Abstract
Background
Water buffalo and goats are natural hosts for S. japonicum in endemic areas of China. The susceptibility of these two hosts to schistosome infection is different, as water buffalo are less conducive to S. japonicum growth and development. To identify genes that may affect schistosome development and survival, we compared gene expression profiles of schistosomes derived from these two natural hosts using high-throughput microarray technology.
Results
The worm recovery rate was lower and the length and width of worms from water buffalo were smaller compared to those from goats following S. japonicum infection for 7 weeks. Besides obvious morphological difference between the schistosomes derived from the two hosts, differences were also observed by scanning and transmission electron microscopy. Microarray analysis showed differentially expressed gene patterns for parasites from the two hosts, which revealed that genes related to lipid and nucleotide metabolism, as well as protein folding, sorting, and degradation were upregulated, while others associated with signal transduction, endocrine function, development, immune function, endocytosis, and amino acid/carbohydrate/glycan metabolism were downregulated in schistosomes from water buffalo. KEGG pathway analysis deduced that the differentially expressed genes mainly involved lipid metabolism, the MAPK and ErbB signaling pathways, progesterone-mediated oocyte maturation, dorso-ventral axis formation, reproduction, and endocytosis, etc.
Conclusion
The microarray gene analysis in schistosomes derived from water buffalo and goats provide a useful platform to disclose differences determining S. japonicum host compatibility to better understand the interplay between natural hosts and parasites, and identify schistosome target genes associated with susceptibility to screen vaccine candidates.
Introduction
Schistosomiasis japonica is caused by the trematode Schistosoma japonicum and is one of the most prevalent zoonotic diseases in many Asian countries. In China, there are currently 365,770 human schistosomiasis cases, thus S. japonicum remains an important public health concern [1], [2]. Despite more than a half century of control efforts, there is no effective means for the control of this disease and no substantial progress in S. japonicum vaccine development has been made, thus treatment is mainly dependent on the drug praziquantel to kill the adult worm within the host. There is a wide range of hosts for S. japonicum that include at least 46 mammalian species, including humans and a variety of domestic and wild animals, such as rats, rabbits, dogs, cats, horses, yellow cattle, goats, sheep, donkeys, and monkeys, among others [3]. Previous studies have revealed that susceptibility of different types of hosts is varied, as mice, goats, and yellow cattle are more sensitive than rats and water buffalo for S. japonicum (Chinese strain) infections [1].
In China, uncontrolled schistosomiasis endemic areas are mostly distributed in marshland/lake and mountainous regions [4]–[6] and epidemiological surveys have revealed that domestic animals play important roles in schistosomiasis transmission [7]. Water buffalo and goats are major domestic animals reared in endemic areas of China, especially water buffalo can spread more eggs into the environment than humans or other animal hosts and, thus they are considered as primary transmission sources of schistosomiasis in endemic areas [8]–[9]. He et al. [10] infected mice, rats, guinea pigs, rabbits, goats, sheep, pigs, water buffalo, yellow cattle, horses and 12 other kinds of animals with S. japonicum under the same conditions and observed the development of parasites in these hosts for up to 60 weeks. Their results showed that the developmental rate of S. japonicum in these hosts was quite different, with the highest infection rate of 60.3% in goats, 43.6% in yellow cattle, and 1% in water buffalo and horses [10]. Water buffalo and goats act as major natural reservoir hosts for schistosomiasis in China although their susceptibility to schistosome infection is quite different, as goats are more susceptible to S. japonicum and result in higher rates of oviposition and sustain more severe pathological damage than water buffalo; however, the molecular basis of these differences remains unknown.
Several large-scale microarray analyses were recently performed using schistosomes to study gender-, stage-, and strain-specific gene expression of S. japonicum and Schistosoma mansoni [11]–[15]. The results of these studies suggested that gene and protein expression analysis in worms from different susceptible hosts can provide useful information to further elucidate the schistosome/host relationship. Recent studies in our laboratory revealed that schistosomes from a susceptible host (BALB/c mice), a less susceptible host (Wistar rat), and a non-permissive host (Microtus fortis, the reed vole) displayed different mRNA and protein expression profiles, and the gene expression analysis suggested that these three hosts may have different response mechanisms to schistosome infection [16]–[18]. These studies also indicated that the gene or protein expression profiles of schistosomes from natural hosts were different than those from laboratory animals. To better elucidate the susceptibility mechanism of schistosome in natural hosts and to identify molecules which might affect schistosome development, we infected water buffalo and goats with S. japonicum then analyzed and compared differences in gene expression profiles of the parasites obtained from the two hosts using microarray analysis. Our results will aid in screening vaccine candidates or new drug targets for the control of schistosomiasis in natural hosts dwelling within endemic areas.
Materials and Methods
Ethics Statement
All procedures were carried out in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The animal study protocol was approved by the Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, People's Republic of China.
Animals and infection
Male water buffalo and goats (n = 3 each), 15–18 months old, free of parasitic helminths and other infectious diseases were used for experimental infection. All animals were purchased from schistosome non-endemic areas with similar body weights for each host and were housed in covered pens, cared for by trained animal keepers, and fed hay and a commercial pelleted ration. S. japonicum (Chinese strain) cercariae were obtained from the snail maintenance room at Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Shanghai, China). Water buffalo were challenged percutaneously with 2000 cercariae of S. japonicum through the upper back using the cover glass method and goats were challenged percutaneously with 400 cercariae through the inguinal groove [19]. The cercariae were shed at 20–25°C before challenge to ensure maximum vitality.
Sample collection and worm observation
The animals were sacrificed 7 weeks postinfection and the parasites were perfused through the hepatic portal vein. All worms were detached manually, counted, and the length and width of each was measured by the same investigator. The worms were measured using a motorized microscope equipped with auto camera ACT-2U (Nikon, Japan) and controlled by a Nikon image analysis software (NIS-Elements). Next, the worms were washed in phosphate-buffered saline (PBS) and then fixed in 2.5% glutaraldehyde phosphate buffer solution. Some of the worm samples were washed with PBS (pH 7.4) three times for 15–30 min each, fixed for 1.5 h with 1% osmic acid, washed three times as before, dehydrated with gradient alcohol (30%, 50%, 70%, 80%, 90%, 95%, and 100%), vacuum dried, spurted for 3 min using an ion sputtering instrument, and then observed via scanning electron microscopy (SEM) (JSM-6380LV; JEOL Ltd., Tokyo, Japan). The other samples were further dehydrated with acetone twice, embedded in an embedding medium, cut in ultrathin 70-nm sections and then observed via transmission electron microscopy (TEM) (Hitachi H-600; Hitachi Medical Corporation, Tokyo, Japan).
RNA extraction and microarray analysis
Worm samples were collected and stored in RNAlater RNA stabilization reagent (Ambion, Carlsbad, CA, USA). Total RNA was extracted from the parasites(10 pairs each animal) collected from water buffalo and goats using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and purified using the RNAeasy mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. The RNA integrity and quality was evaluated using the Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
The microarrays used to analyze the gene expression profiles in schistosomes from water buffalo and goat were constructed by Agilent Technologies, Inc. and included 13,821 contiguous sequences (contigs) plus proprietary positive and negative controls. Contigs were based on the nucleotide sequences from a recent S. japonicum database. Full details of this schistosome microarray design have been deposited in the Gene Expression Omnibus (GEO) database with the platform accession number GPL10987. Microarrays were printed in an 8×15 k feature format.
A 200-ng aliquot of total RNA from each sample was converted into complementary RNA, labeled with the fluorophore cyanine 3-CTP (CY3c) and hybridized according to the manufacturer's instructions (Agilent Technologies, Inc.; One-Color Microarray-Based Gene Expression Analysis). Samples were examined at wavelengths of A260 and A550 using the ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) to determine yield, concentration, amplification efficiency, and abundance of CY3c. Two technical replicates were performed for each sample and three independent biological replicates were designed for each host.
Feature extraction and data analysis
Microarrays were scanned using an Agilent Microarray Scanner (G2565BA) at a wavelength of 550 nm. Hybridized slides were scanned as tagged image files (TIFF) and processed with the Feature Extraction 9.5.3.1 Image Analysis program (Agilent Technologies, Inc.) to produce standardized data for statistical analysis. All slides were assessed for background evenness by viewing the TIFF image using Feature Extraction. Feature extracted data was analyzed using the GeneSpring GX statistical tool (version 7.3.1; Agilent Technologies, Inc.). Microarray data were normalized using a normalization scenario for ‘Agilent FE one-color,’ which included ‘Data Transformation: Set measurements less than 5.0 to 5.0’, ‘Per Chip: Normalize to 50th percentile’, and ‘Per Gene: Normalize to median’.
Data sets were further analyzed based on one-color experiments that have been published elsewhere [20]. The gProcessed Signal values were determined by the GeneSpring GX statistical tool using Feature Extraction software (Agilent, Inc.), including aspects of signal/noise ratio, spot morphology, and homogeneity. The gProcessed Signal represents signals after localized background subtraction and includes corrections for surface trends. Features were deemed absent when the processed signal intensity was >2-fold of the value of the processed signal error value. Features were deemed marginal when the measured intensity was at a saturated value or if there was a substantial amount of variation in the signal intensity within the pixels of a particular feature. Features that were neither absent nor marginal were deemed present. Data points were included only if they were present or present/absent and probes or contigs were retained if all data points were present or present/absent. All microarray data were submitted to the Gene Expression Omnibus public database under the accession number GSE24615. All statistical analyses were performed using R statistical language software (www.r-project.org/) and the q-value was estimated using the false discovery rate (FDR) as a control [21]–[22]. Heatmap and principal component analysis (PCA) were plotted using Java Treeview software (Stanford University, Stanford, CA, USA) and a multidimensional scaling algorithm [23].
Gene ontology and pathway pattern analysis
Further analysis was performed using Blast2Go Batch BlastX software (6-frame translation protein homology; http://www.blast2go.de) [24] to evaluate differences in annotation between two groups of data. The analysis of gene ontology (GO) terms associated to genes considered differentially expressed in Group B (schistosomes from water buffalo) compared to Group G (schistosomes from goats) was performed using the combined graph function of the software. GO correlations with relative gene expression values were made using ErmineJ software [25]. Kyoto Encyclopedia of Genes and Genomes pathway patterns of differentially expressed genes of interest were analyzed using the SBC Analysis System (http://sas.ebioservice.com/).
Real-time polymerase chain reaction (PCR) validation
Real-time PCR was used to validate a subset of genes predicted to be differentially expressed in the microarray experiment. All gene-specific primers were designed using PRIMER3 software (http://frodo.wi.mit.edu/primer3/input.htm). Purified RNA from mixed parasite samples from each animal in each group was used for reverse transcription in a final volume of 20 µL using the PrimerScript RT kit with gDNA Eraser (Cat# DRR047; Takara Bio, Inc., Shiga, Japan). Products were amplified using the SYBR Premix Ex Taq (Cat#DRR041A; Takara Bio, Inc.) in an ABI 7500 Real-time System (Applied Biosystems) with the following profile: 50°C for 2 min, 95°C for 30 s; 40 cycles of 95°C for 5 s and 60°C for 34 s; 95°C for 15 s, and 60°C for 1 min. Each reaction was performed using 20 µL of cDNA from the RT reaction in a final volume of 50 µL. Expression levels of S. japonicum nicotinamide adenine dinucleotide dehydrogenase (GenBank accession no.: AY812950) were used as endogenous controls within each sample. Relative levels of gene expression were calculated using the 2−ΔΔCT method [26]. The correlation of microarray and qPCR analysis was performed by SPSS 16.0 [27].
Results and Discussion
Morphology of schistosomes derived from two natural hosts
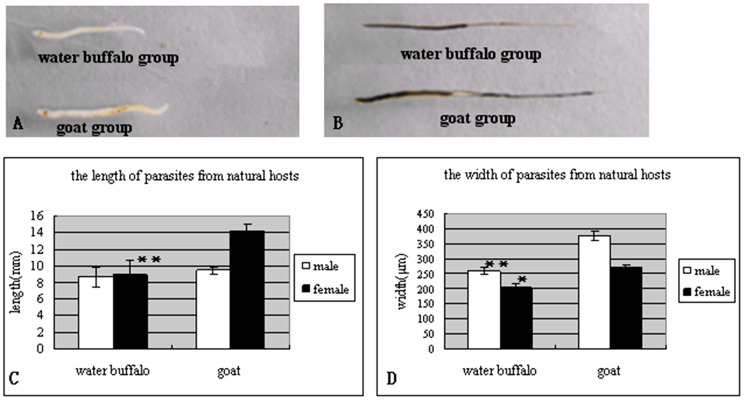
The infected goats displayed more serious disease manifestations than the water buffalo, including diarrhea, athrepsia, egg-deposition, and pathological liver damage [28]. The worm recovery rate in water buffalo was 2.9±1.05%, which was much lower than that in goats (49.50±5.50%). Compared to schistosomes from a third natural host, yellow cattle, schistosomes from water buffalo presented growth and paring retardation [29]. In this study, schistosomes from water buffalo also showed growth retardation compared to parasites from goats (Figure 1A and B). Parasitic length and width from the two hosts were obviously different, as results showed that the female/male worm lengths from water buffalo and goats were 8.86±1.86 mm/8.66±1.23 mm and 14.20±0.84 mm/9.40±0.55 mm, respectively, and the width of female/male worms from water buffalo and goats were 202.98±12.33 µm/259.54±14.57 µm and 267.40±15.24 µm/377.05±15.97 µm, respectively (Figure 1C and D).
Figure 1. Comparison of the length and width of worms from water buffalo and goats at week 7 postinfection.
(A) male worms; (B) female worms; (C) parasite length; (D) parasite width. *, p<0.05; **, p<0.01. The number of worms compared per group was at least n = 50 for females and males, respectively, and included worms from all animals.
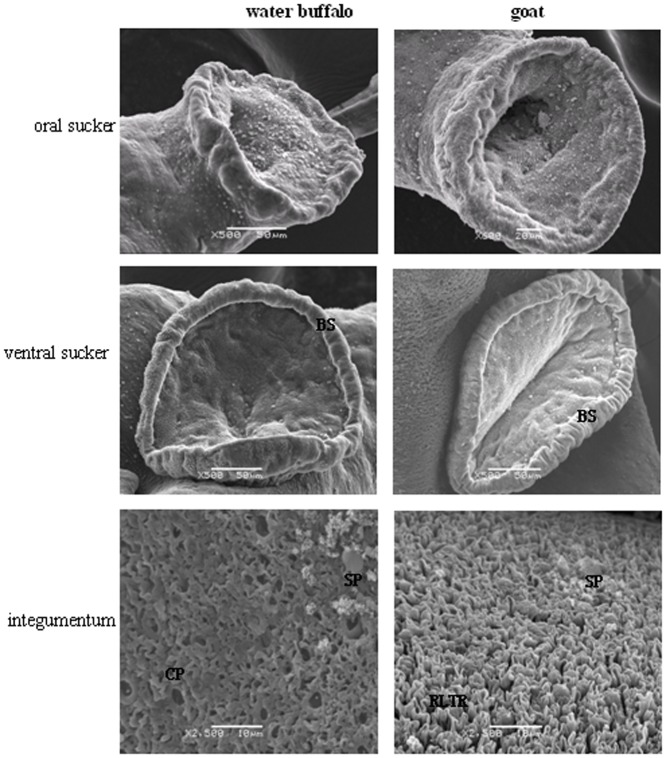
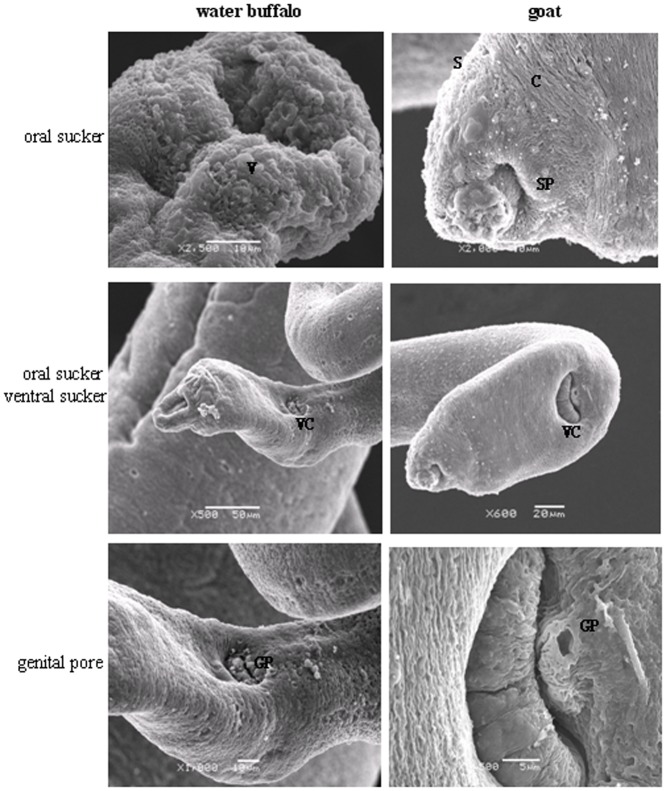
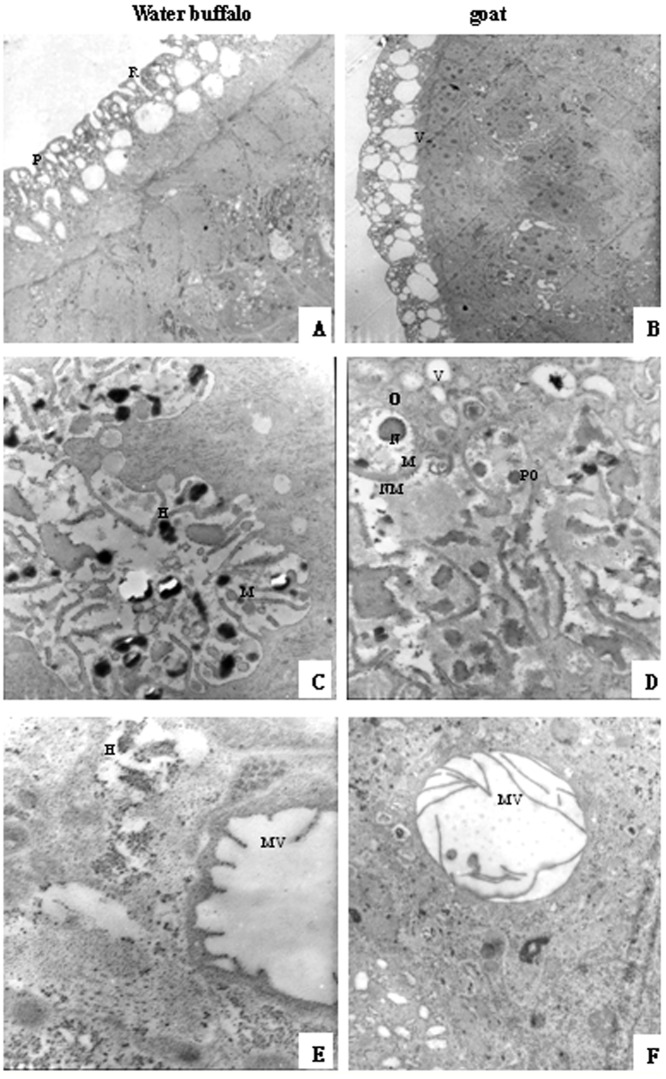
Internal structure analysis at the ultrastructural level by SEM and TEM showed differences between worms from the two natural hosts. Of male schistosomes from water buffalo, the oral and ventral suckers were crimpled, tension loosed, and the esthema mastoid processes in the integument were flattened and vacuolated, while the surface crest and sensory papillae in male worms from goats were affluent, compact, and well arranged (Figure 2). The female schistosomes from water buffalo had collapsed oral suckers on both sides, depressed ventral suckers, and poorly distinguished genital pores beneath the ventral suckers, while female worms from goats were affluent with more esthema mastoid processes in the oral suckers and obvious genital pores next to ventral suckers (Figure 3).
Figure 2. Comparisons of the oral suckers, ventral suckers, and the integument of male worms derived from water buffalo and goats by SEM.
BS, border spine; CP, cortical pore; RLTR, rope-like tegumental ridge; SP, sensory papillae.
Figure 3. Comparisons of the oral suckers, ventral suckers, and the genital pores of female worms derived from water buffalo and goats by SEM.
V, vacuole; C, crest; S, spine; SP, sensory papillae; VC, ventral sucker; GP, genital pore.
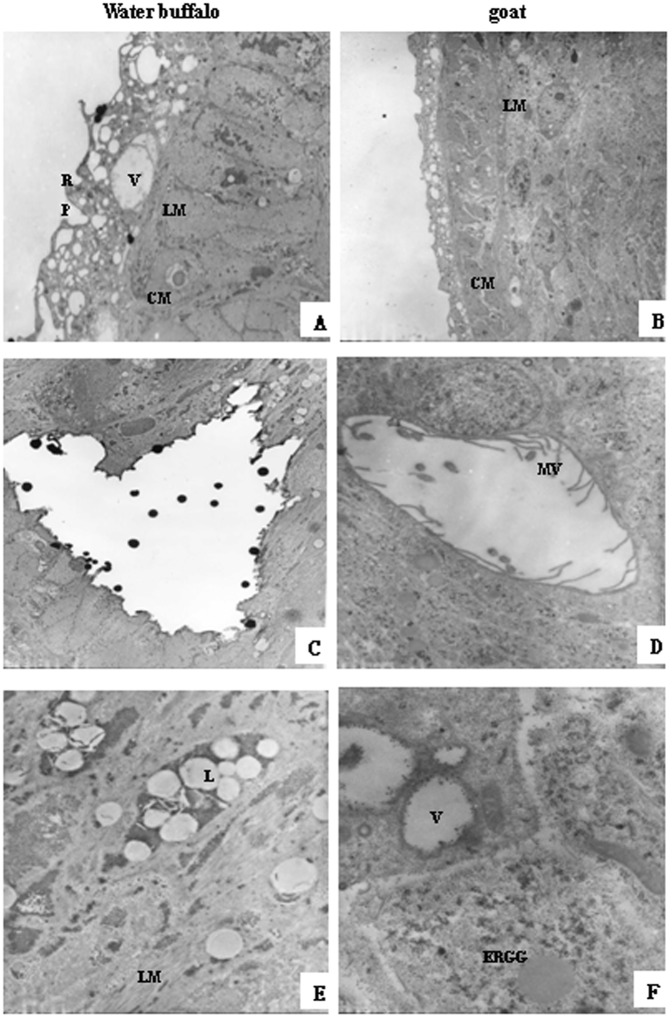
A comparison of male worms from water buffalo and goats using TEM showed that those from water buffalo had more vacuolar structures in the teguments, no internal microvilli, and the cytoplasmic organelles were dissolved (Figure 4). The female worms from water buffalo and goats had no obvious differences in the tegument, but there were more and longer internal microvilli in female worms from goats than from water buffalo (Figure 5).
Figure 4. Comparisons of male worms derived from water buffalo and goats by transmission electron microscopy (TEM).
(A, B) tegument, 2,000× magnification; (C–F) subtegument and inner structures; (C, D) 3,000× magnification; (E, F) 10,000× magnification. R, ridge; P, pore; V, vacuoles; LM, longitudinal muscle; CM, outer circular muscle; MV, microvilli; L, lipid droplet; ERGG, endoplasmic reticulum glycogen granule.
Figure 5. Comparisons of the female worms derived from water buffalo and goats by TEM.
(A, B) tegument, 2000× magnification; (C–F) subtegument and inner structure, 12,000× magnification. H, heterochromatin; M, mitochondria; O, oogonia; N, nuclei; NM, nuclear membrane; PO, primary oocytes.
Global gene expression profiles in schistosomes from water buffalo and goat
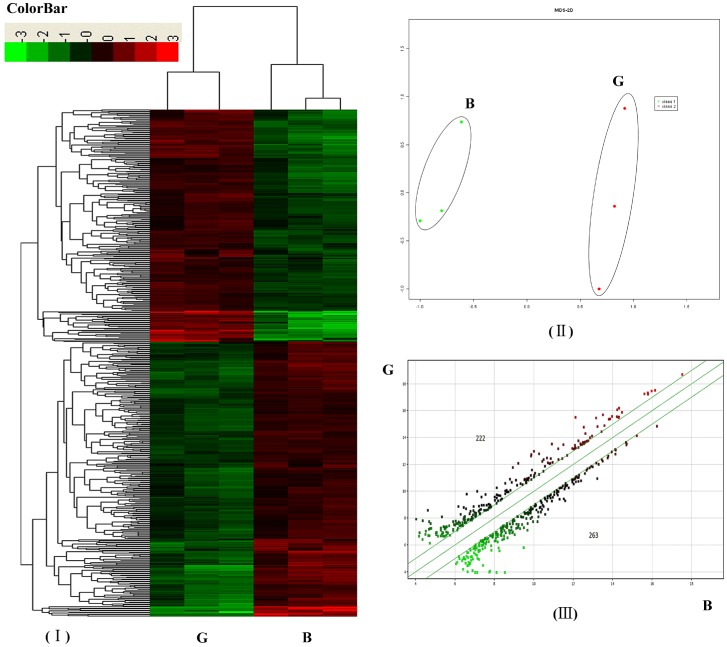
Three biological replicates of schistosomes from each host were evaluated and the correlation between biological replicates was 0.99 for each host. All data have been deposited in the Gene Expression Omnibus database maintained by the National Center for Biotechnology Information (GEO Series accession no.: GSE24615) (http://www.ncbi.nlm.nih.gov/geo/). We used 485 significant transcript expression values of schistosomes from the two natural hosts (p<0.05, fold change (FC)>2) for hierarchical clustering. Different profiles could be clearly identified among schistosomes from water buffalo (Group B) and goats (Group G). Two main clusters separated the schistosome genes among the two different hosts and the gene expression patterns of groups B and G were clustered (Figure 6I). To evaluate the overall data structure, we plotted the first two principal components of a PCA to capture the overall variance of the samples in two dimensions. This analysis clearly separated the data into two subgroups, which clustered the biological replicates together and separated the samples by the host from which the schistosomes were derived (Figure 6II).
Figure 6. Transcription profile analysis of significantly differentiated expressed genes in S. japonicum from water buffalo (group B) and goats (group G).
(A) Hierarchical clustering using differentially expressed genes (probe sets) (p<0.05; FC>2); (B) Principal component analysis (PCA) of transcript profiles from groups B and G; (C) A scatter plot comparing groups G and B. The number at the upper left denotes up-regulated genes and the number at the lower right denotes down-regulated genes (p<0.05; FC>2).
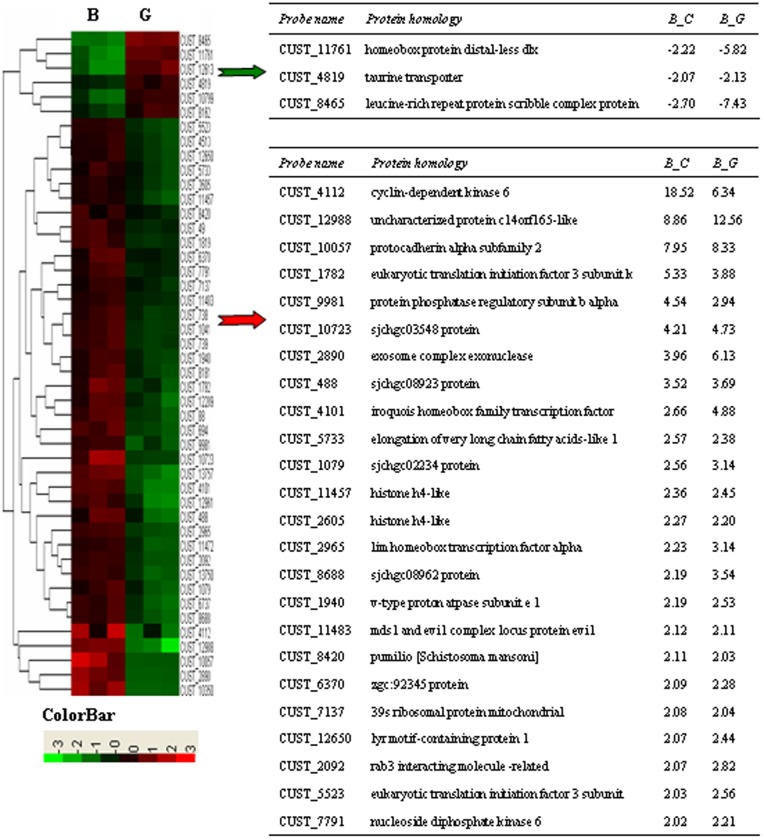
For genes to be considered differentially expressed, a ≥2 FC in gene expression was required with a t-test p-value <0.05. Compared to parasites from water buffalo, the distribution of the up- and down-regulated genes are shown in the scatter plot in Figure 6-III(222 up-regulated and 263 down-regulated). More differentially expressed genes were found in schistosomes between water buffalo and goats than between water buffalo and yellow cattle (485 vs. 69) because the members of the latter pair were more closely related to each other phylogenetically. Most differentially expressed schistosome genes between water buffalo and yellow cattle are also found in schistosomes between water buffalo and goats (46/69 vs. 46/485). A heatmap is shown in Figure 7 and a gene list is shown in Table S1 and S2 for the common differentially expressed genes. The majority of them were analyzed in detail in our previous report [30]. In this study, we identified several additional important differentially expressed genes between worms from water buffalo and goats.
Figure 7. Heatmap clustering of common differentially expressed genes.
The right tables listed the common differentially expressed genes (p<0.05; FC>2). Schistosomes from water buffalo, yellow cattle, and goats were simplified as groups B, C, and G respectively. “-” indicates downregulation. FC is represented as the mean value. The tables do not contain genes without protein homology descriptions.
Real-time PCR validation of microarray data
Subsets of genes with different expression levels and various biological functions in schistosomes from water buffalo and goats were selected for real-time PCR analysis to validate the microarray transcription results. The primer sequences and validation results are listed in Table 1. The real-time PCR results matched the microarray data very well, including the directionality and fold changes, a significant correlation of 0.817 was observed in our data (Spearman's Rho, p<0.01, n = 9), thereby validating the microarray results.
Table 1. Details of real-time RT-PCR primers and results of confirmation.
| ProbeName | Primer(5′-3′) | Size (bp) | Real-time FCa | Microarry FCb | Description |
| CUST_551 | FP:agatcgcgttcaaacaacaaRP:atctgccggatatgaaccaa | 196 | −2.63 | −6.84 | Expressed protein |
| CUST_12936 | FP:gaacgtgatgctgttgttgcRP:atcctcgacatcccaatcag | 192 | −2.64 | −7.27 | Prostatic spermine-binding protein precursor (SBP) |
| CUST_4819 | FP:ttaagcgggatcaatggaagRP:caccacgacgtgttaattgc | 210 | −2.05 | −2.14 | Solute carrier family 6 (ko:K05045) |
| CUST_12988 | FP:ctgctgtggagggaatgtttRP:tggaggattccaggtttcag | 219 | 50 | 12.56 | Putative uncharacterized protein C14orf165 |
| CUST_4112 | FR:gttattggatttcccgctcaPR:atggcaatgaaagtgcatca | 199 | 3.34 | 6.34 | Cyclin-dependent kinase 6, CDK6 |
| CUST_10350 | FR:ggattgattccgccattacPR:gaatggcagtattggttgacg | 198 | 4.75 | 6.88 | Asparagine-rich protein (Ag319) (ARP) |
| CUST_10723 | FR:tgtgccgttattgcgtttagPR:attatcgcttttgccgtcag | 178 | 2.11 | 4.73 | Expressed protein |
| CUST_4101 | FR:atactggtgagcggcctatgPR:gcattcgcaaaccatgtaga | 230 | 3.52 | 4.88 | Iroquois homeobox protein 3, IRX-3 |
| CUST_5523 | FR:gcgttcgccaattgaattatPR:tgttgtattgggtggggatt | 184 | 5.2 | 2.56 | Putative eukaryotic translation initiation factor 3 subunit (eIF-3) |
Fold change (FC) is the ratio of gene expression in schistosomes from water buffalo compared to those from goats; p<0.05, FDR<0.1;
Mean FC in real-time PCR results for validation.
GO functional distribution and pathway pattern analysis of differentially expressed genes in schistosomes from water buffalo and goats
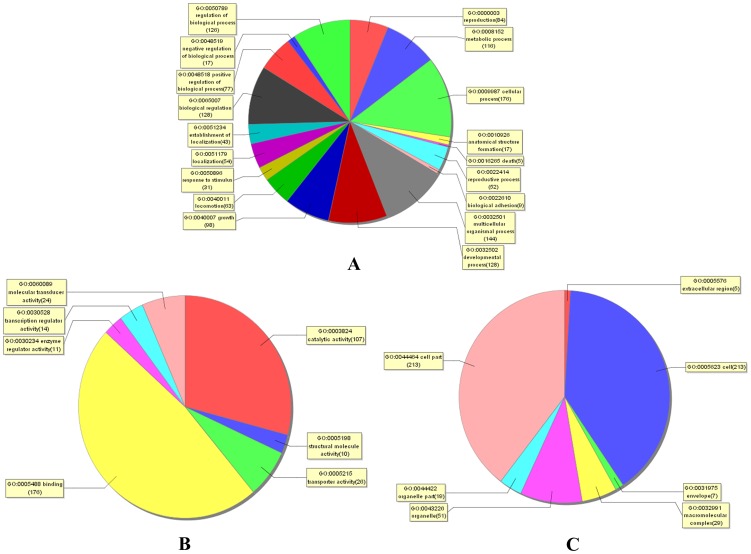
The differentially expressed genes were mainly involved in biological regulation, developmental processes, growth, metabolic processes, cellular processes, and reproduction, (Figure 8A). Molecular functional analysis revealed that most of these molecules were involved in binding, catalytic activity, transportation, molecular transduction, transcription regulation, and enzymatic regulation (Figure 8B). Cellular component analysis showed that most of these molecules were components of the cell, cell parts, organelles, organelle parts, and envelope (Figure 8C). GO analysis and other bioinformatic techniques were further applied to predict/analyze possible functions of the identified bioactive molecules.
Figure 8. Distribution of gene ontology (GO) terms for the differentially expressed genes in schistosomes from water buffalo and goats.
A pie plot showing GO classifications of the biological processes (A), the molecular functions (B), and the cellular components (C). The graph excluded data without assigned GO terms.
Up-regulated genes in schistosomes from water buffalo compared with those from goats
Compared with schistosomes from goats, many genes related to lipid metabolism (prostaglandin synthase 1(PGS-1), acyltransferase-like family member 8 (ACL-8), acetylcholinesterase 1 (AChE-1), and degenerative spermatocyte homolog 1(DEGS-1)), genetic information processing (DNA primase, nucleoside diphosphate kinase (NDK), trans-inducing factor, etc.), folding, sorting and degradation (26S proteasome, bromodomain and tryptophan-aspartic acid (WD) repeat domain containing 3 (BRWD3), and ribosomal RNA-processing protein 4 (RrP4)) were significantly up-regulated in schistosomes from the less-susceptible host water buffalo (Table 2).
Table 2. Selected genes overexpressed in schistosomes from water buffalo compared to those from goats.a .
| Pathway | Probe name | Accession | Gene and/or protein homology | Fold change | p value | FDR |
| Lipid Metabolism | Glycerophospholipid metabolism | |||||
| CUST_3764 | CNUS0000098059 | Prostaglandin synthase 1 | 2.19 | 0.021 | 0.007 | |
| CUST_7726 | CNUS0000102024 | Acetylcholinesterase 1 | 2.71 | 0.036 | 0.007 | |
| Glycerophospholipid metabolism & Glycerolipid metabolism | ||||||
| CUST_8880 | CNUS0000103178 | Acyltransferase-like family member 8 | 2.57 | 0.026 | 0.008 | |
| Sphingolipid metabolism | ||||||
| CUST_6882 | CNUS0000101178 | Degenerative spermatocyte homolog 1 | 2.45 | 0.021 | 0.008 | |
| genetic information processing | Purine metabolism, Pyrimidine metabolism | |||||
| CUST_3246 | CNUS0000097541 | DNA primase small subunit | 2.02 | 0.023 | 0.01 | |
| CUST_9849 | CNUS0000104147 | Nucleoside-diphosphate kinase | 2.08 | 0.001 | 0 | |
| Transcription | ||||||
| CUST_9094 | CNUS0000103392 | Transcription initiation factor TFIIE alpha | 2.34 | 0.018 | 0.008 | |
| Spliceosome | ||||||
| CUST_11072 | CNUS0000105370 | Hypothetical protein | 2.62 | 0.036 | 0.01 | |
| CUST_488 | FN317168 | SJFCE0697 | 3.69 | 0.022 | 0.008 | |
| CUST_7382 | CNUS0000101679 | Expressed protein | 4.53 | 0.019 | 0.007 | |
| Translation | ||||||
| CUST_6052 | CNUS0000100348 | Small subunit ribosomal protein S27Ae | 4.71 | 0.002 | 0.007 | |
| Folding, Sorting and Degradation | Proteasome | |||||
| CUST_3409 | CNUS0000097704 | 26S proteasome regulatory subunit N12 | 3.26 | 0.001 | 0.006 | |
| Ubiquitin mediated proteolysis | ||||||
| CUST_9175 | CNUS0000103473 | Bromodomain and WD repeat domain containing 3 | 2.55 | 0.001 | 0.006 | |
| RNA degradation | ||||||
| CUST_2890 | CNUS0000097185 | RNA-binding protein Rrp4 and related proteins | 6.13 | 0.020 | 0.007 | |
This list includes the probe name, gene accession number, fold change, as well as p- and q-values for protein homology (BlastX). In the gene accession columns, data with identifiers such as CNUS0000098407 were retrieved from the S. japonicum genome project database (LSBI; http://lifecenter.sgst.cn/schistosoma/cn/genomeProject.do; in Japanese) and those with identifiers FN326902 were retrieved from The European Molecular Biology Laboratory database (http://www.ebi.ac.uk/ena/). The FDR (q-value) was determined using the false discovery rate as a control using R statistical language (http://www.bioconductor.org/packages/release/bioc/html/qvalue.html).
Lipid metabolism
The genes associated with lipid metabolism were overexpressed in schistosomes from water buffalo compared to those from goats, but the differentially expressed genes varied among the hosts. The up-regulated genes in schistosomes from water buffalo included PGS-1, ACL-8, AChE-1, and DEGS-1. Prostaglandins are found in most tissues and organs, and synthesized by almost all nucleated cells from essential fatty acids as autocrine and paracrine lipid mediators. Prostaglandins ligate G-protein-coupled receptors, a sub-family of cell surface seven-transmembrane receptors. Ten prostaglandin receptors on various cell types were reported recently and the diversity of the receptors indicates that prostaglandins act on an array of cells and have a wide variety of effects, such as inducing constriction or dilation in vascular smooth muscle cells, aggregation or disaggregation of platelets, and controlling hormone regulation, cell growth, and other processes [31]. Acylation and deacylation are the most common eukaryotic protein post-translational modifications. Acyltransferase functions as an acylation/deacylation catalytic protein and may have multiple functions in regulation of protein biological activity and gene expression.
The parasites derived from water buffalo were stunted and increased expression of nervous system- and transport-related genes when compared to those from yellow cattle [29]. In this study, the AChE-1 and DEGS-1 genes were up-regulated in worms from water buffalo compared to those from goats. AChE-1 is a serine protease that belongs to the carboxylesterase enzyme family and hydrolyzes the neurotransmitter acetylcholine at the synapses and can also hydrolyze butyrylthiocholine. AchE-1 is found mainly at neuromuscular junctions and cholinergic brain synapses where it serves to terminate synaptic transmission. Although AChE-1 is detected at all growth stages of Caenorhabditis elegans, it is more abundant in larval stages than in embryos or adults [32]. DEGS-1 encodes a membrane-bound fatty acid desaturase, which is responsible for forming double bonds into specific positions in fatty acid molecules. DEGS-1 can up-regulate cyclin D1 expression and activation of the transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells [33]. Cyclin D1 plays a pivotal role in cell-cycle transition through the G1 phase and regulates cyclin-dependent kinase 6, which, in turn, regulates schistosome embryogenesis and worm paring through the transforming growth factor beta signaling pathway [34], [35]. DEGS1 overexpression inhibited biosynthesis of the epidermal growth factor receptor (EGFR) [36] and in this study, the EGFR gene was down-regulated (CUST_3717, FC −4.52) in schistosomes from water buffalo, which might be attributable to the DEGS1 over-expression.
Genetic information processing
Biological phenotypes were determined according to gene expression. The genetic information processing-associated genes showed significant differential expression in schistosomes among the hosts. Schistosomes cannot synthesize purines alone, thus they are dependent on a supply from the host [37]. Our results determined that purine and pyrimidine metabolism-associated genes were overexpressed in schistosomes from water buffalo compared to than those from goats, including DNA primase (CUST_3246) and nucleoside-diphosphate kinase (CUST_9849). In addition to its catalytic activity of phosphorylation, NDK reportedly regulates growth and development [38] and is considered a housekeeping enzyme for DNA and RNA synthesis. However, NDK, as well as other transcription-, splicesome-, and translation-associated genes were upregulated in the less susceptible host water buffalo, suggesting that they are important for parasite retardation (Table 2).
Folding, Sorting, and Degradation
The differentially expressed molecules found in this class included posttranslational modification molecules, as 26S proteasome (CUST_3409), BRWD3 (CUST_9175), and RrP4 (CUST_9175) were up-regulated in schistosomes from water buffalo. The 26S proteasome is at the executive end of the ubiquitin proteasome pathway (UPP) for the controlled degradation of intracellular proteins. In eukaryotes, the UPP is essential for proteostasis, in which misfolded or otherwise defective proteins as well as short-lived regulatory proteins are eliminated by degradation [39]. The UPP regulates many fundamental cellular processes, such as protein quality control, DNA repair, cell cycle regulation, antigen processing, and signal transduction [40]. Lysine acetylation is similar to protein phosphorylation in its prevalence in posttranslational modifications and also has a large effect on the physicochemical properties of the modified residues. Protein recruitment to macromolecular complexes by acetylated lysine residues is mediated by bromodomains (BRDs), which are evolutionarily highly conserved protein interaction modules that recognize ε-N-lysine acetylation motifs [41]. The WD repeat proteins, BRWD1 and BRWD3, also contain tandem BRDs. Members of this family are involved in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation [42]; however, little is known regarding the biological function of BRWD3. In Drosophila, BRWD3 function has been genetically linked to the janus kinase- signal transducer and activator of transcription pathway [43]. Mutations in mice revealed a role for BRWD1 in spermiogenesis and oocyte–embryo transition [44]. RNA-binding proteins (RBPs) bind to either double-strand or single-strand RNA molecules through RNA recognition motifs. Thus, RBPs may regulate RNA translation and post-transcriptional events, such as RNA splicing and editing.
Down-regulated genes in schistosomes from water buffalo compared with those from goats
Compared with schistosomes from goats, many genes related to signal transduction (i.e., Akt, ribosomal s6-p90 kinase (RSKN-1), mitogen-activated protein kinase kinase 1 (MEK-1), MEK-6, protein kinase-18 (KIN-18), EGFR, 70-kDa heat shock protein (HSP70), dishevelled (Dsh), etc.), endocrine function (Akt, RSKN-1, MEK-1), development (MEK-1, EGFR), reproduction (SBP), immune function (MEK-1), endocytosis (MEK-1, HSP70, EGFR, partitioning-defective 3 homolog (PAR-3), etc.), amino acid metabolism (aspartate aminotransferase, dopamine beta-hydroxylase, and aromatic-L-amino-acid decarboxylase), carbohydrate metabolism (PI3K-C2α, PI4Kα, pyruvate carboxylase, and galactokinase 2 (GALK2)), glycan biosynthesis, and metabolism (glycosylphosphatidylinositol anchor attachment protein 1 (GPAA1) and glycoprotein 2 (GLY-2)) were significantly down-regulated in schistosomes from the less-susceptible host water buffalo (Table 3).
Table 3. Selected downregulated genes in schistosomes from water buffalo compared with those from goats.a .
| Pathway | Probe name | Accession | Gene and/or protein homology | Fold change | p value | FDR |
| Signal transtruction | MAPK signaling pathway | |||||
| CUST_11642 | CNUS000010594 | Akt1;RAC serine/threonine-protein kinase | −6.92 | 0.022 | 0.005 | |
| CUST_12915 | CNUS0000107219 | Protein RSKN-1 | −2.48 | 0.018 | 0.005 | |
| CUST_3675 | CNUS0000097970 | MAPK/ERK kinase 1(MEK-1) | −4.15 | 0.0002 | 0 | |
| CUST_9010 | CNUS0000103308 | MAPK/ERK kinase 6(MEK-6) | −2.91 | 0.003 | 0 | |
| CUST_9047 | CNUS0000103345 | Protein KIN-18 | −15.09 | 0.0003 | 0 | |
| CUST_3717 | CNUS0000098012 | Epidermal growth factor receptor | −4.52 | 0.004 | 0.005 | |
| CUST_9487 | CNUS0000103785 | Probable beta-arrestin | −2.33 | 0.040 | 0.007 | |
| CUST_5301 | CNUS0000099597 | Heat shock 70 kDa protein cognate 4 | −2.24 | 0.024 | 0.01 | |
| ErbB signaling pathway | ||||||
| CUST_11642 | CNUS0000105941 | Akt1;RAC serine/threonine-protein kinase | −6.92 | 0.022 | 0.005 | |
| CUST_3675 | CNUS0000097970 | MAPK/ERK kinase 1 | −4.15 | 0.0002 | 0 | |
| CUST_5301 | CNUS0000099597 | Heat shock 70 kDa protein cognate 4 | −2.24 | 0.024 | 0.01 | |
| Wnt signaling pathway, Notch signaling pathway | ||||||
| CUST_5485 | CNUS0000099781 | Dishevelled protein(Dsh) | −4.48 | 0.001 | 0.005 | |
| Endocrine System | Progesterone-mediated oocyte maturation | |||||
| CUST_11642 | CNUS0000105941 | Akt1;RAC serine/threonine-protein kinase | −6.92 | 0.022 | 0.005 | |
| CUST_12915 | CNUS0000107219 | Protein RSKN-1 | −2.48 | 0.018 | 0.005 | |
| CUST_3675 | CNUS0000097970 | MAPK/ERK kinase 1 | −4.15 | 0.0002 | 0 | |
| Development/Reproduction | Dorso-ventral axis formation | |||||
| CUST_3675 | CNUS0000097970 | MAPK/ERK kinase 1 | −4.15 | 0.0002 | 0 | |
| CUST_3717 | CNUS0000098012 | Epidermal growth factor receptor | −4.52 | 0.004 | 0.005 | |
| Reproduction | ||||||
| CUST_12936 | CNUS0000107240 | Prostatic spermine-binding protein (SBP) | −7.27 | 0.012 | 0.005 | |
| Immune System | Natural killer cell mediated cytotoxicity | |||||
| CUST_3675 | CNUS0000097970 | MAPK/ERK kinase 1 | −4.15 | 0.0002 | 0 | |
| Endocytosis | CUST_10516 | CNUS0000104814 | beta-adrenergic-receptor kinase | −2.05 | 0.0033 | 0 |
| CUST_3717 | CNUS0000098012 | Epidermal growth factor receptor | −4.52 | 0.004 | 0.005 | |
| CUST_5301 | CNUS0000099597 | Heat shock 70 kDa protein cognate 4 | −2.24 | 0.024 | 0.01 | |
| CUST_8672 | CNUS0000102970 | Partitioning-defective 3 homolog (PAR-3) | −3.85 | 0.032 | 0.005 | |
| CUST_9487 | CNUS0000103785 | Probable beta-arrestin | −2.33 | 0.040 | 0.007 | |
| Amino Acid Metabolism | CUST_4493 | CNUS0000098788 | Aspartate aminotransferase | −2.11 | 0.001 | 0.005 |
| CUST_6397 | CNUS0000100693 | Dopamine beta-hydroxylase | −3.02 | 0.037 | 0.012 | |
| CUST_6212 | CNUS0000100508 | Aromatic-L-amino-acid decarboxylase | −4.70 | 0.029 | 0.01 | |
| Carbohydrate Metabolism | Inositol phosphate metabolism | |||||
| CUST_10542 | CNUS0000104840 | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha (PI3K-C2α) | −2.43 | 0.025 | 0.008 | |
| CUST_13086 | CNUS0000107391 | Phosphatidylinositol 4-kinase alpha (PI4Kα) | −2.03 | 0.027 | 0.01 | |
| Citrate cycle (TCA cycle),Pyruvate metabolism | ||||||
| CUST_6418 | CNUS0000100714 | Pyruvate carboxylase | −2.61 | 0.002 | 0.005 | |
| Galactose metabolism, Amino sugar and nucleotide sugar metabolism | ||||||
| CUST_7443 | CNUS0000101740 | Galactokinase 2(GALK2) | −3.99 | 0.019 | 0.008 | |
| Glycan Biosynthesis and Metabolism | Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | |||||
| CUST_7658 | CNUS0000101956 | Glycosylphosphatidylinositol anchor attachment protein 1(GPAA1) | −3.07 | 0.0016 | 0.005 | |
| N-Glycan biosynthesis | ||||||
| CUST_9370 | CNUS0000103668 | Glycoprotein(Protein GLY-2) | −2.00 | 0.007 | 0.006 | |
Signal transduction/Endocrine system/Development/Reproduction
Mitogen-activated protein kinases (MAPKs) are intracellular serine/threonine protein kinases. The study confirmed that the MAPK signal transduction pathway was present in most cells and transduction of extracellular signals to the cell and its nucleus played a crucial role in cellular biological responses, such as cell proliferation, differentiation, transformation, and apoptosis. MAPK signal transduction pathway was highly conserved evolutionarily both in prokaryotic and eukaryotic cells. The serine-threonine kinase Akt, also known as protein kinase B (PKB), is an important effector for phosphatidylinositol 3-kinase signaling initiated by numerous growth factors and hormones, and has also been implicated in many metabolic functions, such as protein and lipid synthesis, carbohydrate metabolism, and transcription [45]. The serine-threonine kinase Akt represents an important mediator of insulin action in worms and flies. In C. elegans, mutations in Akt result in alterations in development and aging, and Akt/PKB has been implicated as a critical regulator of growth and longevity [46], [47]. Other studies have reported that Akt1−/− mice displayed a conspicuous growth impairment and demonstrated defects in both fetal and postnatal growth, which persisted into adulthood [48]. Akt1−/− mice were smaller when compared to wild-type littermates. In addition, Akt1−/− mouse embryo fibroblasts are more susceptible to apoptosis induced by the tumor necrosis factor receptor superfamily, UV irradiation, and serum withdrawal [49]. Nonetheless, much uncertainty remains concerning how this signaling pathway diverges to allow independent regulation of such disparate biological processes as metabolism, aging, and growth. In schistosomes from water buffalo, the Akt gene was significantly down-regulated (CUST_11642, FC −6.92), suggesting that Akt may be involved in growth retardation via MAPK signal transduction or the endocrine system.
The RSKN-1 protein is a downstream effector of MPK-1/ERK and is critical for dedifferentiation. Rskn-1 RNAi suppressed spermatocyte dedifferentiation and instead induced meiotic division in the C. elegans germline. These regulators are broadly conserved, suggesting that similar molecular circuitry may control cellular dedifferentiation in other organisms as well [50]. The Rskn-1 gene was underexpressed in schistosomes from water buffalo (CUST_12915, FC −2.48) and might also be associated with cellular dedifferentiation; however, this hypothesis requires further investigations.
KIN-18 encodes a previously uncharacterized protein in C. elegans and its catalytic domain shares over 60% identities with TAO kinase 1 (TAO1) and TAO2, which have been recently characterized with a MAPK/ERK kinase role during stress response. Berman et al. [51] reported that expression of constitutively active forms of TAO1/KIN-18 affect the physiology of intact worms and demonstrated that KIN-18 was a 120-kDa protein and its promoter was active in the pharynx and intestine of C. elegans. These worms grow more slowly, lay fewer eggs, and phenotypes could result from reduced feeding. Other research has suggested a role for MEK-1 in stress responses, with a focus in the pharynx and/or intestine [52]. The KIN-18 (CUST_9047, −15.09) and MEK-1 (CUST_3675, −4.15) genes were significantly downregulated in schistosomes from water buffalo, which may be attributed to the morphological defection in worms from water buffalo compared to those from goats, as reduced nutritional intake can retard worm growth.
EGFR belongs to the ErbB family of receptor tyrosine kinases, which possess protein tyrosine kinase activity and are found only in metazoans [53]. The downstream signaling proteins initiate several signal transduction cascades, principally the MAPK, Akt, and mitogen-activated protein kinase 8 isoform beta 2 pathways, leading to DNA synthesis and cell proliferation. EGFR plays an important role in development and cell differentiation, and EGFR homologues have been identified in a broad range of vertebrate and invertebrate organisms [54]. Here, we found that EGFR was differentially underexpressed in schistosomes from water buffalo (CUST_3717, −4.52). EGFR can act as a target regulated by host factors, resulting in different worm compatibility within different hosts, thereby supporting the hypothesis that host EGF can regulate S. mansoni development [55].
Hsp70 proteins are ubiquitously expressed and exist in virtually all living organisms. The Hsp70 proteins are an important part of the cellular machinery for protein folding and help to protect cells from thermal or oxidative stress [56]. In addition to improving overall protein integrity, Hsp70 directly inhibits apoptosis by blocking the recruitment of procaspase-9 to the Apaf-1/dATP/cytochrome c apoptosome complex [57]. Given that water buffalo is the less suitable host, the Hsp70 gene of schistosomes from water buffalo was down-regulated (CUST_5301 FC,-2.24), therefore Hsp70 is predicted to promote parasitic apoptosis in vivo [58].
Dsh is a family of proteins involved in canonical and non-canonical integration 1/wingless (Wnt) signaling pathways. Dsh is a cytoplasmic phosphoprotein that acts directly downstream of frizzled receptors [59] and plays important roles in both the embryo and the adult, ranging from cellular differentiation and cell polarity to social behavior [60]. However, the role of Dsh downregulation in schistosomes from water buffalo (CUST_5485, −4.48) has not yet been investigated.
Prostatic spermine-binding protein (SBP) binds to forkhead protein A1 (FoxA1) and FoxA1 is critical for the androgenic regulation of prostate-specific promoters. Androgen plays a key role in the normal prostate development and physiology [61]. Female worms require stimulation from male worms to achieve and maintain a mature reproductive state. The SBP gene was significantly underexpressed in schistosomes from water buffalo (CUST_12936,−7.27), which was predicted to have a role in reproduction processes by influencing interactions among male and female worms, resulting in parasitic retardation and less egg deposition in the liver [28].
Endocytosis/Amino acid/Carbohydrate metabolism/Glycan biosynthesis, and metabolism
Endocytosis is a process by which cells absorb biological macromolecules, including hormones, growth factors, lymphokines, and some nutrients, by engulfing them and is used by all cells of the body because most important substances are large polar molecules that cannot pass through the hydrophobic plasma or cell membrane [62]. Normal parasitic growth must be accompanied by basic metabolism and since schistosome survival and development in water buffalo was hindered, genes associated with endocytosis of nutrients, amino acid metabolism, carbohydrate metabolism, and glycan biosynthesis/metabolism-associated genes were accordingly downregulated.
A comparison of schistosomes from the less susceptible host (water buffalo) with those from the more susceptible host (goats) at the phenotype and gene expression levels. Except the length and width difference, SEM and TEM showed that the ultrastructure of worms from different hosts were very different. A total of 485 differentially expressed genes were identified and their gene expression patterns in schistosomes from water buffalo and goat hosts were identified here. Our results revealed that, compared with schistosomes from goats, genes involved in lipid metabolism, genetic information processing, folding, sorting and degradation were upregulated in schistosomes from water buffalo, whereas other genes associated with signal transduction, endocrine function, development, reproduction, endocytosis, amino acid/carbohydrate metabolism, immune function, etc. were downregulated. These events were deduced to be key differences for the survival and development of schistosomes in different compatible natural host environments.
The microarray analysis of gene expression differences in schistosomes derived from water buffalo and goat provided a useful platform to discover the differences in host compatibilities, and furthered our understanding of the interplay between natural hosts and parasites, and identified several schistosome target genes associated with susceptibility for the screening of vaccine candidates.
Supporting Information
The common up-regulated genes in schistosomes from water buffalo compared with those from yellow cattle and goat.
(DOC)
The common down-regulated genes in schistosomes from water buffalo compared with those from yellow cattle and goat.
(DOC)
Acknowledgments
We thank Hua Yu and Liang He (Animal Schistosomiasis Control Station, Agriculture Department of Jiangxi Province) for help with the animal experiment. We acknowledge Yanli Zhang for help in animal infection and parasites collection, and Naiying Yang(Shanghai Normal University) for technological assistance in ultrastructural observation.
Funding Statement
This work was supported by research grants from the National Science & Technology Major Project (No. 2012ZX10004220-008, No. 2008ZX10004-011), the National Basic Research Program (No. 2007CB513108), Basic Foundation for Scientific Research of State-level Public Welfare Institutes of China (No. 2012JB17), and Special Fund for Agro-scientific Research in the Public Interest (No. 200903036). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. He YX, Salafsky B, Ramaswamy K (2001) Host-parasite relationships of schistosoma japonicum in mammalian hosts. Trends Parasitol 17: 320–324. [DOI] [PubMed] [Google Scholar]
- 2.TDR: TDR Strategic Direction: Schistosomiasis. (2002) World Health Organization.
- 3. McGarvey ST, Zhou XN, Willingham AL 3rd, Feng Z, Olveda R (1999) The epidemiology and host-parasite relationships of Schistosoma japonicum in definitive hosts. Parasitol Today 15: 214–215. [DOI] [PubMed] [Google Scholar]
- 4. Dai ZJ, Yan JP (1992) Study on the epidemiology of schistosomiasis japonica of mountainous area animals in Sichuan Province. Acta Veterinaria et Zootechnica Sinica 23: 87–91. [Google Scholar]
- 5. Wu SW, Liu ZD, Pu KM, Hu GH, Zhou SJ, et al. (1992) Role of human and domestic animal reservoirs of schistosomiasis japonica in Dongting and Boyang Lake regions. Chin J Parasitol Parasitic Dis 10: 194–197. [PubMed] [Google Scholar]
- 6. Zheng J, Zheng QS, Wang XF, Zheng HH (1997) Influence of livestock husbandry on schistosomiasis transmission in mountainous regions of Yunnan Province. Southeast Asian J Trop Med Pub Health 28: 291–295. [PubMed] [Google Scholar]
- 7. Zheng J, Guo JG (2000) The position of animal hosts in schistosomiasis transmission in China. Chin J Zoonoses 16: 87–88. [Google Scholar]
- 8. Shen W (1992) Review and suggestion of control of animal schistosomiasis in China. Chin J Schisto Control 4: 82–84. [Google Scholar]
- 9. Liu SX, He YK, Song GC, Luo XS, Xu YX, et al. (1997) Anti-fecundity immunity to Schistosoma japonicum induced in Chinese water buffaloes (Bos buffelus) after vaccination with recombinant 26 kDa glutathione-S-transferase (reSjc26GST). Vet Parasitol 69: 39–47. [DOI] [PubMed] [Google Scholar]
- 10. He YX, Yang HZ, Mao SB (1960) Host specificity of schistosomiasis japonica I: The developmental rates, distribution and survival in mammals. Chin Med J 46: 470–475. [Google Scholar]
- 11. Fitzpatrick JM, Johansen MV, Johnston DA, Dunne DW, Hoffmann KF (2004) Gender-associated gene expression in two related strains of Schistosoma japonicum . Mol Biochem Parasitol 136: 191–209. [DOI] [PubMed] [Google Scholar]
- 12. Dillon GP, Feltwell T, Skelton JP, Ashton PD, Coulson PS, et al. (2006) Microarray analysis identifies genes preferentially expressed in the lung schistosomulum of Schistosoma mansoni . Int J Parasitol 36: 1–8. [DOI] [PubMed] [Google Scholar]
- 13. Vermeire JJ, Taft AS, Hoffmann KF, Fitzpatrick JM, Yoshino TP (2006) Schistosoma mansoni: DNA microarray gene expression profiling during the miracidium-to-mother sporocyst transformation. Mol Biochem Parasitol 147: 39–47. [DOI] [PubMed] [Google Scholar]
- 14. Chai M, McManus DP, McInnes R, Moertel L, Tran M, et al. (2006) Transcriptome profiling of lung schistosomula, in vitro cultured schistosomula and adult Schistosoma japonicum . Cell Mol Life Sci 63: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moertel L, McManus DP, Piva TJ, Young L, McInnes RL, et al. (2006) Oligonucleotide microarray analysis of strain- and gender-associated gene expression in the human blood fluke, Schistosoma japonicum. Mol Cell Probes 20: 280–289. [DOI] [PubMed] [Google Scholar]
- 16. Peng JB, Han HX, Gobert GN, Hong Y, Jiang WB, et al. (2011) Differential gene expression in Schistosoma japonicum schistosomula from Wistar rats and BALB/c mice. Parasites & Vectors 4: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang W, Hong Y, Peng J, Fu Z, Feng X, et al. (2010) Study on differences in the pathology, T cell subsets and gene expression in susceptible and non-susceptible hosts infected with Schistosoma japonicum . PLoS ONE 5: e13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong Y, Peng J, Jiang W, Fu Z, Liu J, et al. (2011) Proteomic analysis of schistosoma japonicum schistosomulum proteins that are differentially expressed among hosts differing in their susceptibility to the infection. Mol Cell Proteomics 10: M110.006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Da'Dara KA, Li YS, Xiong T, Zou J, Williams GA, et al. (2008) DNA-based vaccines protect against zoonotic schistosomiasis in water buffalo. Vaccine 26: 3617–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, et al. (2006) Performance comparison of one-color and two-color platforms within the Microarray Quality Control (MAQC) project. Nat Biotechnol 24: 1140–1150. [DOI] [PubMed] [Google Scholar]
- 21. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Storey JD, Tibshirani R (2003) Statistical significance for genome-wide studies. Proc Natl Acad Sci U S A 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saldanha AJ (2004) Java Treeview—extensible visualization of microarray data, Bioinformatics. 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- 24. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 25. Lee HK, Braynen W, Keshav K, Pavlidis P (2005) ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics 6: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)). Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 27. Morey JS, Ryan JC, Van Dolah FM (2006) Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online 8: 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang JM, Yuan CX, Feng XG, Fu ZQ, Shi YJ, et al. (2012) Comparative study on Schistosoma japonicum infection in different permissive animal hosts. Chinese Journal of Zoonoses 28: 1207–1211. [Google Scholar]
- 29. Yang JM, Fu ZQ, Feng XQ, Shi YJ, Yuan CX, et al. (2012) Comparison of worm development and host immune responses in natural hosts of schistosoma japonicum, yellow cattle and water buffalo. BMC Veterinary Research 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang JM, Feng XG, Fu ZQ, Yuan CX, Hong Y, et al. (2012) Ultrastructural observation and gene expression profiling of Schistosoma japonicum derived from two natural reservoir hosts, water buffalo and yellow cattle. PLoS ONE 7: e47660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsuboi K, Sugimoto Y, Ichikawa A (2002) Prostanoid receptor subtypes. Prostaglandins Other Lipid Mediat 68–69: 535–56. [DOI] [PubMed] [Google Scholar]
- 32. Kaji H, Kamiie J, Kawakami H, Kido K, Yamauchi Y, et al. (2007) Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol Cell Proteomics 6: 2100–2109. [DOI] [PubMed] [Google Scholar]
- 33. Zhou W, Ye XL, Sun ZJ, Ji XD, Chen HX, et al. (2009) Overexpression of degenerative spermatocyte homolog 1 up-regulates the expression of cyclin D1 and enhances metastatic efficiency in esophageal carcinoma Eca109 cells. Mol Carcinog 48: 886–94. [DOI] [PubMed] [Google Scholar]
- 34. Freitas TC, Jung E, Pearce EJ (2007) TGF-b signaling controls embryo development in the parasitic flatworm Schistosoma mansoni . PLoS Pathog 3: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osman A, Niles EG, Verjovski-Almeida S, LoVerde PT (2006) Schistosoma mansoni TGF-β receptor II: Role in host ligand-induced regulation of a schistosome target gene. PLoS Pathog 2: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cadena DL, Kurten RC, Gill GN (1997) The product of the MLD gene is a member of the membrane fatty acid desaturase family: overexpression of MLD inhibits EGF receptor biosynthesis. Biochemistry 36: 6960–6967. [DOI] [PubMed] [Google Scholar]
- 37. Liu F, Lu J, Hu W, Wang SY, Cui SJ, et al. (2006) New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum . PLoS Pathog 2: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krejcova R, Horska K (1997) Nucleoside diphosphate kinases. Chem Listy 91: 466–476. [Google Scholar]
- 39. Hershko A, Ciechanover A, Varshavsky A (2000) Basic Medical Research Award. The ubiquitin system. Nat Med 6: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 40. Tanaka K (2009) The proteasome: Overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 85: 12–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mujtaba S, Zeng L, Zhou MM (2007) Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26: 5521–5527. [DOI] [PubMed] [Google Scholar]
- 42. Ramos VC, Vidal-Taboada J, Berqonon S, Eqeo A, Fisher EM, et al. (2002) Characterisation and expression analysis of the WDR9 gene, located in the Down critical region-2 of the human chromosome 21. Biochim Biophys Acta 1577: 377–383. [DOI] [PubMed] [Google Scholar]
- 43. Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M (2005) Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436: 871–875. [DOI] [PubMed] [Google Scholar]
- 44. Philipps DL, Wigglesworth K, Hartford SA, Sun FY, Pattabiraman S, et al. (2008) The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev Biol 317: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kandel ES, Hay N (1999) The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Exp Cell Res 253: 210–229. [DOI] [PubMed] [Google Scholar]
- 46. Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ (1999) Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol 1: 500–506. [DOI] [PubMed] [Google Scholar]
- 47. Scanga SE, Ruel L, Binari RC, Snow B, Stambolic V, et al. (2000) The conserved PI3'K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 19: 3971–3977. [DOI] [PubMed] [Google Scholar]
- 48. Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ (2001) Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem 276: 38349–38352. [DOI] [PubMed] [Google Scholar]
- 49. Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, et al. (2001) Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev 15: 2203–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cha DS, Datla US, Hollis SE, Kimble J, Lee MH (2012) The Ras-ERK MAPK regulatory network controls dedifferentiation in Caenorhabditis elegans germline. Biochim Biophys Acta 1823: 1847–1855. [DOI] [PubMed] [Google Scholar]
- 51. Berman KS, Hutchison M, Avery L, Cobb MH (2001) Kin-18, a C. elegans protein kinase involved in feeding. Gene 279: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koga M, Zwaal R, Guan KL, Avery L, Ohshima Y (2000) A Caenorhabditis elegans MAP kinase kinase, MEK-1, is involved in stress responses. EMBO J 2000 19: 5148–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stein RA, Staros JV (2000) Evolutionary analysis of the ErbB receptor and ligand families. J Mol Evol 50: 397–412. [DOI] [PubMed] [Google Scholar]
- 54. Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1: 0010 DOI: 10.1038/msb4100014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vicogne J, Cailliau K, Tulasne D, Browaeys E, Yan YT, et al. (2004) Conservation of epidermal growth factor receptor function in the human parasitic helminth schistosoma mansoni . J Biol Chem 279: 37407–37414. [DOI] [PubMed] [Google Scholar]
- 56. Morano KA (2007) New tricks for an old dog: the evolving world of Hsp70. Ann N Y Acad Sci 1113: 1–14. [DOI] [PubMed] [Google Scholar]
- 57. Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, et al. (2000) Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol 2: 469–75. [DOI] [PubMed] [Google Scholar]
- 58. Peng J, Gobert GN, Hong Y, Jiang W, Han H, et al. (2011) Apoptosis governs the elimination of Schistosoma japonicum from the non-permissive host Microtus fortis . PLoS One 6: e21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Penton A, Wodarz A, Nusse R (2002) A mutational analysis of dishevelled in Drosophila defines novel domains in the dishevelled protein as well as novel suppressing alleles of axin. Genetics 161: 747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wallingford JB, Habas R (2005) The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development 132: 4421–4436. [DOI] [PubMed] [Google Scholar]
- 61. Sun Q, Yu X, Degraff DJ, Matusik RJ (2009) Upstream stimulatory factor 2, a novel FoxA1-interacting protein, is involved in prostate-specific gene expression. Mol Endocrinol 23: 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Parton RG, Simons K (2007) The multiple faces of caveolae. Na Rev Mo Cell Biol 8: 185–194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The common up-regulated genes in schistosomes from water buffalo compared with those from yellow cattle and goat.
(DOC)
The common down-regulated genes in schistosomes from water buffalo compared with those from yellow cattle and goat.
(DOC)