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Navarro JM, Casatorres J, Jorcano JL. Elements controlling the expression and induction of the skin hyperproliferation-associated keratin K6. J Biol Chem. 1995;270:21362–21367. doi: 10.1074/jbc.270.36.21362. [DOI] [PubMed] [Google Scholar]
- Ness SL, Edelmann W, Jenkins TD, Liedtke W, Rustgi AK, Kucherlapati R. Mouse keratin 4 is necessary for internal epithelial integrity. J Biol Chem. 1998;273:23904–23911. doi: 10.1074/jbc.273.37.23904. [DOI] [PubMed] [Google Scholar]
- Paladini RD, Coulombe PA. The functional diversity of epidermal keratins revealed by the partial rescue of the keratin 14 null phenotype by keratin 16. J Cell Biol. 1999;146:1185–1201. doi: 10.1083/jcb.146.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladini RD, Takahashi K, Bravo NS, Coulombe PA. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J Cell Biol. 1996;132:381–397. doi: 10.1083/jcb.132.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng MD, Cairns L, van den Ijssel IJ, Prescott A, Hutcheson AM, Quinlan RA. Intermediate filament interactions can be altered by HSP27 and alphaB-crystallin. J Cell Sci. 1999;112:2099–2112. doi: 10.1242/jcs.112.13.2099. [DOI] [PubMed] [Google Scholar]
- Porter RM, Hutcheson AM, Rugg EL, Quinlan RA, Lane EB. cDNA cloning, expression, and assembly characteristics of mouse keratin 16. J Biol Chem. 1998;273:32265–32272. doi: 10.1074/jbc.273.48.32265. [DOI] [PubMed] [Google Scholar]
- Porter RM, Leitgeb S, Melton DW, Swensson O, Eady RAJ, Magin TM. Gene targeting at the mouse cytokeratin 10 locus: severe skin fragility and changes of cytokeratin expression in the epidermis. J Cell Biol. 1996;132:925–936. doi: 10.1083/jcb.132.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnagel JA, Seki T, Ogo M, Longley MA, Wojcik SM, Bundman DS, Bickenbach JR, Roop DR. The mouse keratin 6 isoforms are differentially expressed in the hair follicle, footpad, tongue and activated epidermis. Differentiation. 1999;65:119–130. doi: 10.1046/j.1432-0436.1999.6520119.x. [DOI] [PubMed] [Google Scholar]
- Rothnagel JA, Wojcik S, Liefer KM, Dominey AM, Huber M, Hohl D, Roop DR. Mutations in the 1A domain of keratin 9 in patients with epidermolytic palmoplantar keratoderma. J Invest Dermatol. 1995;104:430–433. doi: 10.1111/1523-1747.ep12666018. [DOI] [PubMed] [Google Scholar]
- Rugg EL, McLean WH, Lane EB, Pitera R, McMillan JR, Dopping HP, Navsaria HA, Leigh IM, Eady RA. A functional “knockout” of human keratin 14. Genes Dev. 1994;8:2563–2573. doi: 10.1101/gad.8.21.2563. [DOI] [PubMed] [Google Scholar]
- Schroder R, Warlo I, Herrmann H, van der Ven PF, Klasen C, Blumcke I, Mundegar RR, Furst DO, Goebel HH, Magin TM. Immunogold EM reveals a close association of plectin and the desmin cytoskeleton in human skeletal muscle. Eur J Cell Biol. 1999;78:288–295. doi: 10.1016/S0171-9335(99)80062-4. [DOI] [PubMed] [Google Scholar]
- Sellheyer K, Bickenbach JR, Rothnagel JA, Bundman D, Longley MA, Krieg T, Roche NS, Roberts AB, Roop DR. Inhibition of skin development by overexpression of transforming growth factor beta 1 in the epidermis of transgenic mice. Proc Natl Acad Sci USA. 1993;90:5237–5241. doi: 10.1073/pnas.90.11.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Idler WW, Zimmerman SB. Self-assembly of bovine epidermal keratin filaments in vitro. J Mol Biol. 1976;108:547–567. doi: 10.1016/s0022-2836(76)80136-2. [DOI] [PubMed] [Google Scholar]
- Stoler A, Kopan R, Duvic M, Fuchs E. Use of monospecific antisera and cRNA probes to localize the major changes in keratin expression during normal and abnormal epidermal differentiation. J Cell Biol. 1988;107:427–446. doi: 10.1083/jcb.107.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Coulombe PA. A transgenic mouse model with an inducible skin blistering disease phenotype. Proc Natl Acad Sci USA. 1996;93:14776–14781. doi: 10.1073/pnas.93.25.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yan B, Yamanishi K, Imamura S, Coulombe PA. The two functional keratin 6 genes of mouse are differentially regulated and evolved independently from their human orthologs. Genomics. 1998;53:170–183. doi: 10.1006/geno.1998.5476. [DOI] [PubMed] [Google Scholar]
- Troy TC, Turksen K. In vitro characteristics of early epidermal progenitors isolated from keratin 14 (K14)-deficient mice: insights into the role of keratin 17 in mouse keratinocytes. J Cell Physiol. 1999;180:409–421. doi: 10.1002/(SICI)1097-4652(199909)180:3<409::AID-JCP12>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Vassar R, Coulombe PA, Degenstein L, Albers K, Fuchs E. Mutant keratin expression in transgenic mice causes marked abnormalities resembling a human genetic skin disease. Cell. 1991;64:365–380. doi: 10.1016/0092-8674(91)90645-f. [DOI] [PubMed] [Google Scholar]
- Weiss RA, Eichner R, Sun TT. Monoclonal antibody analysis of keratin expression in epidermal diseases: a 48- and 56-kdalton keratin as molecular markers for hyperproliferative keratinocytes. J Cell Biol. 1984;98:1397–1406. doi: 10.1083/jcb.98.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Dean D, Lane EB, Dawber RP, Leigh IM. Keratinocyte differentiation in psoriatic scalp: morphology and expression of epithelial keratins. Br J Dermatol. 1994;131:191–200. doi: 10.1111/j.1365-2133.1994.tb08490.x. [DOI] [PubMed] [Google Scholar]
- Wojcik SM, Imakado S, Longley MA, Petherbridge L, Bundman DS, Bickenbach JR, Rothnagel JA, Roop DR. Expression of MK6a dominant-negative and C-terminal mutant transgenes in mice has distinct phenotypic consequences in the epidermis and hair follicle. Differentiation. 1999;65:97–112. doi: 10.1046/j.1432-0436.1999.6520097.x. [DOI] [PubMed] [Google Scholar]
- Wong P, Colucci-Guyon E, Takahashi K, Gu C, Babinet C, Coulombe PA. Introducing a null mutation in the mouse k6alpha and k6beta genes reveals their essential structural role in the oral mucosa. J Cell Biol. 2000;150:921–928. doi: 10.1083/jcb.150.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]