Tubulin glutamylating enzymes are important for beating behavior of ependymal cilia in the brain, whereas glycylating enzymes are critical for stability and maintenance of these cilia.
Abstract
Microtubules are subject to a variety of posttranslational modifications that potentially regulate cytoskeletal functions. Two modifications, glutamylation and glycylation, are highly enriched in the axonemes of most eukaryotes, and might therefore play particularly important roles in cilia and flagella. Here we systematically analyze the dynamics of glutamylation and glycylation in developing mouse ependymal cilia and the expression of the corresponding enzymes in the brain. By systematically screening enzymes of the TTLL family for specific functions in ependymal cilia, we demonstrate that the glycylating enzymes TTLL3 and TTLL8 were required for stability and maintenance of ependymal cilia, whereas the polyglutamylase TTLL6 was necessary for coordinated beating behavior. Our work provides evidence for a functional separation of glutamylating and glycylating enzymes in mammalian ependymal cilia. It further advances the elucidation of the functions of tubulin posttranslational modifications in motile cilia of the mammalian brain and their potential importance in brain development and disease.
Introduction
Microtubules (MTs) are key components of the cytoskeleton involved in many essential cellular functions. The functional diversification of MTs is controlled by interactions with a large variety of MT-associated proteins. Less is known about the regulation of MT functions by posttranslational modifications (PTMs) of tubulin. Glutamylation and glycylation generate side chains consisting of one or several glutamates or glycines on tubulin. The possibility of generating different lengths of side chains on either α- or β-tubulin, and their positioning at the outer surfaces of the MTs, make them ideal regulators of MT-associated protein interactions (Janke and Bulinski, 2011).
Cilia are MT-based eukaryotic organelles projecting from the surface of cells that fulfill a number of important cellular functions. In mammalian brain, primary cilia are present on most cells and function in several physiological and developmental processes (Han and Alvarez-Buylla, 2010). Motile cilia are present only in epithelial cells lining the ventricles, the ependyma. Each ependymal cell extends ∼50 motile cilia whose coordinated beating is responsible for the cerebrospinal fluid (CSF) flow in brain ventricles. This is crucial for brain functions and homeostasis. Mutations disrupting motile cilia structure and functions disturb their beating and the CSF flow and lead to neurodevelopmental disorders (Ibañez-Tallon et al., 2004; Sawamoto et al., 2006; Lechtreck et al., 2008; Tissir et al., 2010; Ihrie and Alvarez-Buylla, 2011). Multiciliated ependymal cells are generated from monociliated progenitors during postnatal development (Spassky et al., 2005); however, the mechanisms controlling motile cilia formation and stability are mostly unknown.
Given the roles of tubulin PTMs in cilia structure and stability in several model organisms (Janke and Bulinski, 2011), we have studied the dynamics of tubulin glutamylation and glycylation in the development of mouse ependymal cilia. We have investigated the expression, distribution, and functional roles of the eight glutamylases and three glycylases, which are members of the tubulin tyrosine ligase-like (TTLL) family (Fig. S1 A). Our data reveal a highly specific expression of TTLL enzymes in the brain, and we identify a subset of enzymes specific to ependymal cells. We further demonstrate that the glycylating enzymes TTLL3 and TTLL8 are important for ciliary maintenance, whereas the polyglutamylase TTLL6 plays a key role in the regulation of ciliary beating.
Results and discussion
Dynamics of posttranslational glutamylation and glycylation during ependymal cell development
Multiciliated ependymal cells develop in newborn mice from radial glia progenitors and line the entire walls of the brain ventricles. During differentiation, ependymal progenitors expand their apical surfaces, lose their primary cilia, and generate multiple basal bodies from which they assemble motile cilia (Spassky et al., 2005; Mirzadeh et al., 2010b). Here, we investigated the dynamics of tubulin glutamylation and glycylation by studying their appearance and distribution in primary and motile cilia of developing ependymal cells.
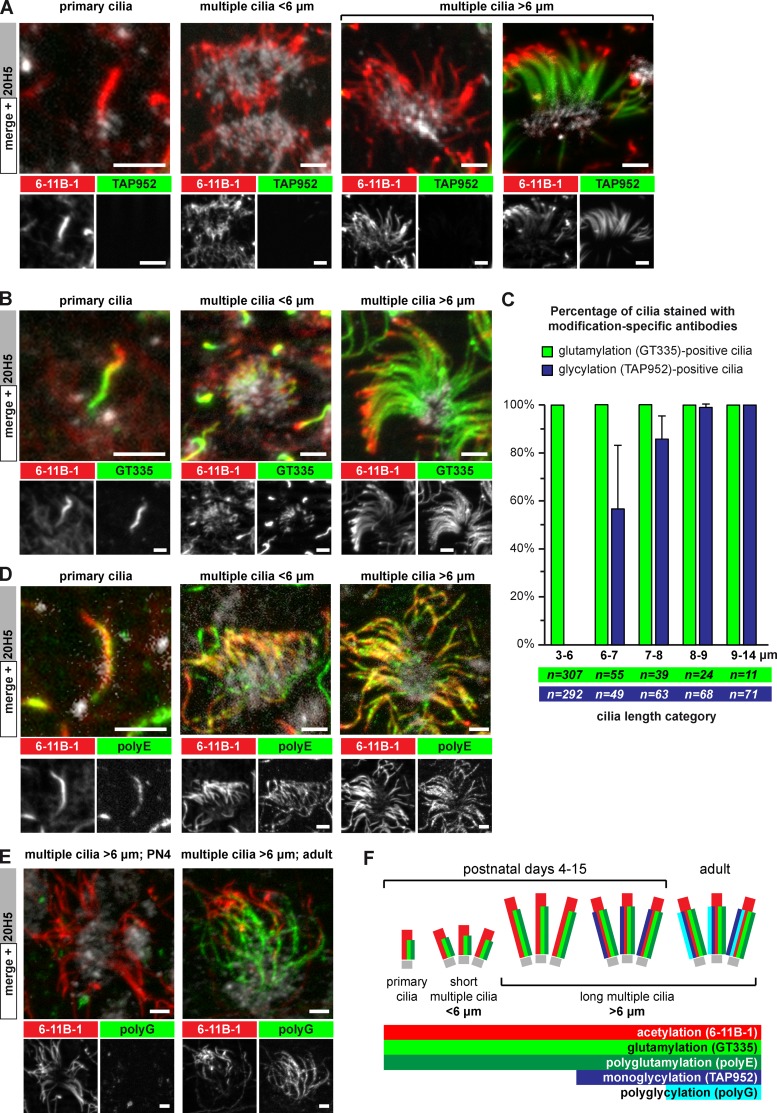
Lateral ventricular walls were isolated at postnatal day (PN) 4, as all stages of ependymal cell differentiation are present at that stage (Spassky et al., 2005). The ventricular walls were triple-immunostained with antibodies specific to either monoglycylation (TAP952; Bré et al., 1996), polyglycylation (polyG; Rogowski et al., 2009), glutamylation (GT335; Wolff et al., 1992), or polyglutamylation (polyE, side chains longer than three glutamates; Rogowski et al., 2010), together with a basal body marker (centrin-20H5; Sanders and Salisbury, 1994) and a pan-cilia marker (6-11B-1, specific to α-tubulin acetylation; Piperno and Fuller, 1985). Primary and motile cilia of all stages of development appeared strongly polyglutamylated (Fig. 1, B and D). In contrast, monoglycylation is barely detected on primary cilia of radial glia cells, as well as on a remarkable share of motile cilia (Fig. 1 A). Polyglycylation is solely detected in motile cilia of adult mice (older than 2 months; Fig. 1 E).
Figure 1.
Glycylation and glutamylation are dynamically regulated in developing mouse ependymal cilia. (A) Maximum projection of image stacks taken from whole-mount ventricles at postnatal day (PN) 4. Basal bodies are labeled with 20H5, acetylated tubulin and thus cilia with 6-11B-1, and monoglycylation with TAP952. (B) Ependymal cells as in A; green channel shows glutamylation (GT335). (C) Quantitative analysis of the presence of monoglycylation (A; TAP952) and glutamylation (B; GT335) as a function of total ciliary length (6-11B-1). Average values with standard deviation are represented for four animals. Total sample numbers are indicated. (D) Ependymal cells as in A; green channel shows polyglutamylation (polyE). (E) Ependymal cells with long (>6 µm) motile cilia at PN4 and in adult mice. Polyglycylation (polyG) was restricted to cilia in adult mice. (F) Schematic representation of the distributions of PTMs in developing ependymal cilia. Bars, 2.5 µm.
Quantification of the monoglycylation signal and ciliary length on multiple motile cilia showed that cilia shorter than 6 µm (indicative of growing cilia) were never positive, while this signal appeared with further elongation of cilia. Above 8 µm of length, most motile cilia were monoglycylated (Fig. 1 C).
These observations demonstrate that polyglutamylation is generated concurrently to ciliary assembly of both primary and motile cilia, and might therefore be important for ciliogenesis. In contrast, glycylation becomes prominent only in mature motile cilia, suggesting that this modification might not be required for ciliogenesis, but could be important for the stability (maintenance) of the axoneme (Fig. 1 F).
Expression analysis of TTLL genes
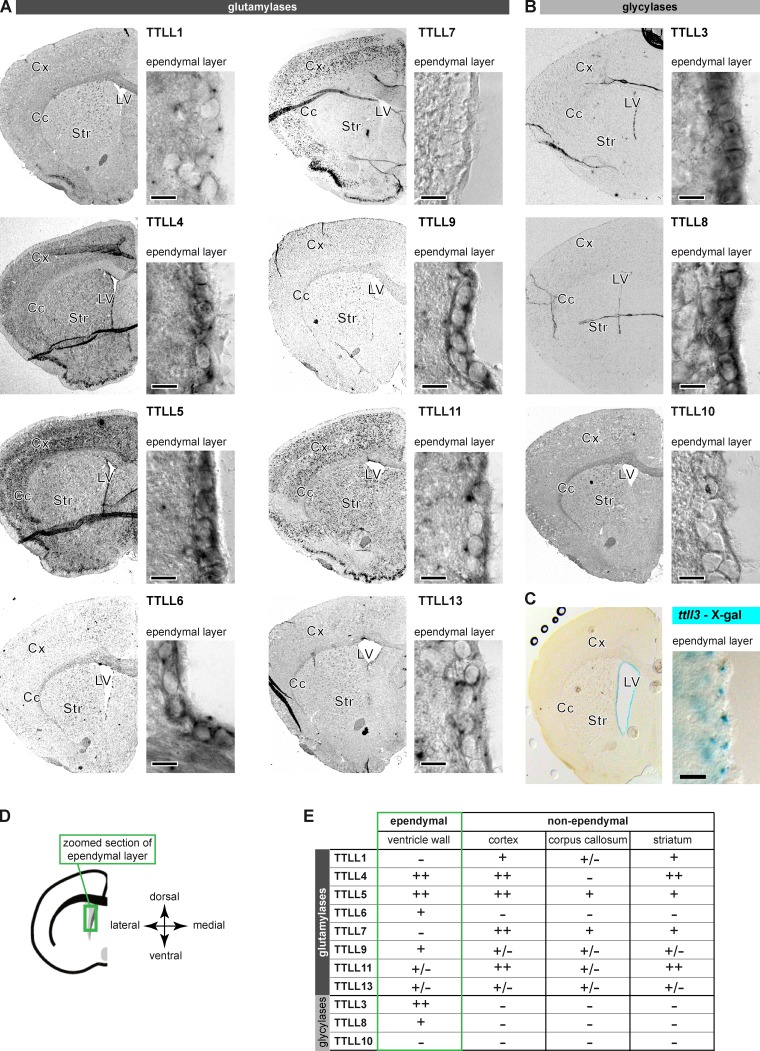
To identify the individual glutamylating and glycylating TTLL enzymes that carry out specific ciliary functions in ependymal cells, we characterized the expression of the TTLL genes in adult mouse brains with a focus on ventricle walls by RNA in situ hybridization and β-galactosidase staining (Fig. 2; Fig. S1). Within the family of glutamylases, only the polyglutamylase TTLL6 was specific for the ependymal layer. The glutamylases TTLL4, TTLL5, and TTLL9 were found in the ependymal layer but also in other regions of the brain. No ependymal-specific signal was detected for TTLL1, TTLL7, TTLL11, and TTLL13, whereas most of these enzymes are clearly detected in other brain regions (Fig. 2, A and E). The monoglycylases TTLL3 and TTLL8 were specifically detected in the ependymal layer (Fig. 2, B and E), which was confirmed for TTLL3 with a specific β-galactosidase staining in ttll3−/− mice (EUCOMM consortium; Fig. 2 C) and by the presence of the modification by co-labeling with the TAP952 antibody (Fig. S1, D and E). To relate the delayed appearance of glycylation (Fig. 1, A and C) with the temporal expression pattern of the glycylases TTLL3 and TTLL8, we performed using RT-PCR, whole-transcriptome sequencing (RNA-Seq) and in situ hybridization at different stages of ependymal development in cell culture and in the brain (Fig. S2). Both approaches showed that TTLL3 and TTLL8 were up-regulated concurrently with ependymal development, which could explain the late appearance of the modification in ciliogenesis. Expression of TTLL10 was not detectable despite a strong expression in testes (Fig. S1 C), in which the highly polyglycylated sperm tails are assembled. As TTLL10 has been shown to be the unique enzyme able to carry out polyglycylation in mammals (Rogowski et al., 2009), our data suggest that its very low expression in the ventricular wall is the key reason for the slow polyglycylation kinetics of the ependymal axonemes.
Figure 2.
A subset of TTLL enzymes is expressed in adult mouse ependymal cells. In situ hybridization revealed the expression patterns of TTLL genes encoding glutamylases (A) or glycylases (B) in coronal brain sections (Cc, corpus callosum; Cx, cortex; LV, lateral ventricle; Str, striatum). Controls in Fig. S1, B and C. (C) TTLL3 expression visualized by X-Gal staining in the ttll3−/− mouse. (D) Schematic representation of the right hemisphere with the green box indicating the localization of zoom images showing the ependymal layer in A–C. (E) Summary of expression analysis of the TTLL genes. Only weak (+) or strong (++) expression levels were considered specific. Bars (A–C): 10 µm.
Identification of TTLL enzymes required for multiciliation of ependymal cells
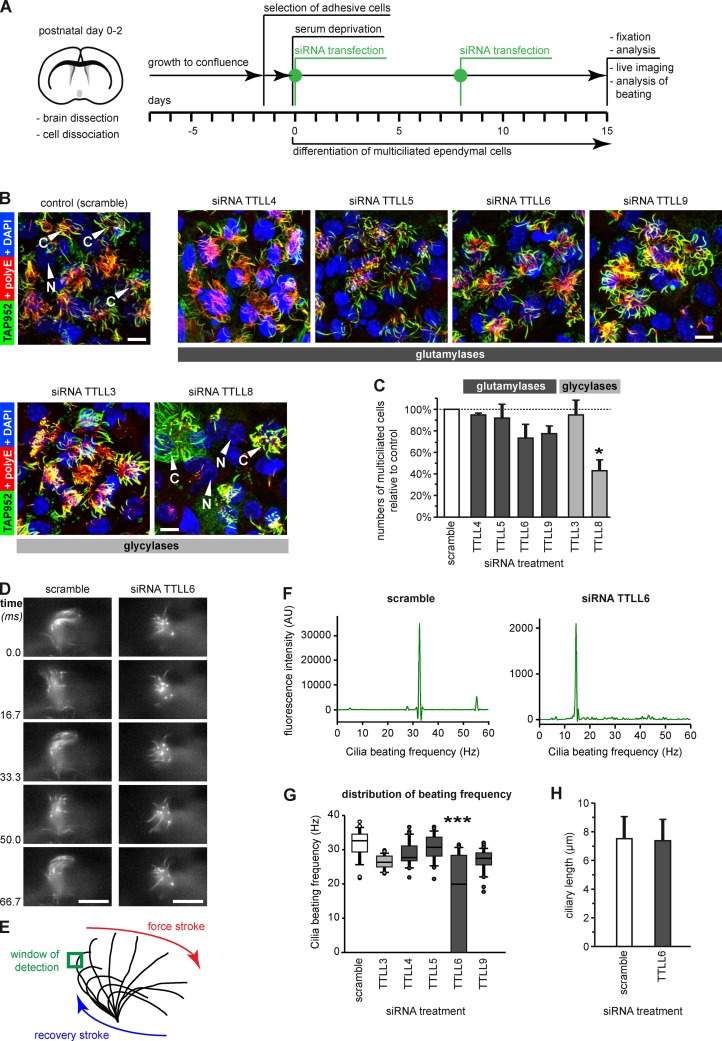
To identify the functional roles of the TTLL enzymes in ependymal epithelium, we systematically depleted the enzymes that were expressed in the ependymal layer (Fig. 2 E) with siRNA. One of three siRNAs for each relevant TTLL gene was selected (Fig. S3 A), and developing ependymal cells were transfected in culture (Guirao et al., 2010; experimental scheme in Fig. 3 A). Cilia were co-labeled with monoglycylation-specific (TAP952) and polyglutamylation-specific (polyE) antibodies to assure that they are stained, even if one of the two corresponding PTMs would be depleted (Fig. 3 B). To determine TTLL enzymes with a role in ciliogenesis or ciliary maintenance, we counted the number of cells carrying a tuft of motile cilia. To avoid that the heterogeneity of the ependymal cell culture leads to misinterpretations, we counted cells only in regions with high cell density.
Figure 3.
Depletion of ependymal-specific glutamylases and glycylases induces different ciliary phenotypes in ependymal cells. (A) Flow scheme of the experimental paradigm of all siRNA experiments. (B) Analysis of multiciliated ependymal cells after siRNA. Cilia were co-labeled in fixed cells for polyglutamylation (polyE) and monoglycylation (TAP952). Bars, 10 µm. (C) Quantification of the relative numbers of multiciliated ependymal cells in areas with high cell density after siRNA treatment. The total number of cells (nuclei, DAPI) was related to the number of multiciliated cells polyE/TAP952 (B). Three independent experiments with more than 1,000 cells were analyzed, and controls (scramble siRNA) were set as 100%. Error bars represent SEM. After one-way ANOVA with Tukey’s post-hoc analysis, differences with P < 0.05 (*) were considered significant. (D) Image sequence of beating cilia after treatment with siRNA (A; 15 d), and labeling with Tubulin Tracker green. Ciliary beating was recorded at 120 frames per second (frame series of 75 ms; Videos 1 and 2). Bar, 10 µm. (E) Schematic representation of ciliary beating with the region of interest (green box) used for measurements. (F) Beating frequency distribution obtained by Fourier transformation of the beating frequency recording (E) of the cilia shown in D. (G) Box plot of the distribution of ciliary beating frequencies after siRNA (A). For each siRNA, three independent experiments, each with more than 25 cells, were recorded. Error bars show SEM; P < 10−6 (***) in Fisher variance test was considered significant. (H) The length of motile cilia after siRNA treatment measured on fixed cells (B) showed no difference between scramble and TTLL6 siRNA (Welch’s t test).
None of the ependymal-specific glutamylases appeared essential on its own for cilia formation and/or maintenance. In contrast, depletion of the glycylase TTLL8 led to a significant reduction in the number of multi-ciliated cells (Fig. 3 C), showing that this enzyme is important for proper ciliogenesis or for ciliary maintenance.
Identification of TTLL enzymes important for ciliary beating in ependymal cells
To identify TTLL enzymes that are critical for ciliary beating, we focused on ependymal-specific TTLLs (Fig. 2 E) that showed no impact on ciliogenesis or ciliary maintenance (Fig. 3 C). After depletion of the relevant TTLLs (Fig. 3 A), ciliary beating was recorded with a high-speed confocal microscope, and the beating frequency of a single cilium was determined within a ROI (Fig. 3, D and E). Using Fourier transformation, we obtained the beat-frequency distributions (Fig. 3 F), which were strikingly narrow for each of the observed cilia (representative cells for scramble- and TTLL6-siRNA treated cells: Fig. 3, D and F; Videos 1 and 2).
Analysis of average beat frequencies of at least 25 cells per siRNA showed that only the depletion of TTLL6 significantly reduced ciliary beating frequency in multiciliated ependymal cells (Fig. 3 G), whereas it did not affect ciliary length (Fig. 3 H). Thus, the polyglutamylase TTLL6 appears to regulate axonemal motility, and cannot be replaced by any of the other glutamylases expressed in ependymal cells. This unique function could be related either to the generation of a highly specific subtype of polyglutamylation (e.g., occupation of a specific modification site on tubulin, generation of a particular side chain length), or to a specific localization of the enzyme within the axoneme.
Two glycylating enzymes are crucial for maintenance of motile ependymal cilia in vivo
The observation that the depletion of one out of two glycylases (TTLL8; Fig. 3 C) leads to decreased number of ciliated cells in cultured ependymal cells points toward an important role of glycylation in ciliary assembly and/or stability. However, depletion of the second glycylase, TTLL3, which shows a similar expression profile (Fig. 2; Fig. S1, D and E; Fig. S2), did not affect the ciliation of ependymal cells (Fig. 3 C). Moreover, a transgenic mouse model in which the expression of TTLL3 had been knocked down (ttll3−/−; Fig. S3 B) by the insertion of a cassette containing the β-galactosidase gene between exons 5 and 6 showed no ciliary defects in the ependymal layer (Fig. S3 C).
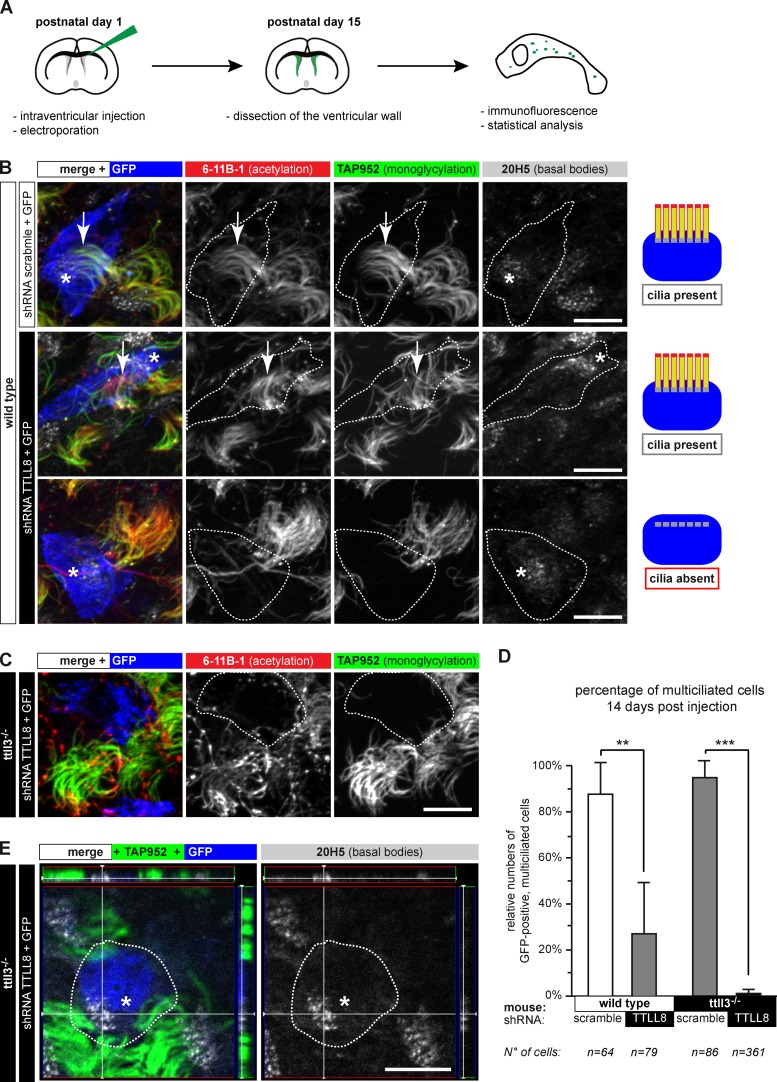
To investigate the respective roles of TTLL3 and TTLL8 in vivo, we electroporated DNA vectors expressing CFP as well as shRNAs (Fig. S3 D) into the ventricles of PN1 mice (Figs. 4 A and 5 A; Boutin et al., 2008). Ciliogenesis was analyzed on whole-mount preparations of PN15 ventricular walls by immunodetection with anti-GFP (shRNA-expressing cells), TAP952 (monoglycylation), 6-11B-1 (acetylated tubulin), and 20H5 (basal bodies; Fig. 4). For quantitative analysis, we focused on CFP-expressing cells with a typical ependymal morphology, namely a large apical process in contact with the lateral ventricle and the presence of multiple motile cilia and multiple basal bodies in the neighboring cells of the same plane (Mirzadeh et al., 2010b).
Figure 4.
Long-term ablation of glycylases leads to nonciliated ependymal cells. (A) Flow scheme of the experimental paradigm used for shRNA-mediated depletion of TTLL8 in vivo. (B) Ependymal layer of wild-type mice. GFP-positive cells (blue or contours) express shRNA. Motile cilia were labeled for acetylation (6-11B-1), monoglycylation (TAP952), and basal bodies (20H5). Expression of TTLL8-shRNA partially led to loss of motile cilia; cells still contain multiple basal bodies. Quantification in D. (C) Expression of TTLL8-shRNA in the ependymal epithelium of ttll3−/− mice. Transfected cells (blue or contours) have no cilia. (D) Quantification of multiple motile cilia on shRNA-expressing cells 14 d after electroporation (B, C, and E). Three independent experiments per condition were performed (total number of counted cells are given below). Mean values with SEM and statistics (Welch t test) are represented (**, P < 0.01; ***, P < 0.001). (E) 3D images of TTLL8-depleted cells in the ependymal layer show fully developed and correctly arranged multiple basal bodies in cells without motile cilia. Panels on the top and the right represent the Z-stack of the image. (B, C, and E) Arrows, cilia; asterisks, basal bodies in GFP-positive cells. Bars, 10 µm.
Figure 5.
Normal ciliogenesis precedes loss of cilia in ependymal cells depleted of glycylases. (A) Scheme of the experiment. (B) Lateral ventricle walls with ependymal cells 3 d after electroporation (A), expressing either scramble or TTLL8-shRNA (blue or contours). Cilia were visualized with acetylated tubulin (6-11B-1) and basal bodies (20H5). Arrows, cilia; asterisks, basal bodies of GFP-positive (blue) cells. Bars, 10 µm. (C) Quantification of GFP-positive cells with motile cilia 3 d after electroporation (B). Analysis was done as in Fig. 4 D. No significant differences between control and TTLL8 shRNA were detected (Welch’s t test).
Depletion of TTLL8 in wild-type mice led to the absence of motile cilia in more than half of the GFP-positive cells (Fig. 4, B and D), corroborating siRNA experiments in Fig. 3, B and C. The fact that the remaining TTLL8-depleted cells still extend monoglycylated cilia suggested a partial functional redundancy with the other glycylating enzyme, TTLL3. This was confirmed by depleting TTLL8 in ttll3−/− mice, which led to an almost complete absence of motile cilia in the GFP-positive cells (Fig. 4, C and D). It demonstrates that cilia formation and/or maintenance requires the redundant action of both TTLL3 and TTLL8 in mammalian ependymal cells. Because TTLL3 and TTLL8 are the unique monoglycylating enzymes in mammals, and the polyglycylases TTLL10 cannot modify tubulin in the absence of these two enzymes (Rogowski et al., 2009), our results suggested that the glycylation itself is essential for formation and/or maintenance of cilia.
We observed that cells without cilia contained multiple basal bodies, which were normally localized near the plasma membrane of the ttll3−/−/TTLL8-shRNA cells (Fig. 4 E). This suggested that the depletion of glycylases could lead to loss of cilia after normal development of multiple basal bodies. To verify this possibility, we repeated the same type of experiments, but quantified multiciliated cells 3 d after TTLL8-shRNA transfection (Fig. 5). Remarkably, at this early stage, most of the GFP-positive cells are multiciliated in both scramble- and TTLL8-shRNA (Fig. 5, B and C). This suggested that cilia are normally assembled in ttll3−/−/TTLL8-depleted cells, but are later disassembled. Together with the earlier observation that ependymal cilia cannot regrow after disassembly (Kuo et al., 2006; Carlén et al., 2009), our data implicate that absence of TTLL3 and TTLL8, and thus most likely the reduction or absence of glycylation on axonemes, destabilizes motile ependymal cilia and results in ciliary disassembly.
Conclusions
Our study highlights a differential timing of two tubulin PTMs, glutamylation and glycylation, in developing mammalian ependymal cilia. Although polyglutamylation is detected from the onset of ciliogenesis, glycylation lags behind and strongly increases with ciliary maturation. We have further identified the enzymes that generate these two PTMs in ependymal cells, and systematically studied their functions. Our data show that glycylation, redundantly generated by the enzymes TTLL3 and TTLL8 (Fig. S1 A), is required for motile cilia stability. Similar conclusions on the role of glycylation for axonemal stability have been obtained in other model organisms, suggesting a conserved function as axoneme stabilizer for glycylation in evolution. In contrast, the number and subtypes of enzymes involved in this modification have been demonstrated to be highly different between species (Rogowski et al., 2009; Wloga et al., 2009).
Glutamylases exist in larger numbers compared with glycylases in most organisms. This might reflect their higher functional diversity (Janke and Bulinski, 2011), but also increases the probability of functional redundancies of those enzymes. One way to limit redundancy in vivo is spatial and temporal control. We show here that in the ependymal cells, only some members of the glutamylase family are expressed. Our observation that depletion of neither of these enzymes resulted in decreased ciliary polyglutamylation suggests that they are redundantly involved in glutamylation of axonemes; however, their functions were not entirely redundant. We further demonstrated that TTLL6, an α-tubulin–specific glutamylase generating long side chains (Fig. S1 A; van Dijk et al., 2007), is an important regulator of ciliary beating and cannot be replaced by any of the other glutamylases expressed in ependymal cells.
Comparison with studies in other organisms suggests that the enzymes involved in the regulation of ciliary beating are rather flexible in evolution. In mouse ependymal cilia, ciliary beating is regulated by the α-tubulin–specific polyglutamylase TTLL6, whereas in Tetrahymena a similar function has been found for TTLL6A (specific for β-tubulin; van Dijk et al., 2007) and TTLL6F (Suryavanshi et al., 2010). In Chlamydomonas reinhardtii, flagellar beating is regulated by a homologue of TTLL9 (Kubo et al., 2010), which has no function in ciliary beating in mouse ependymal cells. Strikingly, TTLL1, a glutamylase important for motile airway cilia in mice (Ikegami et al., 2010; Vogel et al., 2010), is not detected in the ependymal layer. Similarly, TTLL5, which has no unique function for ciliary stability or beating in ependymal cilia, has been found to be essential for the correct function of sperm flagella (Lee et al., 2013), and plays an important role in the polyglutamylation of primary cilia in hTERT-RPE1 cells (Backer et al., 2012).
Understanding the roles of MT PTMs and the biochemical and functional specification of the modifying enzymes in ependymal cilia thus provides a strong basis for deciphering the factors modulating ciliary beating in the brain. The beating of ependymal cilia is crucial for brain functions and homeostasis and leads to human pathologies frequently associated with morbidity and mortality when defective (Zhang et al., 2006). The key roles of the glycylating and glutamylating enzymes described here suggest important repercussions of MT PTMs for human neuropathologies.
Materials and methods
Plasmids, siRNA, and shRNA
The cloning of plasmids for the expression of the TTLL-EYFP fusion genes in mammalian cells has been described previously (van Dijk et al., 2007). In brief, TTLL genes were amplified from mouse brain or testes cDNA and inserted into a cloning vector containing a C-terminal EYFP tag. The same genes were also inserted into the pCS2+ vector for generation of probes using the T7 (antisense) and the SP6 (sense) promotors. Small hairpin RNA (shRNA) was expressed from a plasmid under the control of an H1 promoter, together with CFP, which was expressed using a cytomegalovirus (CMV) promoter from the same plasmid. Plasmids were purified by using an EndoFree Plasmid kit (Maxiprep kit; QIAGEN) and resuspended in TE buffer.
For each TTLL gene, three different short interfering RNA (siRNA) dimers (Eurogentech; sequences listed in Table S1) were tested for silencing of their target TTLL genes. Cells were transfected with siRNA and grown for 24 h before plasmids encoding for TTLLs-EYFP were transfected. A second siRNA transfection was performed 2 h after the transfection of the TTLL-EYFP expression plasmids, and cells were harvested 22 h later. The efficiency of siRNA was determined by evaluating its effect on the expression of the TTLL-EYFP fusion proteins using anti-GFP immunoblotting (Fig. S3 A).
Animals
Experiments were performed on OF1 mice (Janvier-Europe) or TTLL3 mutant mice (ttll3−/−) bearing loxP sites flanking exon 6 of the TTLL3 gene (obtained from European Mouse Mutant Archive [EMMA]; mouse strain B6N;B6N-Ttll3<tm1a(EUCOMM)Wtsi>/Wtsi). Animals were maintained with access to food and water ad libitum in a colony room kept at constant temperature (19–22°C) and humidity (40–50%) on a 12-h light/12-h dark cycle. Genotyping was performed by routine PCR technique according to the EMMA protocols (EM03775).
All experimental procedures were performed in strict accordance with the guidelines of the European Community (86/609/EEC) and the French National Committee (87/848) for care and use of laboratory animals.
In vivo electroporation in neonatal mice
Postnatal electroporation was performed as described previously (Boutin et al., 2008). In brief, the scramble or TTLL8-shRNA plasmids were diluted in PBS containing 0.1% fast green as a tracer to a concentration of 0.5–2.0 mg/ml. 2 µl of plasmid solution was injected into the lateral ventricle using a pulled glass pipette (beveled to a <50-µm diameter) into the ventricles of 2-d-old mouse pups anesthetized on ice. Positive pressure for plasmid injection was applied by expiration. After plasmid injection, 5-mm tweezer-type electrodes (model CUY650P5; NepaGene) coated with conductive gel (Control Graphique Medical) were placed on the heads of the pups, and 5 square-pulses of 50-ms duration with 850-ms intervals at 99 V were applied using a pulse CUY21SC generator (NepaGene). Animals were reanimated by warming them on a heating pad under light, and were then placed back in the cage with the mother.
Whole-mount dissection of the lateral ventricular wall and immunohistochemistry
Electroporated mice were sacrificed either at PN4 or at PN15. Brains were removed from the skulls after cervical dislocation and cut in the midline. The overlying cerebral cortex, medial ventricular wall, and hippocampus were dissected to reveal the lateral ventricular wall (Mirzadeh et al., 2010a). Whole mounts were fixed in 4% paraformaldehyde (PFA) at 4°C overnight, followed by a wash in phosphate-buffered saline (PBS), and then incubated for 1 h in 10% FCS with 0.1% Triton X-100 in PBS at room temperature. Next, whole mounts were incubated for 18 h in primary antibodies in PBS containing 5% FCS and 2% Triton X-100 at 4°C. After rinsing in PBS, the whole mounts were incubated for 2 h with secondary antibodies (conjugated with Alexa Fluor 350, 488, 594, and/or 647; Invitrogen) at room temperature, followed by a PBS wash, DAPI (0.02 µg/ml; Sigma-Aldrich), and a final PBS wash. Whole mounts were trimmed to 200–300-µm sections and mounted on Superfrost slides and Fluoromount-G mounting medium (SouthernBiotech). For 20H5 immunostaining, samples were pretreated with 0.2% Triton X-100 in PBS for 2 min before fixation with 4% PFA. Antibodies used are listed in Table S2. Specificity controls omitting primary antibodies were routinely performed.
Optical stack images were taken using either a fluorescence microscope (AxioImager Z1/Apotom; Carl Zeiss) or a laser confocal scanning microscope (LSM510; Carl Zeiss). Analysis of ependymal cells was done in whole-mount reconstructions of the ependymal epithelium in 50 serial en face ultrathin optical sections (0.24 µm per section), cut in the plane parallel to the ventricular surface. Transfected ependymal cells were identified by GFP staining (chicken GFP antibody). We quantified the number of CFP-expressing cells with a typical ependymal morphology, namely a large apical process in contact with the lateral ventricle and the presence of multiple motile cilia and multiple basal bodies in the neighboring cells of the same plane (Mirzadeh et al., 2010b).
Quantification of ciliary length and modification state
Whole-mount preparations of lateral ependymal walls were fixed at PN0, PN4, PN10, and after 3 months (adult). Immunostaining was performed using TAP952 to label monoglycylation, GT335 for glutamylation, and acetylated α-tubulin antibody (6-11-B1) to identify cilia and measure total cilium length. Images were analyzed using the ObjectJ plugin (Norbert Vischer and Stelian Nastase, University of Amsterdam, Amsterdam, Netherlands) for ImageJ software (National Institutes of Health) on n ≥ 30 cells from each category.
In situ hybridization
Digoxigenin-labeled RNA probes were generated by in vitro transcription with the DIG labeling mix (Roche) and T7 or SP6 polymerase (Promega). As a template, pCS2-TTLL plasmids were used. The plasmids were cut with restriction enzymes in a way that allowed the generation of 600–800-bp probes.
Animals were deeply anaesthetized with xylazin/ketamine and transcardially perfused with PBS followed by 4% PFA in PBS. Brains were dissected and post-fixed overnight in 4% PFA, cryo-protected in 30% sucrose-PBS, and cut on a cryostat in 15-µm coronal sections.
In situ hybridizations were performed using a standard protocol (Schaeren-Wiemers and Gerfin-Moser, 1993). In brief, digoxigenin-labeled RNA probes were incubated (0.1–1 µg/ml; DIG labeling mix; Roche) at 65°C for 16 h. After several high-stringency washes, the presence of DIG was revealed using an alkaline phosphatase–coupled anti-DIG antibody (1:2,000 dilution in 20% sheep serum; Roche) and visualized with NBT-BCIP mixture (Roche). Color development was performed for 3–16 h, depending upon the abundance of the target mRNA.
β-Galactosidase staining
Animals were perfused with 4% PFA in PBS. Dissected brains were postfixed for 30 min at room temperature, sliced at 80-µm thickness using a vibratome, and stained with a solution containing 1 mg/ml X-Gal (5-bromo-4-chloro-indolyl-β-d-galactopyranoside), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 2 mM MgCl2 in PBS, for 16–24 h at 37°C. Samples were washed in PBS and mounted with Fluoromount-G mounting medium (SouthernBiotech).
Cell culture, transfection, and immunofluorescence
HEK293T and HeLa cells were cultured on plastic dishes under standard conditions. Expression plasmids were transfected using JetPEI (PolyPlus) and siRNA using Oligofectamine (Invitrogen).
For primary ependymal culture, cells from the subventricular zone of newborn mice were dissociated, resuspended in fresh medium, plated at high density in culture medium containing 10% FCS, and grown to confluence. Pure confluent astroglial monolayers were replated into new dishes at a density of 104 cells/µl and maintained in serum-free medium for ependymal differentiation for up to 15 d (Fig. 3 A, experimental scheme). Transfection of specific siRNA for endogenous TTLL silencing was performed twice, at 0 and 8 d after serum starvation, using the jetSI-ENDO cationic transfection reagent (PolyPlus).
The ependymal cells were fixed 15 d after serum starvation, using a protocol for the preservation of cytoskeletal structures (Bell and Safiejko-Mroczka, 1995). In brief, cells were incubated for 10 min at room temperature in 1 mM dithiobis(succinimidyl propionate; DSP) in Hank’s balanced salt solution, followed by 10 min in 1 mM DSP in microtubule-stabilizing buffer (MTSB). Cells were then washed for 5 min in 0.5% Triton X-100 in MTSB, and subsequently fixed in 4% PFA in MTSB. After a 5-min wash in PBS, cells were incubated for 5 min in 100 mM glycin in PBS, and again washed in PBS.
Fixed cells were incubated in PBS containing 0.1% Triton X-100 and 5% FCS for 30 min at room temperature, and subsequently incubated with polyE and TAP952 antibodies in the same buffer for 1 h, followed by 45 min with anti–mouse or anti–rabbit Alexa Fluor 568, or anti–mouse or anti–rabbit Alexa Fluor 488 antibodies. DNA was visualized by DAPI staining (0.02 µg/ml). Coverslips were mounted with Mowiol polyvinyl alcohol 4-88 (Fluka). No specific labeling was observed after omission of primary antibodies.
Microscopy
All acquisitions were performed at room temperature. We used an epifluorescence microscope (AxioImager Z1/Apotom; Carl Zeiss) with 40× (NA 1.3) and 63× (NA 1.4) oil objectives. Images were acquired using an ORCA-R2 camera (Hammamatsu Photonics) and AxioVision software (Carl Zeiss). For 3D reconstruction and image analysis, image stacks were processed with ImageJ 1.46f. Image assembly was performed using Zen 2011 software (Carl Zeiss).
Whole coronal brain images (in situ hybridization, β-galactosidase staining) were acquired using a stereomicroscope (model M205FA; Leica) with a camera (model DFC310FX; Leica). Images were mounted using Adobe Photoshop CS3 (Adobe Systems, Inc.). Only linear adjustments of max/min levels were performed. Colors in the merged panels representing multiple antibody labeling do not always correspond to the fluorophore used for antibody staining. False colors have been chosen to increase readability of the merge images.
Quantification of multiciliated cells in ependymal cultures
For quantification, all cells present in the field of view from the eyepiece of a microscope (Zeiss AxioImager Z1/Apotom; Carl Zeiss) with the 40× (NA 1.3) oil objective were analyzed. The percentage of multiciliated ependymal cells was determined by counting all cells bearing a tuft of cilia, stained with polyE and TAP952. The number of ciliated cells was then related to the total number of cells per field, determined by counting nuclei labeled with DAPI. Each series of experiments was normalized to control experiments with scramble siRNA. Because the density of cells is highly important for ciliogenesis in this system, only experiments with equivalent cell densities were quantified.
Quantification of ciliary beating frequency
Ependymal cell cultures were processed 15 d after serum deprivation, corresponding to late stage of differentiation. Ependymal cells seeded on 36-mm-diameter dishes with glass bottoms were incubated for 30 min at 37°C with DMEM + FCS 10% and 250 nM Tubulin Tracker green reagent (Invitrogen) to stain ciliary axonemes in living cells. After three washes with medium, the cells were immediately recorded using an inverse confocal microscope (Axio Observer Z1/LSM 5 LIVE DUO; Carl Zeiss) equipped with a 63×/1.4 NA Plan Apochromatic oil objective, a digital microscope camera (AxioCam) and an incubator chamber with controlled temperature and CO2.
A square ROI including a region of single beating cilia was selected within the microscopy field (Fig. 3 E), and the alterations of green fluorescence within this ROI were recorded at 120 frames/s at 512 × 256 pixel resolution during 2.5 s to 5 s, using the LSM Image Browser software. The beating frequency distribution (Fig. 3 F) of each measured cilium was determined by Fourier transformation of the measured beating frequency, using ImageJ with the Ciliary Beating Analyzer 514 plug-in, developed by N. Bonnet (INSERM/UMRS 903; Reims, France). At least 25 cells were analyzed for each condition in 3 independent experiments.
RNA isolation and RT-PCR
Total RNA was isolated from ependymal cell cultures at different time points after the induction of differentiation (−1, 0, 2, 4, 8, 19, 21, and 24 d after serum deprivation) with the RNeasy MICRO kit (QIAGEN). Quality and concentration of total RNA was examined with a Nanodrop Spectrophotometer (Thermo Fisher Scientific).
For RT-PCR, cDNA was synthesized with the First-Strand cDNA Synthesis kit (GE Healthcare). PCR amplification was performed on a Thermocycler (Mastercycler; Eppendorf).
TTLL3 was amplified with the primers TTLL3-F1263 (5′-CCTGTGTAACAACTCCATCCAG-3′) and TTLL3-R2105 (5′-GTCATCAAGGCCTTGCCTGTGG-3′), 33 cycles. TTLL8 was amplified with TTLL8-F598 (5′-GCCTGACCTGTGACCAGATGC-3′) and TTLL8-R1295 (5′-CGATGTACTTCTGGACCACCC-3′), 35 cycles (Fig. S2 A), or with TTLL8-F793 (5′-CTGCCAGCATCCTCAAGTGGG-3′) and TTLL8-R1746 (5′-GTGGGGCTGGAGTTGATCTCG-3′), 35 cycles (Fig. S3 B). As a control, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified with mGAPDH-F37 (5′-GCGCCTGGTCACCAGGGCTGC-3′) and mGAPDH-R1013 (5′-TCCACCACCCTGTTGCTGTAGCC-3′), 20 cycles.
Whole-transcriptome sequencing (RNA-Seq)
A primary ependymal cell culture was prepared as described above. Following the scheme in Fig. 3 A, RNA was extracted as described above from cultures just before serum starvation (day 0) and 5 d afterward (day 5). RNA quality was assessed using a Nanodrop photometer and a Bioanalyzer. Directional libraries were constructed (ScriptSeq mRNA-Seq Epicentre) after depletion of ribosomal RNAs, and libraries were sequenced (Illumina Technology). The quality of the reads was controlled (FastQ 0.10). The reads were filtered to avoid bad quality reads and aligned using UCSC Mm10 as a reference genome (Bowtie 0.12.9). Alignments were filtered to avoid nonaligned reads and multiple alignments reads (Picard, Samtools). Abundance was estimated at gene level using HTSeq-count 0.5.3, Eoulsan version (Jourdren et al., 2012).
Protein electrophoresis and immunoblot
SDS-PAGE was performed using standard protocols. Proteins were transferred to nitrocellulose membranes (EMD Millipore) and detected with antibodies. Membranes were incubated with a rabbit anti-GFP antibody. Protein bands were visualized with HRP-labeled donkey anti–rabbit, followed by detection with chemoilluminescence (ECL Western blot detection kit; GE Healthcare).
Statistics
Data in Fig. 3 C are expressed as mean ± SEM. Multiple comparisons were performed by one-way ANOVA with Tukey post-hoc analysis (software R version 2.13.1; R Development Core Team, 2011). In Fig. 3 G, data were presented as mean ± SEM analyzed with the Fisher variance test (MATLAB software). Data in Figs. 3 H, 4 D, and 5 C were represented as mean ± SEM and analyzed with Welch’s t test (MATLAB).
Online supplemental material
Online material comprises three supplemental figures showing control experiments as well as additional data supporting the main figures. Two videos represent the image series shown in Fig. 3 D. siRNA sequences are listed in Table S1, and primary antibodies used in this study are detailed in Table S2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305041/DC1.
Supplementary Material
Acknowledgments
We thank C. Alberti, E. Belloir, Y. Bourgeois, I. Grandjean, A. Thadal (Institut Curie animal facility), S. Heurtebise-Chrétien (Institut Curie), A. Aubusson-Fleury (CBM, Gif-sur-Yvette, France), N. Gold, J. Miro (CRBM, Montpellier, France), K. Chebli, M. Hahne, C. Jacquet, E. Jouffre (IGMM, Montpellier, France), V. Bäcker, J. Cau, S. DeRossi, P. Travo, H. Boukhaddaoui (RIO Imaging Montpellier), J. Boutet de Monvel (Institut Pasteur, Paris, France), F. Coulpier, S. Lemoine, and S. Le Crom (IBENS genomic facility) for technical assistance. We are grateful to K. Rogowski, J. van Dijk (CRBM), N. Delgehyr, P. Foerster, A. Meunier (IBENS), and A. Wehenkel (Institut Curie) for instructive discussions. We want to thank A. Aubusson-Fleury and J. Bowen LaRose and M. Gorovsky (University of Rochester, Rochester, NY) for the gift of TAP952 and polyG antibodies; and the EUCOMM consortium for generating and providing the ttll3−/− mouse.
This work was supported by the Institut Curie, the CNRS, the ENS, the INSERM, a FPGG grant, an FRM grant DEQ20081213977 (C. Janke), an FRM startup grant (N. Spassky), the ANR award 08-JCJC-0007 (C. Janke) and CiliaStem (N. Spassky), HFSP grants RGP0023/2008 (C. Janke) and RGY0074/2007 (N. Spassky), a startup grant from the Mairie de Paris (N. Spassky), INCA grant 2009-1-PL BIO-12-IC-1 (C. Janke), and EMBO YIP (C. Janke).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CBF
- cilia beating frequency
- CCP
- cytosolic carboxy peptidase
- CSF
- cerebrospinal fluid
- MT
- microtubule
- PN
- mouse postnatal day of age
- PTM
- posttranslational modification
- ROI
- region of interest
- TTLL
- tubulin tyrosine ligase like
References
- Backer C.B., Gutzman J.H., Pearson C.G., Cheeseman I.M. 2012. CSAP localizes to polyglutamylated microtubules and promotes proper cilia function and zebrafish development. Mol. Biol. Cell. 23:2122–2130 10.1091/mbc.E11-11-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P.B., Jr, Safiejko-Mroczka B. 1995. Improved methods for preserving macromolecular structures and visualizing them by fluorescence and scanning electron microscopy. Scanning Microsc. 9:843–857, discussion :858–860 [PubMed] [Google Scholar]
- Boutin C., Diestel S., Desoeuvre A., Tiveron M.-C., Cremer H. 2008. Efficient in vivo electroporation of the postnatal rodent forebrain. PLoS ONE. 3:e1883 10.1371/journal.pone.0001883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bré M.H., Redeker V., Quibell M., Darmanaden-Delorme J., Bressac C., Cosson J., Huitorel P., Schmitter J.M., Rossler J., Johnson T., et al. 1996. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 109:727–738 [DOI] [PubMed] [Google Scholar]
- Carlén M., Meletis K., Göritz C., Darsalia V., Evergren E., Tanigaki K., Amendola M., Barnabé-Heider F., Yeung M.S.Y., Naldini L., et al. 2009. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat. Neurosci. 12:259–267 10.1038/nn.2268 [DOI] [PubMed] [Google Scholar]
- Guirao B., Meunier A., Mortaud S., Aguilar A., Corsi J.-M., Strehl L., Hirota Y., Desoeuvre A., Boutin C., Han Y.-G., et al. 2010. Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat. Cell Biol. 12:341–350 10.1038/ncb2040 [DOI] [PubMed] [Google Scholar]
- Han Y.-G., Alvarez-Buylla A. 2010. Role of primary cilia in brain development and cancer. Curr. Opin. Neurobiol. 20:58–67 10.1016/j.conb.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez-Tallon I., Pagenstecher A., Fliegauf M., Olbrich H., Kispert A., Ketelsen U.-P., North A., Heintz N., Omran H. 2004. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum. Mol. Genet. 13:2133–2141 10.1093/hmg/ddh219 [DOI] [PubMed] [Google Scholar]
- Ihrie R.A., Alvarez-Buylla A. 2011. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 70:674–686 10.1016/j.neuron.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Sato S., Nakamura K., Ostrowski L.E., Setou M. 2010. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc. Natl. Acad. Sci. USA. 107:10490–10495 10.1073/pnas.1002128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Bulinski J.C. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12:773–786 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- Janke C., Rogowski K., Wloga D., Regnard C., Kajava A.V., Strub J.-M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., et al. 2005. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 308:1758–1762 10.1126/science.1113010 [DOI] [PubMed] [Google Scholar]
- Jourdren L., Bernard M., Dillies M.-A., Le Crom S. 2012. Eoulsan: a cloud computing-based framework facilitating high throughput sequencing analyses. Bioinformatics. 28:1542–1543 10.1093/bioinformatics/bts165 [DOI] [PubMed] [Google Scholar]
- Kubo T., Yanagisawa H.A., Yagi T., Hirono M., Kamiya R. 2010. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 20:441–445 10.1016/j.cub.2009.12.058 [DOI] [PubMed] [Google Scholar]
- Kuo C.T., Mirzadeh Z., Soriano-Navarro M., Rasin M., Wang D., Shen J., Sestan N., Garcia-Verdugo J., Alvarez-Buylla A., Jan L.Y., Jan Y.-N. 2006. Postnatal deletion of Numb/Numblike reveals repair and remodeling capacity in the subventricular neurogenic niche. Cell. 127:1253–1264 10.1016/j.cell.2006.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.-F., Delmotte P., Robinson M.L., Sanderson M.J., Witman G.B. 2008. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180:633–643 10.1083/jcb.200710162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.-S., He Y., Dougherty E.J., Jimenez-Movilla M., Avella M., Grullon S., Sharlin D.S., Guo C., Blackford J.A., Jr, Awasthi S., et al. 2013. Disruption of Ttll5/Stamp gene (Tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2 associated modulatory protein gene) in male mice causes sperm malformation and infertility. J. Biol. Chem. 288:15167–15180 10.1074/jbc.M113.453936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Doetsch F., Sawamoto K., Wichterle H., Alvarez-Buylla A. 2010a. The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 2010:1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Han Y.-G., Soriano-Navarro M., García-Verdugo J.M., Alvarez-Buylla A. 2010b. Cilia organize ependymal planar polarity. J. Neurosci. 30:2600–2610 10.1523/JNEUROSCI.3744-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Fuller M.T. 1985. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101:2085–2094 10.1083/jcb.101.6.2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rogowski K., Juge F., van Dijk J., Wloga D., Strub J.-M., Levilliers N., Thomas D., Bré M.-H., Van Dorsselaer A., Gaertig J., Janke C. 2009. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 137:1076–1087 10.1016/j.cell.2009.05.020 [DOI] [PubMed] [Google Scholar]
- Rogowski K., van Dijk J., Magiera M.M., Bosc C., Deloulme J.-C., Bosson A., Peris L., Gold N.D., Lacroix B., Grau M.B., et al. 2010. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 143:564–578 10.1016/j.cell.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Sanders M.A., Salisbury J.L. 1994. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 124:795–805 10.1083/jcb.124.5.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K., Wichterle H., Gonzalez-Perez O., Cholfin J.A., Yamada M., Spassky N., Murcia N.S., Garcia-Verdugo J.M., Marin O., Rubenstein J.L.R., et al. 2006. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 311:629–632 10.1126/science.1119133 [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N., Gerfin-Moser A. 1993. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 100:431–440 10.1007/BF00267823 [DOI] [PubMed] [Google Scholar]
- Spassky N., Merkle F.T., Flames N., Tramontin A.D., García-Verdugo J.M., Alvarez-Buylla A. 2005. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 25:10–18 10.1523/JNEUROSCI.1108-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S., Eddé B., Fox L.A., Guerrero S., Hard R., Hennessey T., Kabi A., Malison D., Pennock D., Sale W.S., et al. 2010. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 20:435–440 10.1016/j.cub.2009.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F., Qu Y., Montcouquiol M., Zhou L., Komatsu K., Shi D., Fujimori T., Labeau J., Tyteca D., Courtoy P., et al. 2010. Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat. Neurosci. 13:700–707 10.1038/nn.2555 [DOI] [PubMed] [Google Scholar]
- van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., Janke C. 2007. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 26:437–448 10.1016/j.molcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Vogel P., Hansen G., Fontenot G., Read R. 2010. Tubulin tyrosine ligase-like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet. Pathol. 47:703–712 10.1177/0300985810363485 [DOI] [PubMed] [Google Scholar]
- Wloga D., Webster D.M., Rogowski K., Bré M.-H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S.T., Gaertig J. 2009. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell. 16:867–876 10.1016/j.devcel.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., Denoulet P. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432 [PubMed] [Google Scholar]
- Zhang J., Williams M.A., Rigamonti D. 2006. Genetics of human hydrocephalus. J. Neurol. 253:1255–1266 10.1007/s00415-006-0245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.