Abstract
Mechanisms that allow replicative DNA polymerases to attain high processivity are often specific to a given polymerase and cannot be generalized to others. Here we report a protein engineering-based approach to significantly improve the processivity of DNA polymerases by covalently linking the polymerase domain to a sequence non-specific dsDNA binding protein. Using Sso7d from Sulfolobus solfataricus as the DNA binding protein, we demonstrate that the processivity of both family A and family B polymerases can be significantly enhanced. By introducing point mutations in Sso7d, we show that the dsDNA binding property of Sso7d is essential for the enhancement. We present evidence supporting two novel conclusions. First, the fusion of a heterologous dsDNA binding protein to a polymerase can increase processivity without compromising catalytic activity and enzyme stability. Second, polymerase processivity is limiting for the efficiency of PCR, such that the fusion enzymes exhibit profound advantages over unmodified enzymes in PCR applications. This technology has the potential to broadly improve the performance of nucleic acid modifying enzymes.
INTRODUCTION
The template-directed synthesis activity of DNA polymerases is essential to the survival of all species. In most cells, multiple classes of DNA polymerases exist to accomplish distinct tasks. The predominant role of DNA polymerases in vivo is to replicate the genome during each cell cycle to preserve the genetic information. This task is accomplished by a class of highly processive replicative DNA polymerases that are capable of incorporating thousands of nucleotides without dissociating from the template DNA (1,2). The non-replicative polymerases, which serve to fill in gaps created during DNA replication, recombination and damage repair, need not be, and usually are not, very processive (3).
Nature has developed several ways of maintaining a high processivity for the replicative DNA polymerases in vivo. With the exception of certain phage (e.g. Φ29) DNA polymerases (4–7), most replicative polymerases rely on accessory proteins to achieve high processivity. For example, T7 DNA polymerase forms a stable complex with an Escherichia coli protein, thioredoxin, to achieve processive DNA synthesis (2,8). In a more complex system, E.coli polIII, T4 DNA polymerase (gp43) as well as eukaryotic replicative polymerases rely on a ring-shaped multimeric ‘DNA sliding clamp’ to maintain high processivity (9–14). For herpes simplex virus (HSV) DNA polymerase, specific association with a DNA binding protein, UL42, is necessary for processive DNA synthesis (15,16). In these examples, specific protein–protein interactions are essential for recruiting the processivity factor to its polymerase. Therefore, each processivity factor is limited to working with one or a small set of polymerases.
DNA polymerases are widely used in in vitro applications. In particular, use of thermostable polymerase Taq from Thermus aquaticus in PCR has revolutionized modern molecular biology (17). Evolved as a non-replicative polymerase, the DNA synthesis efficiency of Taq as well as other enzymes used in PCR is significantly lower than replicative polymerases. Increasing the processivity of this class of polymerases could lead to an improvement in their performance in vitro.
Approaches taken towards this goal have included the generation of a chimeric protein containing a heterologous motif or domain for binding to a known processivity factor, such as thioredoxin (18,19) or PCNA (20). Although improvement in processivity was observed when the chimeric protein and the corresponding processivity factor were used together, the application of this method to improve PCR has not been established. In a different approach, a replication complex composed of multiple polypeptides isolated from T.thermophilus was reassembled in vitro. Although processive DNA synthesis from single-stranded (ss)DNA template was observed, again, utility in PCR was not confirmed (21). Recently, after the publication of a patent application (22) disclosing the technology described in this report, it was reported that fusion of multiple helix–hairpin–helix motifs identified in DNA topoisomerase V to thermostable polymerases led to a significant increase in processivity (23). However, no benefit in PCR was demonstrated. Thus, a generalizable method to improve the processivity of DNA polymerases to benefit in vitro applications has remained elusive.
Here we report a novel and generalizable strategy to enhance the processivity of a non-replicative DNA polymerase by fusion to a sequence non-specific double-stranded (ds)DNA binding protein. We demonstrate that the processivity of both family A and family B DNA polymerases can be significantly enhanced by this approach. We provide evidence that the catalytic activity of the polymerase in nucleotide incorporation and the thermal stability of the enzyme are not affected by fusion to Sso7d. We demonstrate practical benefits of enhancing the processivity of polymerases in PCR applications.
MATERIALS AND METHODS
General reagents and equipment
Restriction enzymes and DNA ligase were purchased from New England Biolabs. Oligonucleotides were from Operon Technologies, Sigma-Genosys or Integrated DNA Technologies. DNA Engine Thermal Cyclers (MJ Research) were used for PCR. ÄKTAprime (Amersham Pharmacia Biotech) was used for protein purification.
Construction of Sso7d fusions
Construction of the Sso7d gene. Overlapping oligonucleotides (Table 1) with codons optimized for E.coli expression were used to reconstruct the Sso7d gene based on the published amino acid sequence (24). Oligonucleotides Sso-W1, Sso-W2, Sso-C1, Sso-C2 and Sso-C3 were annealed at 10 µM each in the presence of 100 mM KOAc. The annealing mix was then added to a ligation mix to ligate Sso-C1, Sso-C2 and Sso-C3 with the aid of the bridging oligonucleotides Sso-W1 and Sso-W2. Oligonucleotides Sso-R1 and Sso-Xba were used to amplify the full-length ligated product in a PCR. The amplified DNA encodes the Sso7d gene juxtaposed by the appropriate restriction sites for the subsequent steps.
Table 1. Oligonucleotides used.
| Oligo name | Oligo sequence |
|---|---|
| Reconstruction of the Sso7d gene | |
| Sso-W1 |
5′-GTATGGCGTGTGGGCAAGATGAT-3′ |
| Sso-W2 |
5′-GAAAAGGACGCGCCGAAGGAGCTG-3′ |
| Sso-C1 |
5′-CACACGCCATACTTTCTTGATCTTGGAGATGTCTACCTCTTTTTCTTCGCCTTTGTACTTGAACTTTACGGTTGC-3′ |
| Sso-C2 |
5′-GCGCGTCCTTTTCGCTTACCGCACCGCGGCCGGTCTTGCCACCGCCCTCGTCGTAGGTGAAGGAGATCATCTTGCC-3′ |
| Sso-C3 |
5′-GATTAATCCAGTGCCTTGGGACTAGTGCCACCGCCGCCCTTTTTCTGCTTCTCCAGCATCTGCAGCAGCTCCTTCG-3′ |
| Sso-R1 |
5′-ACGAATTCGAGCGCAACCGTAAAGTTCAAG-3′ |
| Sso-Xba |
5′-GCGCTCTAGATTAATCTTTTTCTGCTTCTCCAGCAT-3′ |
| Primers used for activity and processivity assays | |
| –47M13L |
5′-CGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCC-3′ |
| –40M13LFF |
5′-FAM-GTTTTCCCAGTCACGACGTTGTAAAACGACGGCC-3′ |
| Primers used for extension efficiency assay | |
| L30350F |
5′-CCTGCTCTGCCGCTTCACGC-3′ |
| L71-0.5R (0.5 kb) |
5′-TCCGGATAAAAACGTCGATGACATTTGC-3′ |
| L71-1R (1 kb) |
5′-GATGACGCATCCTCACGATAATATCCGG-3′ |
| L72-2R (2 kb) |
5′-CCATGATTCAGTGTGCCCGTCTGG-3′ |
| L72-5R (5 kb) |
5′-CGAACGTCGCGCAGAGAAACAGG-3′ |
| L72-8R (8 kb) |
5′-GCCTCGTTGCGTTTGTTTGCACG-3′ |
| L71-10R (10 kb) |
5′-GCACAGAAGCTATTATGCGTCCCCAGG-3′ |
| L71-12R (12 kb) |
5′-TCTTCCTCGTGCATCGAGCTATTCGG-3′ |
| L71-15R (15 kb) |
5′-CTTGTTCCTTTGCCGCGAGAATGG-3′ |
| Primers used for salt dependence PCR assay | |
| L30350F |
5′-CCTGCTCTGCCGCTTCACGC-3′ |
| L31240R | 5′-GCACAGCGGCTGGCTGAGGA-3′ |
S-Taq(Δ289) fusion. Plasmid pUTAQ was generated by inserting the DNA fragment containing the T.aquaticus PolI gene into vector pUC18 at the polylinker site. The PCR fragment containing the synthetic gene for Sso7d was cloned into pUTAQ to replace the region encoding the first 289 amino acids of Taq polymerase. The resulting construct, pYW1, overexpresses the S-Taq(Δ289) gene from lacI promoter upon induction with IPTG.
S-Taq fusion. A PCR fragment encoding the first 289 amino acids of Taq polymerase was amplified and inserted back into plasmid pYW1 (see above) at the junction between Sso7d and Taq(Δ289). The resulting plasmid, pYW2, allows the expression of a polypeptide (S-Taq) containing the Sso7d protein fused to the N-terminus of full-length Taq.
Mutant Sso7d fusions. An oligonucleotide containing a degenerate codon (GNG) at the position corresponding to Trp24 in Sso7d (Fig. 1A) was used to introduce point mutations via two-step sequential PCR. The final PCR fragment encoding the Sso7d gene with the degenerate codon at position 24 was inserted into pYW1 to replace the wild-type Sso7d gene. The resulting plasmids encode one of the following four amino acids, Gly (GGG), Val (GTG), Glu (GAG) or Ala (GCG), at the position corresponding to Trp24 in the wild-type Sso7d protein.
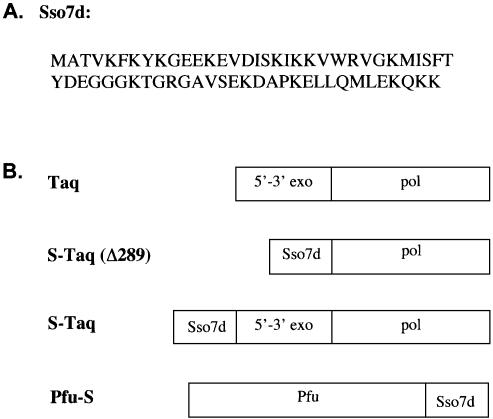
Figure 1.
(A) Amino acid sequence of Sso7d protein. (B) Schematic representation of the domain organization of Taq polymerase and the Sso7d fusion proteins.
All Taq-based Sso7d fusion proteins contained a 6-His affinity tag at the C-terminus to facilitate protein purification.
Pfu-S fusion. A plasmid (pETPFU) carrying the Pfu DNA polymerase gene under the control of the T7 promoter was modified so that unique restriction sites were introduced at the 3′ end of the Pfu gene. The resulting plasmid (pPFKS) expresses a Pfu polymerase (Pfks) with three additional amino acids (Gly-Thr-His) at its C-terminus. No functional difference was observed between Pfks and commercial Pfu polymerase (Stratagene). The Sso7d gene was PCR amplified and inserted into the pPFKS plasmid. The resulting plasmid, pPFS, overexpresses a single polypeptide (Pfu-S) containing Sso7d protein fused to the C-terminus of Pfu polymerase. The identity of the overexpressed protein was determined by both the apparent molecular weight on SDS–PAGE and its cross-reactivity with anti-Sso7d antibodies.
Expression and purification of Sso7d fusion proteins
Plasmid DNAs encoding the Taq-based Sso7d fusion proteins were transformed into E.coli strain BL21 (Stratagene) and grown in 500 ml of 2× YT medium in the presence of carbenicillin to an OD600 of 0.3. IPTG was added to 1 mM to induce expression from the lac promoter. Cells were harvested 3–4 h later. Cleared lysate was prepared as reported previously (25). Solid ammonium sulfate was added to 70% (w/v, 4°C) to precipitate the majority of the soluble proteins in the lysate.
The ammonium sulfate pellet was dissolved in Ni–NTA binding buffer NiB (20 mM Tris–HCl pH 7.9, 5 mM imidazole pH 7.5, 5 mM 2-mercaptoethanol, 0.1% NP40 and 500 mM KCl) and loaded onto a pre-equilibrated Ni–NTA (Qiagen) column (1 ml). The bound protein was step eluted using buffer IM-150 (NiB buffer with 150 mM imidazole). The fractions were pooled and purified on a heparin–agarose column by binding in buffer HP-100 (20 mM Tris-HCl pH 7.9, 0.1% NP40 and 100 mM KCl) and eluting with buffer HP-500 (HP-100 with 500 mM NaCl). The peak fractions were pooled and concentrated via a second Ni–NTA column before dialyzing (Slidelyzer; Pierce) against a pre-storage buffer (50 mM Tris–HCl pH 7.9, 250 mM KCl, 0.25 mM EDTA, 2.5 mM DTT, 0.1% NP40 and 0.1% Tween 20). Glycerol, NP40 and Tween 20 were added to the dialyzed sample so that the final storage buffer contained 20 mM Tris–HCl (pH 7.9), 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5% NP40, 0.5% Tween 20 and 50% glycerol.
Expression of Pfu-S was performed similarly as described above using strain BL21(LysS). Both carbenicillin and chloroamphenicol were added to the growth media. The ammonium sulfate precipitate was resuspended and dialyzed against buffer Q-50 (20 mM Tris–HCl pH 7.0, 50 mM NaCl and 5 mM 2-mercaptoethanol) before loading onto a 1 ml HiTrapQ column (Amersham Pharmacia Biotech). The flow-through was collected and purified on a 1 ml P11 phosphocellulose column (Whatman). The bound protein was eluted with Q-350 buffer (Q-50 buffer with 350 mM NaCl). The peak fractions were dialyzed against Q-50 buffer before loading onto a 1 ml HiTrapSP column (Amersham Pharmacia Biotech). The polymerase was eluted with a 50–800 mM NaCl gradient. The peak fractions, which corresponded to elution with 240–320 mM NaCl, were pooled and dialyzed against a pre-storage buffer (125 mM Tris–HCl pH 8.2, 0.25 mM EDTA, 2.5 mM DTT, 0.1% NP40, and 0.1% Tween 20). Glycerol, NP40 and Tween 20 were added to the dialyzed sample so that the final storage buffer contained 50 mM Tris–HCl pH 8.2, 0.1 mM EDTA, 1 mM DTT, 0.5% NP40, 0.5% Tween 20 and 50% glycerol.
Polymerase activity assay
An oligonucleotide (–47M13L, Table 1) at 0.8 µM was pre-annealed to ssM13mp18 DNA (40 nM) (Bayou Biolab) in a 2.5× reaction buffer (25 mM Tris–HCl pH 8.8, 125 mM KCl and 0.25% Triton X-100) before mixing with dNTPs and DNA polymerase. MgCl2 was added to initiate DNA synthesis at 72°C. The final reaction contained 10 mM Tris–HCl (pH 8.8), 50 mM KCl, 2 mM MgCl2, 200 µM dNTPs, 0.1% Triton X-100 and 16 nM primed template. Samples were taken at various time points and added to a 1:200 dilution of PicoGreen (Molecular Probes) in TE (10 mM Tris–HCl pH 8.0 and 1.0 mM EDTA pH 8.0). The amount of DNA synthesized was quantified using a fluorescence plate reader (BioTek Instruments). The unit activity of the DNA polymerase of interest was determined by comparing its initial rate with that of a control DNA polymerase (e.g. AmpliTaq, from Applied Biosystems). For S-Taq(Δ289), Taq, S-Taq, Pfu-S and the mutant Sso7d fusion proteins, the unit activity determined by this method agreed well (within 2-fold) with that determined by the conventional radioactive unit definition assay (26; T. Tenkanen and T. Soininen, personal communication). When significant differences were observed for Taq(Δ289) and Pfu between the two assays, units were defined using the conventional method.
Processivity assay
A 5′ FAM-labeled primer (50 nM) (–40M13LFF, Table 1) was added to ssM13mp18 DNA (100 nM) in the presence of 10 mM Tris–HCl pH 8.8, 50 mM KCl, 2.5 mM MgCl2, 250 µM dNTPs and 0.1% Triton X-100, unless otherwise indicated. Primer annealing to the DNA template was achieved by heating at 90°C for 5 min, cooling to 72°C at 0.1°C/s, incubating at 72°C for 10 min, cooling again to 4°C at 0.1°C/s, using a DNA Engine thermal cycler (MJ Research). Then 4 μl of DNA polymerase (diluted in 10 mM Tris–HCl pH 8.8, 50 mM KCl and 0.1% Triton X-100) was then added to 16 μl of primed template at a molar ratio of 1:500–1:10 000 to initiate DNA synthesis at 72°C. Samples were diluted in gel loading dye and analyzed on a MJ BaseStation Sequencer (MJ Research). To ensure that no multiple binding/extension occurs on any primer–template complexes, both the polymerase concentration and the reaction time were varied, and the median product length was determined for each reaction. The median product length is defined as the length of the product at which the total fluorescence intensity of all products up to that length equals 50% of the sum fluorescence intensity of all detectable products. In general, a higher ratio of primer–template to polymerase concentration is necessary to achieve processive conditions for low processivity enzymes, whereas for high processivity enzymes a lower ratio is needed to detect longer primer extension products under processive conditions (see corresponding figure legends). When the median product length no longer changes with an increase in reaction time or a decrease in polymerase concentration, the traces of those samples were used to determine the processivity using the analysis method described by von Hippel et al. (27). Each peak with a signal level significantly above the background was integrated to obtain the intensity at each position (nI) and the total peak intensity (nT) of all detectable products. The integration data were plotted as log(nI/nT) versus n – 1, where n is the number of nucleotide residues incorporated, and fitted to the following equation:
log(nI/nT) = (n – 1)logPI + log(1 – PI)
where PI represents the probability of not terminating at position I and is defined as the ‘microscopic processivity parameter’ for this position. The average primer extension length was determined from 1/(1 – PI).
Steady-state kinetic analyses
Steady-state kinetic analyses were performed using the polymerase activity assay described above with the following modifications. Each reaction contained 1.2 nM enzyme. The molar concentration of the enzyme was determined using a combination of the Bradford assay (Bio-Rad) and SDS–PAGE analysis. The pre-annealed primed ssM13 template was used at 1–12 nM for S-Taq(Δ289), Taq and S-Taq, 5–30 nM for Taq(Δ289 and 0.2–6 nM for Pfu and Pfu-S. Buffers with two different salt concentrations were used. For Taq(Δ289), S-Taq(Δ289), Pfu and Pfu-S the buffer contained 10 mM Tris–HCl (pH 8.8), 10 mM KCl, 2 mM MgCl2 and 0.1% Triton X-100. For Taq and S-Taq the buffer contained 50 mM KCl and was otherwise identical to the first buffer. Then, dNTPs were added at 200 µM each. The initial rate of each reaction was plotted against primer–template concentration and fitted to the following equation:
V = kcat × [E] × [D]/(Km (DNA) + [D])
where V is the initial rate, [D] is the primer–template concentration, [E] is the enzyme concentration, kcat is the turnover rate and Km (DNA) is the primer–template concentration at which half of the maximum activity is achieved.
Thermal stability assay
Each enzyme (aliquoted in 30 µl at 40 U/ml) was first incubated at 97.5°C for varied periods of time in the presence of the corresponding optimal reaction buffer as indicated in the figure legends. At each time point, the heated sample was transferred to a 72°C thermal block and allowed to equilibrate for 1 min. Then, 20 µl of the sample was mixed with an equal volume of 2× polymerase activity assay reaction mix pre-heated to 72°C (see above). The remaining activity was plotted versus time spent at 97.5°C. The data were fitted to the following equation:
ln(A) = ln(A0) – kt
where A represents the remaining activity at time t, A0 is the activity at t = 0 and k is the rate constant for enzyme inactivation. The half-life (t1/2) was determined from the following equation:
t1/2 = (ln2)/k
PCR efficiency assay
λ DNA (130 pg/µl) was used as the template to assess the relative efficiency of each polymerase in a PCR. For extension efficiency comparison, a set of primers (see Table 1) was used to amplify amplicons of 0.5, 1, 2, 5, 8, 10, 12 and 15 kb in size from the template in a 20 µl reaction. For the salt dependence comparison experiment, a single pair of primers (see Table 1) that amplifies a 0.9 kb amplicon was used for all reactions. The amount of enzyme, the reaction buffer for each enzyme and the cycling protocol used are indicated in the corresponding figure legends. It was necessary to use a higher amount of the low efficiency polymerases [e.g. Taq(Δ289) and Pfu] so that a sufficient amount of DNA could be generated to allow comparison. Upon completion of the PCR, 5 µl of the PCR was mixed with loading dye and loaded onto a 1% agarose gel. The gel was stained with ethidium bromide. A Kodak gel documentation system was used to photograph the gel.
RESULTS
Strategy to improve the processivity of Taq polymerase
Taq polymerase consists of two distinct structural and functional domains, the 5′→3′ exonuclease domain and the polymerase domain. Mutant Taq lacking the exonuclease domain is significantly less processive than full-length Taq (26,28), suggesting that the exonuclease domain must be involved in maintaining processivity. Previous structural studies of a Taq–DNA complex have revealed possible interactions between the exonuclease domain and the DNA template (29). The reduced processivity of truncated Taq could be the result of losing such interactions. We hypothesized that replacing the exonuclease domain with a dsDNA binding protein might recover the lost processivity of the truncated Taq. The dsDNA binding protein must be thermostable and bind to dsDNA without sequence preference in order to function together with the polymerase domain. We identified Sso7d (Fig. 1A), an abundant 7 kDa protein from hyperthermophilic archaeabacteria Sulfolobus solfataricus (30), as a candidate to replace the Taq exonuclease domain. Sso7d belongs to a family of proteins thought to play the role of stabilizing genomic DNA in hyperthermophilic archaea. This protein binds to dsDNA as a monomer without marked sequence preference (24,31,32). Based on our hypothesis, when covalently linked (or fused) to DNA polymerase, Sso7d could provide additional contact with dsDNA and, consequently, enhance processivity.
To test this hypothesis, we first fused Sso7d to the N-terminus of Taq(Δ289) to replace the 5′→3′ exonuclease domain (Fig. 1B). Sso7d was also fused to the N-terminus of full-length Taq (Fig. 1B) to investigate whether the processivity of full-length Taq can be further improved. Both fusion proteins as well as their non-fusion counterparts were purified to homogeneity and their identities were confirmed using both anti-Taq and anti-Sso7d antibodies (data not shown).
Sso7d fusion increases the processivity of both Taq(Δ289) and full-length Taq
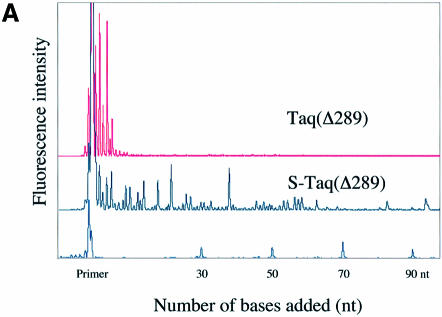
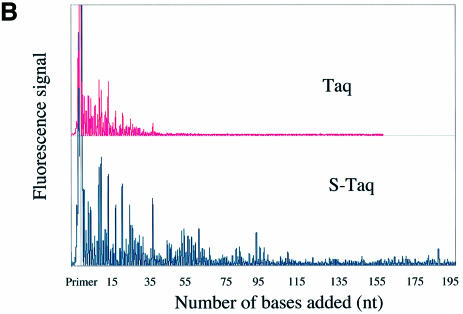
A fluorescence-based highly sensitive processivity assay (Materials and Methods) was used to determine the processivity of Taq(Δ289), S-Taq(Δ289), Taq and S-Taq. Reaction conditions were varied to identify processive conditions for each polymerase under which the median product length was independent of both enzyme concentration and reaction time. Figure 2A and B show the electropherogram traces for Taq(Δ289) and S-Taq(Δ289) and for Taq and S-Taq, respectively, obtained under processive conditions. In von Hippel et al. (27), the processivity of a DNA polymerase is defined as the probability (PI) of the polymerase not terminating at a specific position of the template. The processivity parameter (PI) and the average primer extension length [1/(1 – PI)] for each enzyme were determined (see Materials and Methods) and are summarized in Table 2. The processivity for Taq(Δ289) is 0.64, which correlates to an average primer extension length of 2.9 nt. In contrast, the processivity of S-Taq(Δ289) fusion protein is 0.98, correlating to an average primer extension length of 51 nt, which is significantly higher than that of Taq(Δ289). The processivity of full-length Taq is significantly higher than that of Taq(Δ289). Fusion to Sso7d further increased its processivity, leading to an increase in the average primer extension length from 22 nt for Taq to ∼104 nt for S-Taq. These results demonstrate that for both Taq(Δ289) and full-length Taq polymerases the fusion of Sso7d can significantly enhance the processivity of the polymerase.
Figure 2.
Processivity analyses of Taq-based polymerases. Each trace represents one lane from a sequencing gel and each peak represents a single primer extension product. Reaction time was 5 min for each enzyme. (A) Electropherogram traces of Taq(Δ289) (10 pM) and S-Taq(Δ289) (16 pM). The size marker lane is shown at the bottom and the corresponding primer extension product length is indicated on the x-axis. (B) Electro pherogram traces of Taq (4 pM) and S-Taq (20 pM). The labels on the x-axis indicate the primer extension product length, which is determined based on size markers run on the same gel (trace not shown).
Table 2. Summary of processivity analyses.
| Enzyme | Microscopic processivity (PI)a | Average primer extension length (nt) [1/(1 – PI)]a |
|---|---|---|
| Taq(Δ289) |
0.64 ± 0.09 |
2.9 ± 0.7 |
| S-Taq(Δ289) |
0.980 ± 0.002 |
51 ± 6 |
| Taq |
0.95 ± 0.01 |
22 ± 3 |
| S-Taq |
0.99 ± 0.02 |
104 ± 17 |
| S(G)-Taq(Δ289) |
0.91 ± 0.01 |
11 ± 1 |
| S(V)-Taq(Δ289) |
0.92 ± 0.01 |
12 ± 2 |
| S(E)-Taq(Δ289) |
0.85 ± 0.04 |
7 ± 2 |
| Pfu |
0.84 ± 0.01 |
6.4 ± 0.5 |
| Pfu-S | 0.982 ± 0.001 | 55 ± 3 |
aData were analyzed as described in Materials and Methods. The standard deviation was calculated based on results from multiple sets of data.
Binding of Sso7d to dsDNA is important for enhancing processivity
The interactions between Sso7d and dsDNA have been studied extensively (31,32). One of the key residues in Sso7d, Trp24, was found to play multiple roles in binding to dsDNA (24), but not in the overall thermal stability of Sso7d (33). If the binding interaction between Sso7d and dsDNA is important for the observed enhancement of processivity, mutating the Trp24 residue should reduce the binding affinity of Sso7d for dsDNA and in turn reduce its ability to enhance the processivity of DNA polymerases. To investigate the correlation between the binding of Sso7d to dsDNA and processivity, three mutant fusion proteins were generated, S(V)-Taq(Δ289), S(G)-Taq(Δ289) and S(E)-Taq(Δ289), which contain Val, Gly or Glu, respectively, in place of Trp24 in Sso7d. The processivity of each mutant protein was determined (Table 2). Mutant fusion proteins containing either a Val or a Gly substitution showed a significant reduction in processivity compared to fusion protein containing wild-type Sso7d [S-Taq(Δ289)], but still maintained a higher processivity than Taq(Δ289). The average primer extension lengths for S(V)-Taq(Δ289) and S(G)-Taq(Δ289) are 12 and 11 nt, respectively, which are three to four times longer than that of S-Taq(Δ289). In the case of the Glu substitution, which is the most different from the wild-type residue, the processivity of the mutant fusion protein is further reduced. The average primer extension length of S(E)-Taq(Δ289) is ∼7 nt. These results indicate that mutations in the dsDNA binding motif of Sso7d alters its ability to enhance processivity and establishes a strong correlation between the binding of Sso7d to dsDNA and the enhancement of the processivity of polymerases when fused to Sso7d.
Sso7d fusion increases the processivity of Pfu polymerase
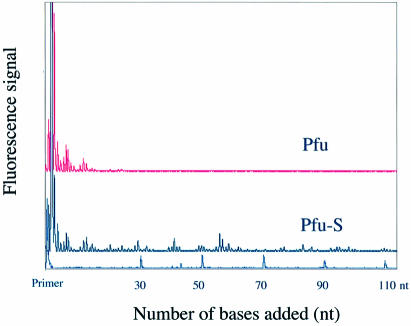
DNA polymerases have been classified into four major families, A, B, C and D, based on alignment of amino acid sequences (34–36). Different families of polymerases have distinct structural and functional properties. Having demonstrated that Sso7d fusion enhances the processivity of Taq, a family A DNA polymerase, we investigated whether the same strategy can be applied to polymerases from other families. We chose the family B polymerase from Pyroccocus furiosis, Pfu polymerase, which is commonly used for high fidelity PCR applications. Based on structural information for a Pfu homolog (37), Sso7d was fused to the C-terminus of Pfu polymerase (Pfu-S) (Fig. 1A) to allow both the polymerase and the Sso7d domain to interact with the primer–template DNA appropriately. Purified Pfu-S was compared with commercial Pfu polymerase (Cloned Pfu from Stratagene) in a processivity assay (Fig. 3). Pfu alone exhibits a processivity of 0.84, which correlates to an average primer extension length of 6 nt (Table 2). In contrast, the fusion protein Pfu-S exhibits significantly higher processivity with a PI of 0.98 and an average extension length of 55 nt. This demonstrates that enhancement of processivity by Sso7d fusion is not limited to one type of polymerase; it can be generally applied to benefit at least both family A and family B DNA polymerases.
Figure 3.
Electropherogram traces of Pfu and Pfu-S for the processivity analysis. Size markers and the corresponding primer extension product length are shown at the bottom. The polymerase concentrations were 16 pM for Pfu and 80 pM for Pfu-S. The reaction buffer for both enzymes contained 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100 and 100 µg/ml BSA. The reaction time was 5 min.
Sso7d fusion does not negatively affect catalytic activity
It is conceivable that increasing the processivity by effectively tethering a polymerase to DNA template might be achieved at the expense of decreased catalytic activity. Naturally existing processivity factors, such as sliding clamps, use a mechanism (11,13) that enhances the processivity without hindering the elongation rate of the polymerase. To investigate whether the fusion of Sso7d to DNA polymerase alters the catalytic rate of nucleotide incorporation, a steady-state kinetic analysis was performed. The rate of primer extension was measured at different primer–template concentrations in the presence of a saturating amount of dNTPs to allow determination of the kcat of nucleotide incorporation with a saturating amount of primer–template (Table 3). Both S-Taq and S-Taq(Δ289) fusion enzymes have nearly identical turnover rates (kcat) in nucleotide incorporation compared to their unmodified counterparts, Taq and Taq(Δ289), respectively. Interestingly, both S-Taq and S-Taq(Δ289) have significantly lower (∼4- to 8-fold) Km (DNA) compared with the corresponding unmodified enzymes (Table 3). Although Km(DNA) is not a direct measurement of the equilibrium binding of these enzymes to the DNA, the difference observed is consistent with the notion that Sso7d fusion has a stabilizing effect on the interactions between the polymerase domain and the DNA template. While the Km (DNA) values cannot be accurately determined for Pfu and Pfu-S due to the very high binding affinity of these enzymes to the primer–template, no decrease in kcat is observed with the fusion protein. The results from the steady-state kinetics analyses demonstrate that, while the fusion of Sso7d increases polymerase processivity significantly, it exhibits no negative affect on the catalytic activity of the polymerase domain.
Table 3. Summary of the steady-state kinetics analyses.
| Enzyme | kcat (s–1) | Km(DNA) (nM) |
|---|---|---|
| Taq(Δ289) |
18 ± 2 |
32 ± 10 |
| S-Taq(Δ289) |
20 ± 1 |
3.9 ± 0.6 |
| Taq |
21 ± 1 |
9.6 ± 0.6 |
| S-Taq |
20 ± 1 |
2.2 ± 0.2 |
| Pfu |
3.2 ± 0.1 |
<1.0 |
| Pfu-S | 5.7 ± 0.2 | <1.0 |
Sso7d fusion does not change polymerase stability
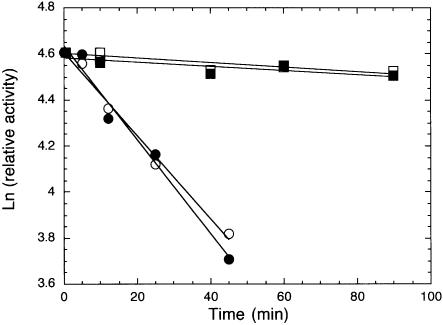
To allow practical application of Sso7d-modified polymerases in PCR, it is essential that the fusion enzymes maintain their original thermal stability. A thermal stability assay based on primer extension activity was used to compare the thermal stability of S-Taq(Δ289) with that of Taq(Δ289) and Pfu-S with that of Pfu (see Materials and Methods). As shown in Figure 4, the effect of incubation at 97.5°C on the remaining primer extension activity is nearly identical for each pair of modified and unmodified polymerases tested. The calculated half-life at 97.5°C for S-Taq(Δ289) and Taq(Δ289) are 35 and 39 min, respectively (Table 4). As expected, Pfu is significantly more stable than the Taq-based enzymes. Nonetheless, the Pfu-S fusion protein showed a nearly identical thermal stability profile to Pfu, with both having a half-life at 97.5°C of the order of 10 h. These results demonstrate that the fusion of Sso7d to polymerases has no negative impact on the thermal stability of the polymerase domain.
Figure 4.
Thermal stability analyses of fusion and non-fusion proteins using primer extension assay. See Materials and Methods for detailed description of reaction conditions and data analyses. The buffers used during the 97.5°C incubation were 10 mM Tris–HCl pH 8.8, 50 mM KCl, 2 mM MgCl2 and 0.1% Triton X-100 for Taq(Δ289) (open circles) and S-Taq(Δ289) (solid circles), 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgCl2, 0.1% Triton X-100 and 100 µg/ml BSA for Pfu (open squares) and Pfu buffer with 60 mM KCl for Pfu-S (solid squares).
Table 4. Summary of thermal stability analyses results.
| Enzyme | k (min–1) | t1/2 (min) |
|---|---|---|
| Taq(Δ289) |
0.018 |
39 |
| S-Taq(Δ289) |
0.020 |
35 |
| Pfu |
0.00095 |
730 |
| Pfu-S | 0.00088 | 789 |
Sso7d fusion proteins are significantly more efficient in PCR amplifications
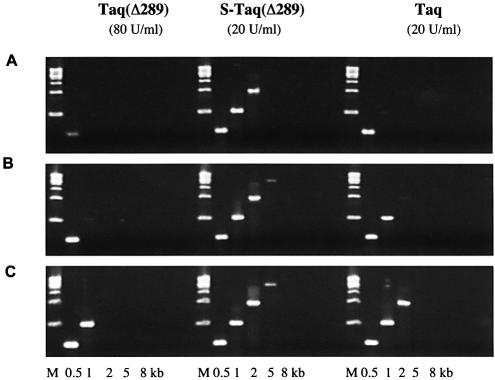
The ultimate goal of improving the processivity of thermostable DNA polymerases is to improve their performance in vitro, such as in PCR or cycle sequencing applications. As an increase in processivity allows the polymerase to incorporate more nucleotides per binding event, it should allow a more efficient in vitro replication of the template strand during each PCR cycle. Consequently, shorter extension time may be required to amplify the same target or a much longer target could be amplified with the same extension time. In this experiment, bacteriophage λ targets (0.5–15 kb) were amplified using a range of extension times to compare the efficiency of the fusion proteins with non-fusion cognates (Fig. 5). With 80 U/ml Taq(Δ289), a 2 min/cycle extension time was required to amplify a 1 kb target. When 20 U/ml Taq polymerase was used, a 1 min/cycle extension time was sufficient for the same target. With the fusion protein S-Taq(Δ289), 20 U/ml enzyme and a 1 min/cycle extension time amplified a 5 kb target, outperforming both its unmodified counterpart, Taq(Δ289), and full-length Taq.
Figure 5.
Comparison of PCR efficiency of Taq, Taq(Δ289) and S-Taq(Δ289). λ DNA (130 pg/µl) was used as the template and the sizes of the amplicons are indicated at the bottom. The PCR buffer contained 10 mM Tris–HCl pH 8.8, 2 mM MgCl2, 200 µM each dNTPs and 0.1% Triton-100 with 10 mM KCl for Taq(Δ289) and 50 mM KCl for S-Taq(Δ289) and Taq. The cycling protocol was: 95°C for 20 s; 20 cycles of 94°C for 5 s and 72°C for 30 s (A) or for 60 s (B) or for 2 min (C); 72°C for 7 min.
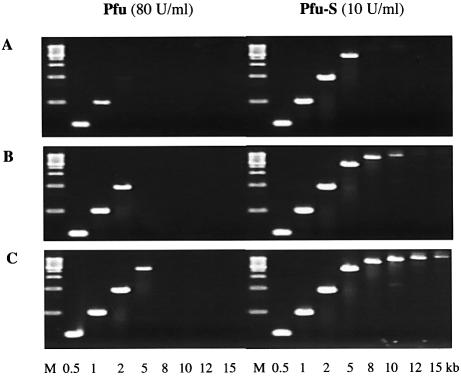
Although Taq polymerase is the most widely used PCR enzyme, its application is limited by a lack of proofreading activity. Pfu polymerase, with a 3′→5′ proofreading exonuclease activity, is a preferred enzyme when high fidelity amplification is required. However, Pfu is rather inefficient in amplifying DNA, which is likely due, at least in part, to its very low processivity. The use of Pfu polymerase in PCR requires high enzyme concentrations and long extension times and often results in low product yields of long amplicons. To assess whether the significant enhancement of the processivity of Pfu by Sso7d fusion could lead to an improvement in its performance in PCR, the same λ targets described above were used to compare the efficiency of Pfu and Pfu-S (Fig. 6). When 80 U/ml Pfu was used, a 2 min/cycle extension time was necessary to amplify a 5 kb target, and no product longer than 5 kb was detected. In contrast, when 10 U/ml Pfu-S was used, the same 5 kb target can be amplified with a 30 s/cycle extension time. When a 2 min/cycle extension time was used, products as long as 15 kb were clearly detected with Pfu-S. These findings establish that the low processivity of a polymerase can be a limiting factor in PCR. By increasing the processivity, Sso7d fusion proteins are significantly more efficient in DNA amplification using PCR, especially for long targets.
Figure 6.
Comparison of PCR efficiency of Pfu and Pfu-S. λ DNA (130 pg/µl) was used as the template and the sizes of the amplicons are indicated at the bottom. M indicates molecular weight marker. PCR buffer contained 20 mM Tris–HCl pH 8.8, 10 mM (NH4)2SO4, 0.1% Triton-100, 2 mM MgCl2 and 200 µM each dNTPs with 10 mM KCl for Pfu and 60 mM KCl for Pfu-S. The cycling protocol was 95°C for 20 s; 20 cycles of 94°C for 5 s and 72°C for 30 s (A) or for 60 s (B) or for 2 min (C); 72°C for 7 min.
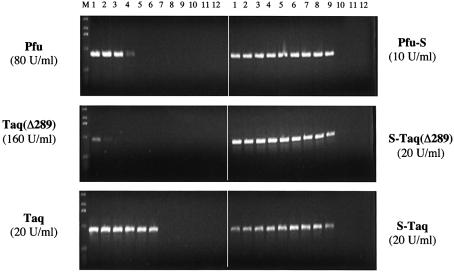
Sso7d fusion proteins have higher and broader salt tolerances in PCR
Impurities in DNA samples, such as salt, could lead to inhibition of PCR amplification. Different thermostable polymerases function optimally under specific salt and buffer conditions. The optimal buffer for the less processive polymerases such as Taq(Δ289) and Pfu contains <10 mM KCl, whereas the more processive Taq polymerase uses 50 mM KCl. There is a general correlation between the use of low salt buffer and a low processivity polymerase, likely due to the influence of ionic strength on the binding interactions between polymerase and DNA template. The enhancement of polymerase processivity by Sso7d fusion may lead to higher tolerance of these enzymes to salt inhibition in PCR. To test this, a 0.9 kb λ target was used to compare the sensitivity of fusion and non-fusion proteins to KCl concentration in PCR (Fig. 7). As expected, both Pfu and Taq(Δ289) preferred low KCl concentrations (<30 mM). In contrast, when Pfu-S and S-Taq(Δ289) were used, efficient amplifications were observed over a broad range of KCl concentrations, and a significant amount of product is observed with as high as 120 mM KCl for both fusion proteins. For Taq polymerase, which alone has a higher tolerance to KCl (10–60 mM) than Pfu and Taq(Δ289), the Sso7d fusion (S-Taq) further broadened the range to 10–120 mM KCl. These results confirm that an increase in processivity by Sso7d fusion leads to an increased tolerance to salt in PCR and allows PCR to succeed in a variety of buffers.
Figure 7.
Comparison of the salt tolerance of Sso7d fusions and the unmodified enzymes in PCR. λ DNA (130 pg/µl) was used as the template and a 0.9 kb amplicon was amplified in PCR buffer with increasing KCl concentrations. Lanes 1–12: 10, 20, 30, 40, 50, 60, 80, 100, 120, 140, 160 and 180 mM KCl. For Pfu and Pfu-S the PCR buffer contained 20 mM Tris–HCl pH 8.8, 10 mM (NH4)2SO4, 2 mM MgCl2, 0.1% Triton-100, 100 µg/ml BSA and 200 µM each dNTPs. For the other enzymes the PCR buffer contained 10 mM Tris–HCl pH 8.8, 2 mM MgCl2, 0.1% Triton-100 and 200 µM each dNTPs. The cycling protocol was 95°C for 20 s; 20 cycles of 94°C for 5 s and 72°C for 60 s; 72°C for 10 min. M indicates molecular weight marker.
DISCUSSION
Use of a sequence non-specific dsDNA binding protein to enhance processivity
The processivity of a DNA polymerase reflects its ability to remain bound to the template during DNA synthesis. The studies reported here demonstrate that covalently linking a sequence non-specific dsDNA binding protein to a non-replicative DNA polymerase can significantly enhance the processivity of the polymerase. The dsDNA binding protein we used here is Sso7d, which belongs to a family of proteins that are highly conserved both in sequence and in size (30,38). This family of proteins exists in abundance in hyperthermophilic archaea and is thought to play the role of stabilizing genomic DNA. To date, there have been no reports of any direct involvement of this family of proteins on DNA replication in vivo.
The Sso7d-based approach to enhancing processivity is novel in its simplicity and generalizability. The naturally existing mechanisms of enhancing processivity often require specific protein factors, such as the ‘DNA sliding clamp’, thioredoxin and UL42, to be recruited by the polymerase (18,39,40). With Sso7d fusion, there is no limitation of specific protein–protein interactions between the DNA binding domain and the polymerase. We have demonstrated that the processivity of both family A and family B types of DNA polymerases can be enhanced effectively by Sso7d. Due to its small size and high thermal stability (41), Sso7d is unlikely to perturb the structural integrity of a fusion partner, which is confirmed by the results of the thermal stability analyses. Most importantly, the presence of Sso7d in cis does not appear to hinder the rate of nucleotide incorporation by the polymerase, as similar kcat values are observed for each pair of fusion and non-fusion proteins.
It is intriguing that the increase in processivity with Sso7d fusion is not achieved at the expense of catalytic activity in nucleotide incorporation. This suggests that the Sso7d portion of the fusion enzyme must have the ability to slide along the template with the DNA polymerase. Neither ‘DNA sliding clamps’ nor thioredoxin, the processivity factor for T7 polymerase, exhibits appreciable affinity for dsDNA (42–45). These processivity factors are thought to enhance processivity by ‘mechanically’ preventing the polymerase from dissociating from its DNA template (13,46). In contrast, UL42, the processivity factor for HSV DNA polymerase, binds directly to dsDNA with a high affinity (Kd = 2 nM) to prevent the polymerase from dissociating (47). In comparison, Sso7d binds weakly to dsDNA with a Kd of 1–2 µM (31), which is 1000-fold weaker than the affinity of Taq polymerase for DNA template (48). Although such a weak interaction is not sufficient to permanently tether the polymerase to its DNA template, it could serve to reduce the probability of complete dissociation of the polymerase from DNA, and in turn increase the processivity of the polymerase. In fact, the weaker affinity of Sso7d for DNA is likely to be beneficial by allowing it to move with the DNA polymerase along the DNA strand without having a negative impact on catalysis. Use of a DNA binding protein with a much higher affinity for dsDNA could be detrimental to the catalytic activity of the polymerase; conversely, use of a DNA binding protein with an even lower affinity for dsDNA may not significantly enhance the processivity of the polymerase. This is consistent with our finding that mutational changes of a key residue (i.e. Trp24) in Sso7d significantly reduces its effectiveness in enhancing processivity. Further studies are needed to identify the optimal range of affinities of the dsDNA binding protein to achieve the ultimate balance between processivity and catalysis.
Enhancement of processivity benefits PCR applications
As PCR has become an indispensable part of modern molecular biology and molecular medicine, the improvement of thermostable DNA polymerase performance has been an ongoing quest for many researchers. The PCR amplification process can be divided into early and late phases. During the early phase, especially when the starting template concentration is low and the target is long, it is crucial to complete replication of the template strands to allow subsequent rounds of exponential amplification. During the late phase, as more DNA template is generated, the molar amount of template exceeds that of the polymerase and the ability of the polymerase to replicate multiple templates in a single cycle is key to achieving a high amplification yield. A polymerase with high processivity is preferred for the early phase. However, a polymerase with excessively high processivity is expected to be less efficient in the late phase, due to its inability to recycle among DNA templates. Therefore, in order to amplify a longer DNA target efficiently, a polymerase with a moderate processivity is preferred.
In our study, when Sso7d is fused to a DNA polymerase, a significant enhancement of processivity is achieved regardless of the starting processivity of the enzyme. When the fusion enzymes are tested in PCR amplifications, a clear advantage over the unmodified enzymes is observed. Not only is much less enzyme required, but a much shorter extension time can be used. The improvement becomes more apparent when long targets are amplified. When both enzyme concentration and reaction time are considered, a combined 16-fold increase in efficiency is observed with S-Taq(Δ289) over Taq(Δ289) and a 32-fold increase with Pfu-S over Pfu. Therefore, a polymerase with a processivity (PI) of 0.98 and an average primer extension length of ∼50 nt is sufficiently processive for replicating long targets (e.g. 15 kb) efficiently in the early cycles of PCR and does not have compromised performance in the late cycles.
Interestingly, even though S-Taq(Δ289) and Pfu-S have comparable processivity, the latter is significantly more efficient in amplifying longer targets. With a 2 min extension time, the longest target amplified by S-Taq(Δ289) was 5 kb, whereas a 15 kb target was amplified by Pfu-S. This is likely due to the difference in the proofreading ability of the two enzymes. Pfu polymerase has a 3′→5′ exonuclease activity that corrects misincorporated nucleotides before continuing DNA synthesis (49). S-Taq(Δ289), on the other hand, is inhibited by a misincorporated nucleotide (30). S-Taq, which has the highest processivity, also shows a limited ability in amplifying long targets (data not shown). Our findings support the notion that both processivity and the ability to proofread are important in achieving efficient amplification of long targets. Most commercial long PCR systems rely on an enzyme mixture containing a high level of non-proofreading polymerase (such as Taq) mixed with a very low level of proofreading enzyme such as Pfu (50,51). Although DNA targets as long as 40 kb have been amplified using this approach, a major downside of using such an enzyme mixture is the fidelity of amplification. While the error frequency of such mixtures is lower than that of non-proofreading polymerase alone, it is still significantly higher than that of the proofreading polymerase alone (52). It has been reported previously that the processivity of a polymerase may be inversely correlated with its accuracy in nucleotide incorporation (53). Fidelity analyses show that Pfu-S maintains the same fidelity as Pfu in PCR (T. Tenkanen et al., unpublished data). Thus, by fusing Sso7d to Pfu, we have generated a unique one enzyme system with both high processivity and high fidelity to allow highly efficient and highly accurate amplification of DNA targets using PCR.
Although the native host of Sso7d, S.solfataricus, has an optimal growth temperature of 86°C, Sso7d is capable of interacting with dsDNA at ambient temperature (31,38). The strategy of enhancing polymerase processivity by Sso7d should be suitable for non-thermostable polymerases as well. In addition, binding to DNA substrates is key to the function of many DNA modifying enzymes. The Sso7d fusion strategy described here has the potential of broadly improving the performance of other types of DNA modifying enzymes, such as DNA ligases, DNA methylases, exonucleases, etc. Furthermore, useful dsDNA binding proteins are not limited to Sso7d, as we have observed that other members of the Sso7d family can serve the same purpose (22). The use of other types of dsDNA binding proteins in place of Sso7d is an interesting possibility that remains to be explored.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Larry Xi and Charles André for helpful discussions and Dr Tuomas Tenkanen and Timo Soininen at Finnzymes Oy for their collaboration in fidelity analyses and polymerase unit definitions. We are grateful to John Finney for his encouragement and support.
REFERENCES
- 1.Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd edn. W.H.Freeman, New York, NY. [Google Scholar]
- 2.Nakai H., Beauchamp,B.B., Bernstein,J., Huber,H.E., Tabor,S. and Richardson,C.C. (1988) Formation and Propagation of the Bacteriophage T7 Replication Fork. American Society of Microbiology, Washington, DC. [Google Scholar]
- 3.Bambara R.A., Uyemura,D. and Choi,T. (1978) On the processive mechanism of Escherichia coli DNA polymerase I. Quantitative assessment of processivity. J. Biol. Chem., 253, 413–423. [PubMed] [Google Scholar]
- 4.Das S.K. and Fujimura,R.K. (1977) Mechanism of T5-induced DNA polymerase. I. Replication of short primer templates. J. Biol. Chem., 252, 8700–8707. [PubMed] [Google Scholar]
- 5.Das S.K. and Fujimura,R.K. (1977) Mechanism of T5-induced DNA polymerase. II. Characterization of the dead-end complex. J. Biol. Chem., 252, 8708–8712. [PubMed] [Google Scholar]
- 6.Das S.K. and Fujimura,R.K. (1979) Processiveness of DNA polymerases. A comparative study using a simple procedure. J. Biol. Chem., 254, 1227–1232. [PubMed] [Google Scholar]
- 7.Salas M. (1991) Protein-priming of DNA replication. Annu. Rev. Biochem., 60, 39–71. [DOI] [PubMed] [Google Scholar]
- 8.Richardson C.C. (1983) Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell, 33, 315–317. [DOI] [PubMed] [Google Scholar]
- 9.Nossal N.G. (1992) Protein–protein interactions at a DNA replication fork: bacteriophage T4 as a model. FASEB J., 6, 871–878. [DOI] [PubMed] [Google Scholar]
- 10.Sexton D.J., Berdis,A.J. and Benkovic,S.J. (1997) Assembly and disassembly of DNA polymerase holoenzyme. Curr. Opin. Chem. Biol., 1, 316–322. [DOI] [PubMed] [Google Scholar]
- 11.Baker T.A. and Bell,S.P. (1998) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- 12.Waga S. and Stillman,B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- 13.Benkovic S.J., Valentine,A.M. and Salinas,F. (2001) Replisome-mediated DNA replication. Annu. Rev. Biochem., 70, 181–208. [DOI] [PubMed] [Google Scholar]
- 14.Von Hippel P.H. (1995) A ring to bind eukaryotic DNA polymerase. Structure, 3, 123–124. [DOI] [PubMed] [Google Scholar]
- 15.Gottlieb J., Marcy,A.I., Coen,D.M. and Challberg,M.D. (1990) The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol., 64, 5976–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisshart K., Chow,C.S. and Coen,D.M. (1999) Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol., 73, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiki R.K., Gelfand,D.H., Stoffel,S., Scharf,S.J., Higuchi,R., Horn,G.T., Mullis,K.B. and Erlich,H.A. (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–491. [DOI] [PubMed] [Google Scholar]
- 18.Bedford E., Tabor,S. and Richardson,C.C. (1997) The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc. Natl Acad. Sci. USA, 94, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson J.E., Fox,R., Harris,D.D., Lyons-Abbott,S. and Loeb,L.A. (2003) Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res., 31, 4702–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motz M., Kober,I., Girardot,C., Loeser,E., Bauer,U., Albers,M., Moeckel,G., Minch,E., Voss,H., Kilger,C. and Koegl,M. (2002) Elucidation of an archaeal replication protein network to generate enhanced PCR enzymes. J. Biol. Chem., 277, 16179–16188. [DOI] [PubMed] [Google Scholar]
- 21.Bullard J.M., Williams,J.C., Acker,W.K., Jacobi,C., Janjic,N. and McHenry,C.S. (2002) DNA polymerase III holoenzyme from Thermus thermophilus identification, expression, purification of components and use to reconstitute a processive replicase. J. Biol. Chem., 277, 13401–13408. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y. (2000) European Union patent no. WO0192501.
- 23.Pavlov A.R., Belova,G.I., Kozyavkin,S.A. and Slesarev,A.I. (2002) Helix–hairpin–helix motifs confer salt resistance and processivity on chimeric DNA polymerases. Proc. Natl Acad. Sci. USA, 99, 13510–13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y.G., Su,S.Y., Robinson,H., Padmanabhan,S., Lim,L., McCrary,B.S., Edmondson,S.P., Shriver,J.W. and Wang,A.H. (1998) The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nature Struct. Biol., 5, 782–786. [DOI] [PubMed] [Google Scholar]
- 25.Desai U.J. and Pfaffle,P.K. (1995) Single-step purification of a thermostable DNA polymerase expressed in Escherichia coli. Biotechniques, 19, 780–782, 784. [PubMed] [Google Scholar]
- 26.Lawyer F.C., Stoffel,S., Saiki,R.K., Chang,S.Y., Landre,P.A., Abramson,R.D. and Gelfand,D.H. (1993) High-level expression, purification and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. PCR Methods Appl., 2, 275–287. [DOI] [PubMed] [Google Scholar]
- 27.Von Hippel P.H., Fairfield,F.R. and Dolejsi,M.K. (1994) On the processivity of polymerases. Ann. NY Acad. Sci., 726, 118–131. [DOI] [PubMed] [Google Scholar]
- 28.Merkens L.S., Bryan,S.K. and Moses,R.E. (1995) Inactivation of the 5′-3′ exonuclease of Thermus aquaticus DNA polymerase. Biochim. Biophys. Acta, 1264, 243–248. [DOI] [PubMed] [Google Scholar]
- 29.Murali R., Sharkey,D.J., Daiss,J.L. and Murthy,H.M. (1998) Crystal structure of Taq DNA polymerase in complex with an inhibitory Fab: the Fab is directed against an intermediate in the helix-coil dynamics of the enzyme. Proc. Natl Acad. Sci. USA, 95, 12562–12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choli T., Henning,P., Wittmann-Liebold,B. and Reinhardt,R. (1988) Isolation, characterization and microsequence analysis of a small basic methylated DNA-binding protein from the archaebacterium, Sulfolobus solfataricus. Biochim. Biophys. Acta, 950, 193–203. [DOI] [PubMed] [Google Scholar]
- 31.Baumann H., Knapp,S., Lundback,T., Ladenstein,R. and Hard,T. (1994) Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nature Struct. Biol., 1, 808–819. [DOI] [PubMed] [Google Scholar]
- 32.Agback P., Baumann,H., Knapp,S., Ladenstein,R. and Hard,T. (1998) Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nature Struct. Biol., 5, 579–584. [DOI] [PubMed] [Google Scholar]
- 33.Catanzano F., Graziano,G., Fusi,P., Tortora,P. and Barone,G. (1998) Differential scanning calorimetry study of the thermodynamic stability of some mutants of Sso7d from Sulfolobus solfataricus. Biochemistry, 37, 10493–10498. [DOI] [PubMed] [Google Scholar]
- 34.Braithwaite D.K. and Ito,J. (1993) Compilation, alignment and phylogenetic relationships of DNA polymerases. Nucleic Acids Res., 21, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito J. and Braithwaite,D.K. (1991) Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res., 19, 4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cann I.K. and Ishino,Y. (1999) Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics, 152, 1249–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopfner K.P., Eichinger,A., Engh,R.A., Laue,F., Ankenbauer,W., Huber,R. and Angerer,B. (1999) Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl Acad. Sci. USA, 96, 3600–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAfee J.G., Edmondson,S.P., Datta,P.K., Shriver,J.W. and Gupta,R. (1995) Gene cloning, expression and characterization of the Sac7 proteins from the hyperthermophile Sulfolobus acidocaldarius. Biochemistry, 34, 10063–10077. [DOI] [PubMed] [Google Scholar]
- 39.Zuccola H.J., Filman,D.J., Coen,D.M. and Hogle,J.M. (2000) The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell, 5, 267–278. [DOI] [PubMed] [Google Scholar]
- 40.Shamoo Y. and Steitz,T.A. (1999) Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell, 99, 155–166. [DOI] [PubMed] [Google Scholar]
- 41.Knapp S., Karshikoff,A., Berndt,K.D., Christova,P., Atanasov,B. and Ladenstein,R. (1996) Thermal unfolding of the DNA-binding protein Sso7d from the hyperthermophile Sulfolobus solfataricus. J. Mol. Biol., 264, 1132–1144. [DOI] [PubMed] [Google Scholar]
- 42.Stukenberg P.T., Studwell-Vaughan,P.S. and O’Donnell,M. (1991) Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J. Biol. Chem., 266, 11328–11334. [PubMed] [Google Scholar]
- 43.Tinker R.L., Kassavetis,G.A. and Geiduschek,E.P. (1994) Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J., 13, 5330–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber H.E., Tabor,S. and Richardson,C.C. (1987) Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem., 262, 16224–16232. [PubMed] [Google Scholar]
- 45.Doublie S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 46.Kelman Z., Hurwitz,J. and O’Donnell,M. (1998) Processivity of DNA polymerases: two mechanisms, one goal. Structure, 6, 121–125. [DOI] [PubMed] [Google Scholar]
- 47.Chow C.S. and Coen,D.M. (1995) Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J. Virol., 69, 6965–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong H., Kucera,R.B. and Jack,W.E. (1993) Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement and exonuclease activities. J. Biol. Chem., 268, 1965–1975. [PubMed] [Google Scholar]
- 49.Perler F.B., Kumar,S. and Kong,H. (1996) Thermostable DNA polymerases. Adv. Protein Chem., 48, 377–435. [DOI] [PubMed] [Google Scholar]
- 50.Cheng S., Fockler,C., Barnes,W.M. and Higuchi,R. (1994) Effective amplification of long targets from cloned inserts and human genomic DNA. Proc. Natl Acad. Sci. USA, 91, 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnes W.M. (1994) PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl Acad. Sci. USA, 91, 2216–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cline J., Braman,J.C. and Hogrefe,H.H. (1996) PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res., 24, 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnes W.M. (1992) The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene, 112, 29–35. [DOI] [PubMed] [Google Scholar]