Abstract
Members of the CUG-BP and ETR-3 like factor (CELF) protein family bind within conserved intronic elements (called MSEs) flanking the cardiac troponin T (cTNT) alternative exon 5 and promote exon inclusion in vivo and in vitro. Here we use a comparative deletion analysis of two family members (ETR-3 and CELF4) to identify separate domains required for RNA binding and splicing activity in vivo. CELF proteins contain two adjacent RNA binding domains (RRM1 and RRM2) near the N-terminus and one RRM (RRM3) near the C-terminus, which are separated by a 160–230 residue divergent domain of unknown function. Either RRM1 or RRM2 of CELF4 are necessary and sufficient for binding MSE RNA and RRM2 plus an additional 66 amino acids of the divergent domain are as effective as full-length protein in activating MSE-dependent splicing in vivo. Non-overlapping N- and C-terminal regions of ETR-3 containing either RRM1 and RRM2 or RRM3 plus segments of the adjacent divergent domain activate MSE-dependent exon inclusion demonstrating an unusual functional redundancy of the N- and C-termini of the protein. These results identify specific regions of ETR-3 and CELF4 that are likely targets of protein–protein interactions required for splicing activation.
INTRODUCTION
The diversity of the human proteome is generated in large part by alternative splicing of pre-mRNAs transcribed from a limited number of genes (1–4). Alternative splicing often results in the expression of different protein isoforms with diverse and even antagonistic activities. Of particular interest are alternative splicing events that are modulated according to developmental and cell-specific regulatory programs. Cis-acting elements that mediate cell-specific regulation have been identified within several pre-mRNAs (4) and some have been shown to bestow cell-specific regulation to heterologous exons (5,6). To understand the mechanisms of cell-specific alternative splicing, it is necessary to identify and characterize the proteins that bind these elements.
An emerging family of proteins that regulate alternative splicing are the CUG-BP and ETR-3 like Factor (CELF) proteins [also known as Bruno-like (Brunol) proteins] (7,8). The human genome contains six known CELF paralogs. While one protein, CUG-BP, is expressed widely, CELF3 and CELF5 are expressed only in brain, and CELF4, ETR-3 and CELF6 are expressed in a subset of tissues (7–11,33). Individual CELF proteins have been shown to regulate splicing of human and chicken cardiac troponin T (cTNT) exon 5, human insulin receptor (IR) exon 11, human muscle-specific chloride channel (ClC-1) intron 2, rat NMDA receptor exons 5 and 21, and rat alpha-actinin mutually exclusive NM and SM exons (7,9,12–17,33). The binding sites associated with regulated splicing have been identified for CUG-BP and ETR-3 and are typically U/G-rich motifs located within the introns adjacent to the regulated exons (12,13,15–17). A neuron-specific CELF ortholog in Caenorhabditis elegans, UNC-75, which is nearly 47% identical to human CELF4 and has a role in modulating neurotransmission, is proposed to regulate neuron-specific alternative splicing (18). Human CELF4 can rescue an unc-75 null mutant phenotype and, like UNC-75, the rescuing human CELF4 protein localizes in nuclear speckles with splicing factors.
Chicken and human cTNT alternative exon 5 are well characterized targets of CELF regulation. cTNT undergoes developmentally regulated alternative splicing conserved in avian and mammalian species such that the exon is predominantly included in embryonic heart and skeletal muscle and is predominantly skipped in the adult (19–24). For chicken cTNT, enhanced exon inclusion in embryonic striated muscle requires four muscle-specific splicing enhancers (MSEs) that are 40–45 nt in length and are located within the introns immediately surrounding exon 5 (5,25). One MSE, MSE2, is sufficient for robust regulation of a heterologous exon in embryonic striated muscle when present in multiple copies located upstream and downstream of the alternative exon (5). CUG-BP, ETR-3 and CELF4 have been shown to bind directly to U/G-rich motifs within MSEs 2 and 3 of chicken cTNT, and CUG-BP binds to a U/G-rich motif 19 nt downstream of human cTNT exon 5 (12,15 and data not shown). All six CELF proteins activate MSE-dependent exon inclusion when co-expressed with human or chicken cTNT minigenes in non-muscle cells (7,33, T. Ho and T. Cooper, unpublished data). Point mutations within the U/G-rich motif of human cTNT that prevent binding of CUG-BP also prevent regulation by all six CELF proteins transiently expressed in vivo as well as activation of exon inclusion in skeletal muscle cultures (15, T. Ho and T. Cooper, unpublished data). Furthermore, exon inclusion of chicken cTNT exon 5 is induced by addition of recombinant ETR-3 to in vitro splicing assays using HeLa nuclear extracts. Point mutations within the U/G motifs in MSEs 2 and 3 that prevent ETR-3 binding also prevent activation by ETR-3 (12). These results demonstrate that CELF proteins bind to U/G motifs within cTNT MSEs and directly activate exon 5 inclusion.
CELF protein domain structure is similar to that of the Elav protein family containing two closely spaced RNA recognition motifs (RRM1 and RRM2) near the N-terminus, a 160–230 residue ‘divergent domain’ and a third RRM (RRM3) near the C-terminus (see Figs 1A and 4A). The CELF RRMs show a high degree of sequence identity among family members, however, there is little sequence identity in the divergent domain. All six genes express multiple isoforms due to alternative splicing generating variability within the N-termini, divergent domain and RRM3 of the protein, and within the mRNA 5′ untranslated region. The six CELF proteins can be separated into two groups based on sequence identity and functional differences. One group contains CUG-BP and ETR-3 which are 78% identical. The second group contains CELF3, CELF4, CELF5 and CELF6 which exhibit at most 43.8% identity to CUG-BP and 62–66% identity to each other. The two protein groups also differ in their ability to regulate splicing of an exon flanked upstream and downstream by three concatamerized copies (six copies total) of MSE2. CELFs 3–6 activate inclusion of this exon while CUG-BP and ETR-3 do not (7,33). Both groups bind MSE2 suggesting that there are differences in either the ability to bind to concatamerized binding sites or in ‘post-binding’ events such as making appropriate protein–protein interactions that are required for splicing activation.
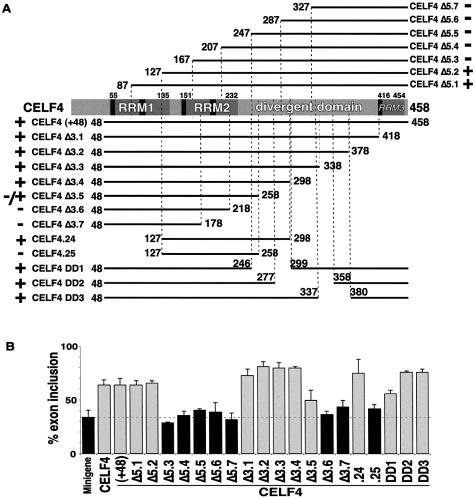
Figure 1.
Splicing activity of human CELF4 deletion mutants. (A) Diagram of full-length human CELF4 protein showing RRM (dark gray) and conserved RNP2 and RNP1 motifs (black). Numbers above the diagram indicate N- and C-terminal positions of the RRMs. Horizontal lines represent the remaining portions of CELF4 deletion mutants. Residue numbers are according to accession number AAK07475. The CELF4 splice form utilized lacks the N-terminal 44 amino acids of RRM3 including the conserved RNP2 hexamer motif. The ability (+) or failure (–) to regulate splicing of an exon flanked by MSEs 1–4 is indicated. (B) Activation of chicken cTNT exon 5 inclusion by transient expression of human CELF4 full-length and truncated proteins in QT35 quail fibroblasts. % = percentage mRNAs including exon 5 as determined by RT–PCR analysis. The percentage of mRNAs including exon 5 is calculated as [CPM exon inclusion band/(CPM exon inclusion band + CPM exon exclusion band)] × 100. Black = minigene alone or not regulated by co-expressed protein; gray = regulated by co-expressed protein (correlates with + or – designation in A).
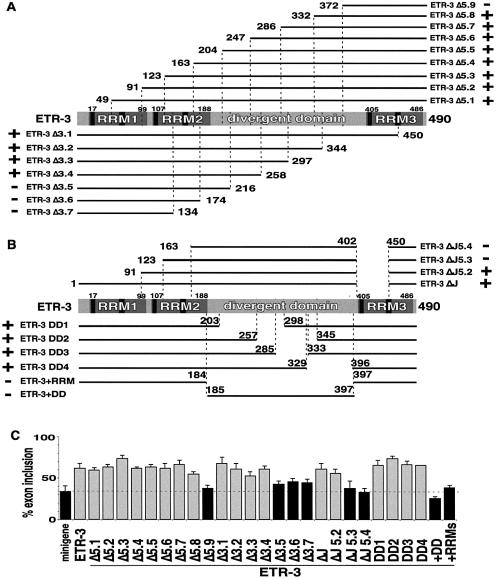
Figure 4.
Splicing activity of human ETR3 deletion mutants. (A, B) Deletion endpoints are comparable to those in CELF4 (Fig. 1A) based on alignment of ETR-3 and CELF4 proteins. The ETR-3 residue numbers are according to accession number AAK92699. The residue numbers differ between ETR-3 and CELF4 due to the longer N-terminus and increased spacing between RRM1 and RRM2 in CELF4, and a longer divergent domain and complete RRM3 in ETR-3. (B) Additional ETR-3 deletion mutants. (C) Activation of chicken cTNT exon 5 inclusion by transient expression of human ETR-3 full-length and truncated proteins in QT35 quail fibroblasts. The evaluation of whether the protein significantly regulates MSE-dependent splicing in fibroblasts is as indicated for Figure 1B.
We have defined the protein domains required for splicing activation in vivo by CELF4 and ETR-3, which were chosen as representatives of the two CELF subgroups. These analyses identified separate domains required for binding and splicing activation for both CELF4 and ETR-3. For CELF4, we found that RRM1 and RRM2 or RRM2 alone plus the adjacent 66 amino acids of the divergent domain activates splicing equivalent to full-length protein. Interestingly, non-overlapping N- and C-terminal segments of ETR-3 activate MSE-dependent exon inclusion, indicating that ETR-3 has functionally redundant segments at the N- and C-termini of the protein. Moreover, an inactive CELF4 deletion mutant lacking functional N-terminal RRMs that does not bind RNA inhibits the ability of active CELF4 proteins to activate exon inclusion in vivo. The dominant-negative activity of this protein is likely to result from its disruption of protein–protein interactions that are required for splicing activation.
MATERIALS AND METHODS
Plasmids
CELF4 and ETR-3 deletion mutants truncated at the positions indicated in Figures 1 and 4 were generated by PCR and cloned using BamHI/XhoI into pcDNA3.1HisC (Invitrogen). All mutants were confirmed by sequencing. cTNT minigene plasmids RTB33.51 and M2/M2TB have been described previously (5).
Cell culture and transfection
Transient transfection of plasmid DNA into quail QT35 fibroblast cultures and primary chicken embryo skeletal muscle cultures, RNA extraction and RT–PCR analysis was performed as described previously (5,7). PCRs included a kinase-labeled oligonucleotide and bands were quantified by phosphoimager analysis. Western blot analysis of expressed proteins was performed as described previously using AntiXpress antibody directed against the N-terminal epitope tag encoded by the pcDNA3.1HisC vector (12).
UV crosslinking and immunoprecipitation of protein expressed in vivo
CELF4 proteins expressed in vivo were tested for RNA binding using a UV crosslink/immunoprecipitation assay as described previously (12) with the following modifications: 5–10 µg of CELF4 expression plasmid was transfected into each of five COS-M6 plates. After 48 h, cells were washed with cold PBS, then 1 ml cold binding buffer (20 mM HEPES, 100 mM KCl, 0.05% Triton X100, 20% glycerol and 1 mM DTT) was added and cells were scraped and centrifuged at 4000 g for 5 min. Binding reactions were performed with 200 µg cytoplasmic extracts, 1 × 107 c.p.m. [32P]GTP and [32P]UTP-labeled RNA following the previously described procedure (12). Western blot analysis of pelleted AntiXpress-tagged proteins was performed using HRP-conjugated AntiXpress (Invitrogen) (to prevent detection of pelleted immunoglobulin) at 1:1000 and bands were visualized with SuperSignal Chemiluminescent Substrate (Pierce).
UV crosslinking using recombinant protein expressed in bacteria
His-ETR3 Δ3.4 and His-ETR3 Δ5.7 were obtained by subcloning ETR3 Δ3.4 and ETR3 Δ5.7 into the pET-28a expression plasmid (Novagen) and were purified using Novagen His-Bind Kits (Novagen). Purified protein was quantitated using the Bio-Rad protein assay kit (Hercules). Recombinant proteins were at least 90% full-length, based on analysis by SDS–PAGE and Coomassie staining. Binding reactions (15 µl) were performed using 500 ng of purified recombinant protein, 90 000 c.p.m. [32P]UTP and [32P]GTP uniformly labeled RNA in a reaction containing 1.5 mM ATP, 1.5 mM magnesium acetate, 1.65 µg yeast tRNA, 150 ng heparin, 10.4 mM HEPES (pH 7.9), 43.68 mM potassium glutamate, 0.104 mM EDTA, 10.4% glycerol, 0.26 mM DTT and 0.26 mM phenylmethylsulfonyl fluoride (PMSF). The RNA was transcribed from T3 promoter of Bluescript KS+ and contained 23 nt from the KS+ vector, the last 15 nt of chicken cTNT exon 5 and the first 142 nt of intron 5 (including MSEs 2–4). Binding was performed for 30 min at 30°C then subjected to UV crosslinking and RNase digestion according to our standard procedure (7). Protein–RNA adducts were separated by SDS–PAGE on a 12% gel then visualized by autoradiography. For competition assays, non-labeled competitor RNAs were synthesized in vitro, gel-purified, quantified by UV absorbance and the indicated amounts were added to the binding reaction 10 min prior to addition of labeled substrate RNA. The MSE competitor RNA was identical to the UV-crosslinking substrate. The non-specific competitor RNA contained the last 96 and first 78 nt of human globin intron 1 flanking a previously described 63 nt artificial exon derived from skeletal troponin I exon 2 (7).
RESULTS
Defining CELF4 domains that are required for splicing activity in vivo
CELF4 promotes MSE-dependent exon 5 inclusion in fibroblasts when co-expressed with a cTNT minigene (7) providing an assay for splicing function in vivo. A deletion analysis of human CELF4 was performed to determine which domains are required for MSE-dependent splicing activation. Activation of MSE-dependent exon inclusion by CELF4 deletion mutants was assayed by transient co-expression with a cTNT minigene in quail QT35 fibroblasts as previously described (7). All CELF4 expression plasmids presented were demonstrated to express proteins of the expected sizes by western blot analysis using AntiXpress antibody (data not shown). Extracts for western blots were from cell cultures transfected in parallel with cultures used for RT–PCR analysis. Therefore, all deletion mutants that do not affect splicing are known to express protein in parallel transfections.
Deletions sequentially removing 40 amino acids from the N- or C-terminus were generated (CELF4 Δ5.1–Δ5.7 and CELF4 Δ3.1–Δ3.7, respectively, Fig. 1A). The CELF4 proteins used in this study are derived from an early cDNA isolate [called CELF4 (+48)] missing the N-terminal 47 residues of full-length CELF4 (7). Splicing activity of the CELF4 (+48) variant is indistinguishable from full-length CELF4 (Fig. 1B). The CELF4 splice variant used predominates in brain and is also found in heart and skeletal muscle (data not shown). This variant contains a remnant of RRM3 that is predicted to be inactive due to loss of its N-terminal 44 residues including the RNP2 motif (26) (depicted in Fig. 1A).
N-terminal deletions partly or completely removing RRM1 retained the ability to activate splicing (CELF Δ5.1 and Δ5.2, Fig. 1A and B). CELF4 lost splicing activity, however, upon deletion of residues 1–166 which removed RRM1 and the RNP2 hexamer motif of RRM2 (CELF4 Δ5.3, Fig. 1A and B). Loss of splicing activity of CELF4 Δ5.3 correlated with a loss of RNA binding (see below). In addition, the CELF4 Δ5.3 and Δ5.4 deletion mutants inhibit splicing activation by full-length CELF4 (see below).
C-terminal deletion mutants containing RRM1 and RRM2 and only 66 or 26 residues of the divergent domain retained full or partial activity, respectively (CELF4 Δ3.4 and Δ3.5, Fig. 1A and B), whereas removal of the entire divergent domain inactivates CELF4 (CELF4 Δ3.6 and Δ3.7, Fig. 1A and B). Next, we combined both N- and C-terminal deletions (CELF4 Δ5.2 + Δ3.4 or Δ3.5) to delineate the minimal domains necessary for CELF4 function. CELF4.24 and CELF4.25 contain one functional RRM (RRM2) and 66 or 26 amino acids of the divergent domain, respectively. While CELF4.25 was completely inactive, CELF4.24 showed activity comparable to full-length CELF4 (Fig. 1A and B), even when the amount of expressed protein was significantly reduced by transfecting only one tenth the amount of CELF4.24 expression plasmid (data not shown). These results demonstrate that RRM2 plus 66 residues of the divergent domain has the same splicing activity as full-length protein in this assay.
Our results (see below) and results of other CELF family members from other labs (see Discussion) indicate that RRM1 and/or RRM2 are required for RNA binding. Therefore, it is likely that the CELF4.24 deletion mutant contains the RNA binding domain (RRM2) plus a domain required for splicing activation located within the first 66 residues of the divergent domain. To determine whether this region of the divergent domain was required for full activity in CELF4 (+48), all but the first 14 residues of this region were removed in construct CELF4 DD1 (Fig. 1A). Deletion of this region slightly reduced CELF4 (+48) splicing activity (Fig. 1B), consistent with this region’s function as an activation domain. Two other deletions within the central and C-terminal regions of the divergent domain did not disrupt splicing activity (CELF4 DD2 and CELF4 DD3, Fig. 1A and B). The fact that CELF4 DD1 retains significant splicing activity suggests either that the remaining 14 residues of the divergent domain retains activity or that other regions of the protein partially compensate for the deleted region. Consistent with the latter possibility, non-overlapping regions of the ETR-3 divergent domain were shown to serve as activation domains (see below).
All CELF4 deletion mutants that activated splicing retained the sequence specificity of full-length protein (7) since they did not activate inclusion of an exon flanked upstream and downstream by the last 76 and first 96 nt, respectively, of human β-globin intron 1 (data not shown).
Either CELF4 RRM1 or RRM2 is sufficient to bind MSE RNA
To determine whether the loss of splicing activity of the CELF4 deletion mutants correlated with loss of RNA binding, we tested binding of proteins expressed in vivo to uniformly labeled in vitro transcribed RNA containing MSEs 1–4 (Fig. 2A). CELF4 expression plasmids were transiently transfected into COS-M6 cells and cytoplasmic and nuclear extracts were prepared 40–48 h later and used in RNA binding/UV crosslinking assays with uniformly labeled MSE RNA. Binding was detected by immunoprecipitation of CELF4-RNA adducts using the Anti-Xpress antibody followed by autoradiography. An equal aliquot of the same pellet was run on a parallel gel for western blot analysis using HRP-conjugated AntiXpress antibody to determine the amount of the immunoprecipitated protein.
Figure 2.
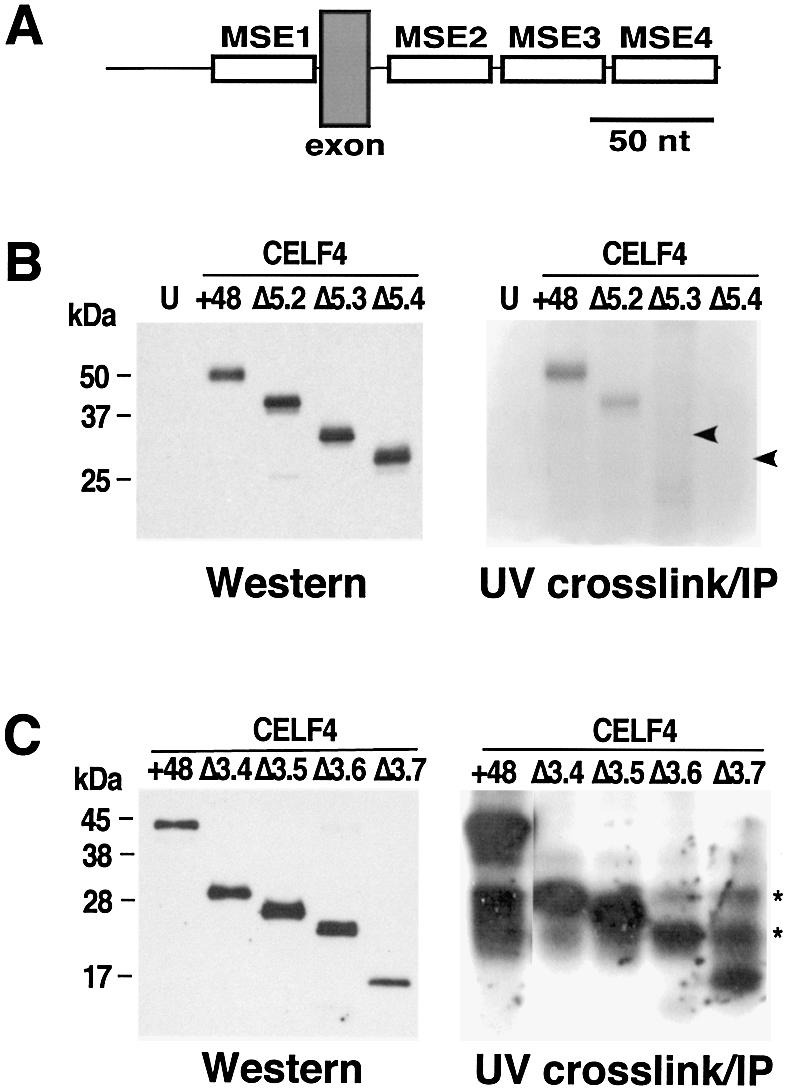
RNA binding of CELF4 deletion mutants. (A) Diagram of RNA containing MSEs 1–4 used for UV crosslinking. (B) RNA binding of N-terminal deletion mutants to a uniformly 32P-labeled cTNT pre-mRNA segment containing MSEs 1–4 (A). Separate aliquots of the same IP pellets were run on separate gels for either detection of immunoprecipitated protein by western blot or binding of the protein to RNA by UV crosslinking. Arrowheads (right) indicate the expected positions of the Δ5.3 and Δ5.4 proteins. U, untransfected; +48, CELF4 (+48), 5.2, 5.3, 5.4 are CELF4 Δ5.2, Δ5.3 and Δ5.4, respectively. (C) RNA binding of C-terminal deletion mutants. Note that the bands detected at ∼31 and ∼23 kDa (indicated by asterisks and particularly apparent in the lane containing CELF4 Δ3.7) are endogenous proteins detected in all lanes.
CELF4 (+48) and CELF4 Δ5.2 which activate splicing in vivo also bind MSE RNA in cytoplasmic extracts from transiently transfected cultures (Fig. 2B, right panel). The same results were obtained using nuclear extracts (data not shown). As preparation of cytoplasmic extracts is rapid compared with nuclear extracts, cytoplasmic extracts were used to reduce concerns of protein degradation and to simplify the procedure for multiple independent repeats. Despite the high level of the CELF4 Δ5.3 and Δ5.4 protein consistently observed in the immunoprecipitate (Fig. 2B, left panel), binding of these proteins to MSE RNA was not detected by UV crosslinking (Fig. 2B, right panel). We conclude that the loss of splicing activity for the N-terminal deletion mutants CELF4 Δ5.3 and Δ5.4 correlates with the loss of RNA binding activity. This result indicates that, as expected, the partial RRM3 present in CELF4 Δ5.3 and Δ5.4 is unable to bind RNA. Based on these binding results and the observation that CELF4 Δ5.2 activates inclusion of exon 5 but not an exon lacking MSEs (data not shown), we also conclude that RRM2 alone is sufficient for sequence-specific binding and MSE-dependent activation.
In contrast to the CELF4 N-terminal deletions, C-terminal deletions that lost the ability to activate splicing retained RNA binding activity. CELF4 Δ3.4 is as active as full-length CELF4, CELF4 Δ3.5 retains partial activity and CELF4 Δ3.6 and Δ3.7 are inactive (Fig. 1B). When these deletion mutants are transiently expressed, both active and inactive proteins still bind MSE RNA (Fig. 2C, right panel). These mutant proteins as well as CELF4 (+48) were in both the nucleus and cytoplasm based on western blot analysis of nuclear and cytoplasmic protein fractions (data not shown) indicating that the failure of CELF4 Δ3.6 and Δ3.7 to activate splicing was not due to their absence from the nucleus. Binding was demonstrated for CELF4 Δ3.7, indicating that RRM1 alone is sufficient for RNA binding. Taken together, these results and the results above indicate that RNA binding activity is localized to RRM1 and/or RRM2 and strongly suggest that at least one region located within the divergent domain immediately downstream from RRM2 is required for splicing activation.
CELF4 N-terminal deletion mutant proteins exhibit dominant-negative activity
We used two criteria to screen the CELF4 deletion series for dominant-negative activity: (i) inhibition of MSE-dependent activation of exon inclusion in primary embryonic skeletal muscle cultures and (ii) inhibition of MSE-dependent exon inclusion in fibroblasts induced by expression of CELF4 (+48). We demonstrated previously that CELF4 Δ5.3 affected only MSE-dependent splicing in muscle cultures and did not affect splicing of an exon flanked by human β-globin introns [CELFΔ in Charlet-B. et al. (12)]. Here we show that CELF4 Δ5.4 also inhibits endogenous CELF activity in primary skeletal muscle cultures (Fig. 3A). CELF4 Δ5.3 was used for subsequent experiments since it consistently displayed a stronger inhibitory effect. Dominant-negative activity of CELF4 Δ5.3 was also demonstrated by its ability to inhibit activation by CELF4 (+48). Co-expression of CELF4 Δ5.3 with CELF4 (+48) prevented activation of exon inclusion despite comparable levels of CELF4 (+48) expressed (Fig. 3B). CELF4 Δ5.3 also inhibited MSE-dependent activation of exon inclusion by ETR-3, indicating an ability to affect activity of other CELF family members (12).
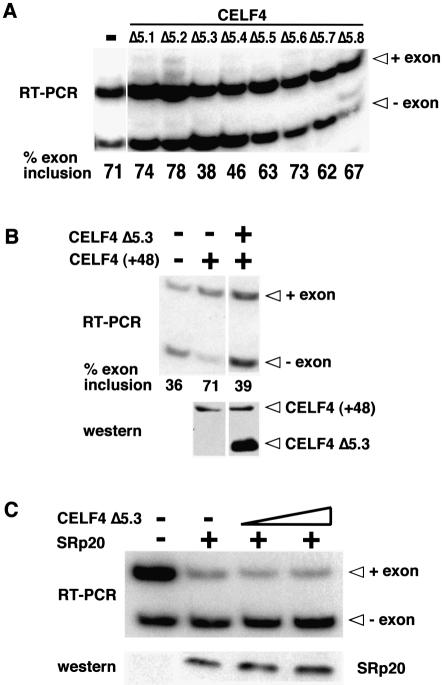
Figure 3.
Characterization of a CELF4 dominant-negative mutant. (A) Co-expression of the series of CELF4 N-terminal deletion mutants with the RTB33.51 cTNT minigene in primary skeletal muscle cultures. In the absence of co-expressed protein, this minigene shows activated inclusion to 70–75% which is MSE-dependent (25, lane 1). CELF4 Δ5.3 and Δ5.4 inhibited the MSE-dependent activation in muscle cultures while other deletion mutants had no effect. These RT–PCR results are representative of five independent experiments. (B) CELF4 Δ5.3 inhibits activation by CELF4 (+48). Western blot analysis using AntiXpress antibody from a parallel transfection showed that overexpression of CELF4 Δ5.3 did not affect expression of cotransfected CELF4 (+48) plasmid. (C) CELF4 Δ5.3 does not affect modulation of splicing by non-CELF splicing factors on non-MSE pre-mRNA substrates. Co-expression of an SRp20 expression vector with a minigene containing clathrin light exons 4–6 in QT35 fibroblasts induces skipping of the alternatively spliced EN exon. Co- expression of CELF4 Δ5.3 has no effect on this modulation by SRp20. SRp20 protein was detected using mAb104 monoclonal antibody and CELF Δ5.3 was detected using Anti-Xpress (data not shown).
To determine whether CELF4 Δ5.3 had a general inhibitory effect on splicing, we tested whether it interfered with the effects of SR proteins on splicing of the clathrin light chain EN exon (exon 5) (27), utilizing a system in which non-CELF proteins regulate splicing of an MSE-independent exon. Co-expression of SRp20 with the clathrin light chain minigene strongly induced skipping of the EN exon (Fig. 3C) and co-expression of CELF4 Δ5.3 did not affect modulation of EN splicing by SRp20 (Fig. 3C). Taken together, our results demonstrate that the effects of the CELF4 Δ5.3 dominant-negative mutant are limited to MSE-driven, CELF-mediated splicing events and are not due to general effects on alternative splicing. Given that CELF4 Δ5.3 protein has MSE-dependent effects (12) without binding MSE-containing RNA (Fig. 2B), we conclude that the mechanism of dominant-negative activity involves interactions of the dominant-negative protein with other proteins that either disrupt the binding of CELF proteins to MSE RNA or the post-binding events that are required to recruit the general splicing machinery (see Discussion).
Non-overlapping N- and C-terminal regions of ETR-3 activate MSE-dependent splicing
Deletion analysis of human ETR-3 was also performed as the representative of the second CELF subgroup. The ETR-3 splice variant used for this analysis (also called NAPOR-1, CUG-BP2 or BRUNOL3) contains a complete RRM3, unlike the CELF4 isoform used above. ETR-3 N- and C-terminal deletions (Fig. 4A) were made at residues exactly comparable to the CELF4 deletion endpoints based on alignment of ETR-3 and CELF4 proteins. As with analysis of CELF4 activity above, protein expression was demonstrated for all ETR-3 mutants by western blot analysis using AntiXpress antibody in cell cultures transfected in parallel with cultures used for RT–PCR analysis (data not shown).
Sequential ETR-3 C-terminal deletions resulted in a loss of splicing activity at a position within the divergent domain that was comparable to that observed for CELF4; between 70 and 28 residues downstream of RRM2 for ETR-3 (ETR-3 Δ3.4 and Δ3.5, Fig. 4A and C) compared with between 66 and 26 residues for CELF4 (see above).
In contrast, ETR-3 N-terminal deletions gave very different results to comparable CELF4 N-terminal deletions. ETR-3 retained splicing activity in the absence of both RRM1 and RRM2. In fact, RRM3 plus the last 119 amino acids of the adjacent divergent domain was sufficient for full MSE-dependent splicing activity (ETR-3 Δ5.7, Fig. 4A and C). Furthermore, the ETR-3 Δ5.7 and Δ3.4 proteins were both able to activate exon 5 inclusion comparably to full-length ETR-3 although they contain non-overlapping N- and C-terminal regions of the protein, respectively. Neither of these mutants activates inclusion of an MSE-minus construct (data not shown) indicating that both ‘ends’ of ETR-3 contain sequence-specific rather than non-specific splicing activity. In addition, the nearly inactive ETR-3 Δ3.5 protein was shown to be abundant in the nucleus (A. Ladd, unpublished results), therefore, loss of splicing activity is due to disruption of intrinsic splicing activity rather than loss of nuclear localization.
The binding sites for full-length ETR-3 have been mapped to MSEs 2 and 3 (12). To determine whether ETR-3 Δ5.7 and Δ3.4 bind MSEs 2–4, bacterially expressed recombinant proteins were used in UV crosslinking assays. As shown in Figure 5B (lanes ‘N’), both proteins bound to uniformly labeled RNA containing MSEs 2–4 (depicted in Fig. 5A). Binding of both proteins was inhibited when challenged with excess non-labeled MSE 2–4 competitor RNA but was only slightly reduced by the same amounts of RNA containing an exon flanked by human β-globin intron 1 (Fig. 5B). Note that splicing of the globin exon in a minigene context is not regulated by either ETR-3 Δ5.7 or Δ3.4 (data not shown). These results demonstrate that the non-overlapping and active N- and C-terminal segments of ETR-3 both bind to MSE RNA in a sequence-specific manner. We conclude that MSE-specific binding and activation of splicing can be mediated by either RRM1 and RRM2 plus 70 residues of the adjacent downstream divergent domain or RRM3 plus 119 residues of the adjacent upstream divergent domain and that activation results from direct binding to the RNA.
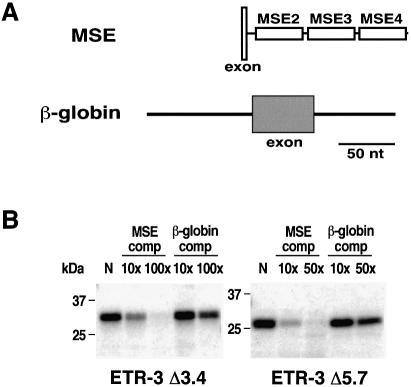
Figure 5.
MSE-specific binding of ETR-3 Δ3.4 and Δ5.7. (A) Diagrams of the RNAs used. UV crosslinking was performed using recombinant ETR-3 Δ3.4 and Δ5.7 proteins and uniformly labeled RNA containing chicken cTNT MSEs 2–4. Globin RNA contains an artificial exon flanked by human β-globin intron 1. (B) Five hundred nanograms of His-tagged bacterially expressed recombinant protein was tested for binding to MSE RNA by UV crosslinking with no competitor RNA (N). Binding of ETR-3 Δ3.4 and Δ5.7 proteins was challenged by the indicated molar excess of non-labeled MSE or globin RNA.
Our characterization of ETR-3 cDNAs expressed in heart and skeletal muscle identified a splice variant missing the 5′-most two thirds of RRM3 comparable to the CELF4 isoform used above (A. Ladd, unpublished results). We tested this ETR-3 isoform (ETR-3 ΔJ, Fig. 4B) for splicing activity and found that it was as strong a splicing activator as full-length ETR-3 (Fig. 4C). N-terminal deletions comparable to those that inactivated CELF4 also inactivated the ETR-3 ΔJ isoform (ETR-3 ΔJ5.3, Fig. 4B and C). This result indicates that the complete RRM3 is required for the splicing activity of the ETR-3 C-terminal region and confirms that at least one RRM is required for activity. Despite the similar structures of CELF4 Δ5.3 and Δ5.4 and ETR-3 ΔJ5.3 and ΔJ5.4, the latter two proteins do not consistently demonstrate dominant-negative activity (data not shown) indicating intrinsic functional differences between CELF4 and ETR-3 (and see below).
Several deletions within the ETR-3 divergent domain were constructed and tested for activity. Complete removal of the divergent domain resulted in a complete loss of activity as did removal of all three RRMs (ETR-3+RRM and ETR-3+DD, respectively, Fig. 4B and C). The ETR-3+RRM construct demonstrated that the divergent domain is required for splicing activity, however, four deletions staggered across nearly the entire divergent domain (ETR-3 DD1–4, Fig. 4B) showed no decrease in the activity of ETR-3 in this assay (Fig. 4C). These results indicate that the divergent domain contains regions that are redundant for splicing activation consistent with the results from above indicating that non-overlapping regions of the divergent domain associated with RRM1 and RRM2 or RRM3 can activate splicing.
Deletion of the N-terminal RRMs allows ETR-3 to activate splicing via a reiterated MSE
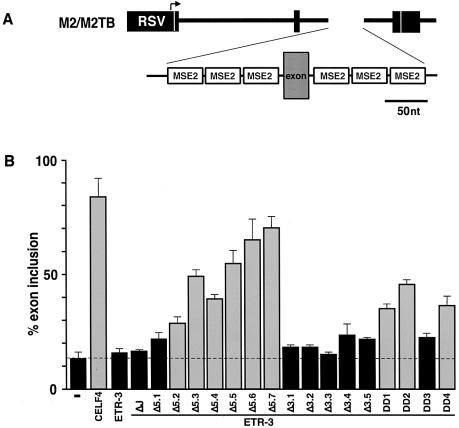
Both ETR-3 and CELF4 activate inclusion of exons flanked by MSEs 1–4. In contrast, CELF4 but not ETR-3 activates splicing of an exon flanked by concatemers of three MSE2 repeats located upstream and downstream [the M2/M2TB minigene (5,7)]. This is surprising given that both CELF4 and ETR-3 bind to MSE2 (12, data not shown), and binding of ETR-3 on MSE2 is required for full activation of exon inclusion in vitro (12). It is possible that activation by ETR-3 requires additional cis-acting elements, such as the one present in MSE3 (12), to which binding of either ETR-3 or additional factors is required for ETR-3-mediated (but not CELF4-mediated) activation. Alternatively, the structure of ETR-3 might differ from that of CELF4 and not be compatible with the artificial spacing of binding sites on concatamerized MSE2, such that binding or post-binding activation cannot take place.
To test whether different deletion mutants of ETR-3 which were active on MSEs 1–4 acquired the ability to activate splicing via concatamerized MSE2 elements, we co-expressed the series of ETR-3 deletion mutants with the M2/M2TB minigene (Fig. 6A). We first tested the ETR-3 ΔJ ETR-3 splice variant since it is the most collinear with the CELF4 variant that activates M2/M2TB exon inclusion, however, it was inactive (Fig. 6B). As the two N-terminal RNA binding domains were deleted, however, ETR-3 gained the ability to activate MSE2-dependent splicing (ETR-3 Δ5.1–5.7, Fig. 6B). Deletion of the C-terminal RRM3 did not have the same effect (ETR-3 Δ3.1–3.5, Fig. 6B). Three of the four internal deletions of the divergent domain also showed some degree of activation. While there are several interpretations of these results (see Discussion), they suggest that there are intrinsic differences in the details of how ETR-3 and CELF4 bind MSE2 RNA and activate exon inclusion.
Figure 6.
The C-terminal but not N-terminal portion of ETR-3 activates splicing via MSE2 alone. (A) The M2/M2TB minigene contains three concatamerized MSE2 elements located upstream and downstream of an artificial alternative exon. (B) M2/M2TB was co-expressed with 1 µg each of the ETR-3 deletion mutant plasmids indicated. Comparable amounts of protein were expressed (data not shown). Splicing of the alternative exon was assayed by RT–PCR and the results are presented as the percent of mRNAs that include the exon. Each expression plasmid was tested in at least three transfections. Bars shaded gray are considered to be a change above background that are worthy of consideration.
DISCUSSION
Binding of CUG-BP and ETR-3 have been characterized in several studies using recombinant CUG-BP and ETR-3 expressed in bacteria or in a yeast three-hybrid assay (8,28–31). Domains of other CELF proteins required for binding have not been characterized. All studies on CUG-BP and ETR-3 show that N-terminal segments containing RRM1 and RRM2 possess RNA binding activity. Results for RRM3 binding vary depending on whether CUG-BP or ETR-3 is used, what species is the source and what RNA substrates are used for binding. Studies on human and zebrafish CUG-BP demonstrated sequence-specific binding of RRM3 alone (28,29) while RRM3 of Xenopus ETR-3 and CUG-BP did not bind RNA (28,31). Our results extend these studies by correlating binding of proteins expressed in mammalian cells with sequence-specific splicing function in vivo. This approach has allowed us to identify separate domains required for RNA binding and splicing function.
Results from the CELF4 deletion series demonstrated that either RRM1 (CELF4 Δ3.7) or RRM2 (CELF4 Δ5.2) alone is sufficient for binding to RNA containing MSEs 1–4 (Fig. 2) and confirmed that the partial RRM3 found in this predominant splice form is unable to bind RNA. These results also identified a region that is required for splicing activation separate from RNA binding (see below). Our deletion analysis of ETR-3 revealed that ETR-3 RRM1 and RRM2 plus 71 amino acids of the adjacent divergent domain, or RRM3 plus the adjacent 119 amino acids of the divergent domain, activate MSE-dependent exon inclusion similarly to full-length protein (ETR-3 Δ3.4 and Δ5.7, respectively, Fig. 4). The two non-overlapping N- and C-terminal segments bind MSE 2–4 RNA in a sequence-specific manner. Thus, we propose that the two sets of RRMs and associated activation domains within the same molecule are able to mediate MSE-dependent activation. This is the first identification of CELF domains required for an RNA processing function beyond RNA binding.
Domains required for CELF splicing activation
A truncated CELF4 protein of 172 amino acids containing only RRM2 and 66 amino acids of the adjacent divergent domain exhibits the same level of splicing activity as full-length CELF4 even when expressed at low levels (CELF4.24, Fig. 1A and B). While either RRM1 or RRM2 is sufficient for RNA binding (CELF4 Δ3.7 and Δ5.2, Fig. 2), the first 66 residues of the divergent domain were required for full splicing activity, suggesting that this region serves as an ‘activation domain’. Consistent with this conclusion, removal of this region from full-length CELF4 resulted in reduced activity (CELF4 DD1). Similarly, the fully active N- and C-terminal ends of ETR-3 (ETR-3 Δ3.4 and Δ5.7) contain either RRM1 and RRM2 or RRM3 plus 71 or 120 residues of the adjacent divergent domain, respectively. In total, three divergent domain regions from the two proteins have been identified as sufficient for splicing activation: CELF4 residues 233–298 and ETR-3 residues 189–258 and 286–404. One key to understanding the mechanism by which CELF proteins regulate splicing is to define how binding of CELF proteins to the MSEs ultimately signals the basal splicing machinery to activate inclusion of exon 5. These regions do not contain obvious homology, however, all three contain residues commonly involved in mediating protein–protein interactions. CELF4 233–298 and ETR-3 189–258 are both rich in glutamines while ETR-3 residues 286–404 share similarly spaced leucines with ETR-3 residues 189–258. In addition, both segments of the ETR-3 divergent domain are rich in serines and threonines, raising a possible scenario in which intrinsic ability to regulate splicing could be modified by phosphorylation. Phosphorylation of the closely related protein, CUG-BP, correlates with its nuclear–cytoplasmic distribution (32); however, phosphorylation of ETR-3 has not been examined beyond showing that phosphatase treatment of cell extracts does not alter ETR-3 mobility on SDS–PAGE (A. Ladd, unpublished observations). As these domains within ETR-3 overlap with regions that are determinative for nuclear and cytoplasmic localization (A. Ladd and T. Cooper, submitted), it is also possible that protein–protein interactions and post-translational modifications of these regions are involved in other aspects of protein metabolism rather than splicing activation. As noted above, proteins that fail to activate splicing are nuclear, indicating that the regions required for splicing regulation reported here have been defined based on intrinsic activity rather than effects secondary to nuclear–cytoplasmic localization. Further analysis is in progress to define the specific role of these regions in splicing activation.
Characterization of a truncated CELF protein with dominant-negative activity
Two CELF4 deletion mutants displayed dominant-negative activity (CELF4 Δ5.3 and Δ5.4). The stronger of the two, CELF4 Δ5.3 was defined as a dominant-negative by several criteria in results presented here and published previously (12). (i) CELF4 Δ5.3 inhibits MSE-dependent exon inclusion observed in primary skeletal muscle cultures in that alternative exons not associated with MSEs are not affected by CELF4 Δ5.3 overexpression (12). (ii) CELF4 Δ5.3 has no effect on splicing modulation mediated by non-CELF proteins and non-MSE cis elements. (iii) It is unable to activate MSE-dependent splicing despite being predominantly nuclear in the QT35 cells used for the analysis of splicing activity. (iv) MSE-dependent splicing activation by ETR-3, CELF3, CELF4, CELF5 and CELF6 is inhibited when co-expressed with CELF4 Δ5.3 (12 and data not shown). Attempts to demonstrate inhibition of CUG-BP activity by CELF4 Δ5.3 have produced variable results (data not shown). (v) CELF4 Δ5.3 does not bind RNA yet its effects are MSE-dependent indicating that it interferes in trans with an MSE-dependent process.
The fact that CELF4 Δ5.3 inhibits MSE-dependent splicing activation without binding MSE RNA indicates that it mediates its effects via disrupting protein–protein interactions required for MSE-dependent effects. One possibility is that CELF Δ5.3 disrupts multimerization of CELF proteins required for RNA binding. EDEN-BP, the Xenopus homolog of CUG-BP, has been shown to dimerize in a yeast two-hybrid assay and binds RNA as a dimer (31). It remains to be determined whether human CELF proteins multimerize and whether multimerization is required for binding. Alternatively, CELF4 Δ5.3 could bind and sequester yet to be identified CELF-binding proteins that are required to activate exon inclusion.
ETR-3 truncations modify its capacity for element-dependent activated splicing
Concatamerized copies of MSE2 in the M2/M2TB minigene mediate MSE-dependent inclusion of a heterologous exon in embryonic striated muscle as well as, if not better than, MSEs 1–4 (5). Despite the fact that both ETR-3 and CUG-BP bind to MSE2 (12 and unpublished data), neither protein can promote exon inclusion via concatamerized MSE2 unlike CELFs 3–6 (7,33). Functional differences between ETR-3 and CELF4 have also been observed in their regulation of splicing of alpha-actinin NM and SM exons (17). To investigate the cis and trans requirements for activation by ETR-3, we tested whether ETR-3 deletion mutants gained the ability to activate splicing of the M2/M2TB minigene. We found that the C-terminal but not N-terminal portions of ETR-3 gained the ability to activate MSE2-dependent exon inclusion. These results illustrate several points: (i) ETR-3 (albeit a truncated form) does not require cis elements in addition to MSE2 to activate splicing; binding of ETR-3 to MSE2 alone is sufficient. (ii) There are intrinsic differences between the mechanistic details of MSE-dependent activation by ETR-3 and CELF4. The C-terminal portion of ETR-3 activates M2/M2TB splicing while the N-terminal region of ETR-3 is essentially inactive, but it is the N-terminal region of CELF4 that activates M2/M2TB exon inclusion. This latter point was confirmed by demonstrating that CELF4.24 containing only RRM2 and 66 amino acids of adjacent divergent domain also activates inclusion of M2/M2TB (data not shown). (iii) It is likely that these results reflect a limited flexibility in the intrinsic ‘architecture’ of full-length ETR-3 protein domains. For example, the ETR3 ΔJ protein which is essentially collinear with CELF4 except for a divergent domain that is 33 amino acids longer and has eight fewer residues between RRM1 and RRM2, is unable to activate M2/M2TB exon inclusion (Fig. 6). It is unclear whether ETR-3 is unable to bind to the concatamerized MSE2 elements or whether it binds but bound ETR-3 is unable to form the protein–protein contacts relevant to promoting binding of the basal splicing machinery to the flanking splice site(s). For example, ETR-3 might require additional coactivators that bind outside MSE2 and that are not required by CELF4. One interesting feature is that the C-terminal ETR-3 portion is inactive in the presence of the N-terminal portion in the full-length protein. This is not the case when segments of the divergent domain are removed (ETR-3 DD1, 2 and 4, which show at least some activation). These results indicate that specific spatial requirements must be fulfilled for assembly of a competent activation complex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Andrea Ladd for unpublished results, Nicole Nguyen for constructing the full-length CELF4 expression plasmid and Andrea Ladd and Andre Faustino for helpful comments on the manuscript. This work was supported by NIH R01HL45565 to T.A.C. and a postdoctoral fellowship from the American Heart Association, Western States Affiliate to J.H.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Graveley B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- 3.Modrek B. and Lee,C. (2002) A genomic view of alternative splicing. Nature Genet., 30, 13–19. [DOI] [PubMed] [Google Scholar]
- 4.Black D.L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem., 27, 27–48. [DOI] [PubMed] [Google Scholar]
- 5.Cooper T.A. (1998) Muscle-specific splicing of a heterologous exon mediated by a single muscle-specific splicing enhancer from the cardiac troponin T gene. Mol. Cell. Biol., 18, 4519–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modafferi E.F. and Black,D.L. (1997) A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol., 17, 6537–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladd A.N., Charlet-B.,N. and Cooper,T.A. (2001) The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol., 21, 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Good P.J., Chen,Q., Warner,S.J. and Herring,D.C. (2000) A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem., 275, 28583–28592. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W., Liu,H., Han,K. and Grabowski,P.J. (2002) Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA, 8, 671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu X., Timchenko,N.A. and Timchenko,L.T. (1999) Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum. Mol. Genet., 8, 53–60. [DOI] [PubMed] [Google Scholar]
- 11.Li D., Bachinski,L.L. and Roberts,R. (2001) Genomic organization and isoform-specific tissue expression of human NAPOR (CUG-BP2) as a candidate gene for familial arrhythmogenic right ventricular dysplasia. Genomics, 74, 396–401. [DOI] [PubMed] [Google Scholar]
- 12.Charlet-B. N., Logan,P., Singh,G. and Cooper,T.A. (2002) Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell, 9, 649–658. [DOI] [PubMed] [Google Scholar]
- 13.Charlet-B. N., Savkur,R.S., Singh,G., Philips,A.V., Grice,E.A. and Cooper,T.A. (2002) Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell, 10, 45–53. [DOI] [PubMed] [Google Scholar]
- 14.Savkur R.S., Philips,A.V. and Cooper,T.A. (2001) Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nature Genet., 29, 40–47. [DOI] [PubMed] [Google Scholar]
- 15.Philips A.V., Timchenko,L.T. and Cooper,T.A. (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki H., Jin,Y., Otani,H., Yasuda,K. and Inoue,K. (2002) Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells, 7, 133–141. [DOI] [PubMed] [Google Scholar]
- 17.Gromak N., Matlin,A.J., Cooper,T.A. and Smith,C.W. (2003) Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA, 9, 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loria P.M., Duke,A., Rand,J.B. and Hobert,O. (2003) Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr. Biol., 13, 1317–1323. [DOI] [PubMed] [Google Scholar]
- 19.Cooper T.A. and Ordahl,C.P. (1985) A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J. Biol. Chem., 260, 11140–11148. [PubMed] [Google Scholar]
- 20.Greig A., Hirschberg,Y., Anderson,P.A.W., Hainsworth,C., Malouf,N.N., Oakeley,A.E. and Kay,B.K. (1994) Molecular basis of cardiac troponin T isoform heterogeneity in rabbit heart. Circ. Res., 74, 41–47. [DOI] [PubMed] [Google Scholar]
- 21.McAuliffe J.J. (1994) Delineation of the cardiac troponin T expression pattern during murine development. J. Cell. Biochem., 18D (Suppl), W347. [Google Scholar]
- 22.Saggin L., Gorza,L., Ausoni,S. and Schiaffino,S. (1990) Cardiac troponin T in developing, regenerating and denervated rat skeletal muscle. Development, 110, 547–554. [DOI] [PubMed] [Google Scholar]
- 23.Townsend P.J., Farza,H., Macgeoch,C., Spurr,N.K., Wade,R., Gahlmann,R., Yacoub,M.H. and Barton,P.J.R. (1994) Human cardiac troponin T: Identification of fetal isoforms and assignment of the TNNT2 locus to chromosome 1q. Genomics, 21, 311–316. [DOI] [PubMed] [Google Scholar]
- 24.Jin J.P. and Lin,J.J. (1989) Isolation and characterization of cDNA clones encoding embryonic and adult isoforms of rat cardiac troponin T. J. Biol. Chem., 264, 14471–14477. [PubMed] [Google Scholar]
- 25.Ryan K.J. and Cooper,T.A. (1996) Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol. Cell. Biol., 16, 4014–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 27.Stamm S., Casper,D., Dinsmore,J., Kaufmann,C.A., Brosius,J. and Helfman,D.M. (1992) Clathrin light chain-B—gene structure and neuron-specific splicing. Nucleic Acids Res., 20, 5097–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timchenko N.A., Welm,A.L., Lu,X. and Timchenko,L.T. (1999) CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res., 27, 4517–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki H., Maegawa,S., Nishibu,T., Sugiyama,T., Yasuda,K. and Inoue,K. (2000) Vegetal localization of the maternal mRNA encoding an EDEN-BP/Bruno-like protein in zebrafish. Mech. Dev., 93, 205–209. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi N., Sasagawa,N., Suzuki,K. and Ishiura,S. (2000) The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun., 277, 518–523. [DOI] [PubMed] [Google Scholar]
- 31.Bonnet-Corven S., Audic,Y., Omilli,F. and Osborne,H.B. (2002) An analysis of the sequence requirements of EDEN-BP for specific RNA binding. Nucleic Acids Res., 30, 4667–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts R., Timchenko,N.A., Miller,J.W., Reddy,S., Caskey,C.T., Swanson,M.S. and Timchenko,L.T. (1997) Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc. Natl Acad. Sci. USA, 94, 13221–13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladd A.N., Nguyen,H.N., Malhotra,K. and Cooper,T.A. (2003) CELF6, a member of the CELF6 family of RNA binding proteins, regulates MSE-dependent alternative splicng. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]