Abstract
The quantification of single nucleotide polymorphism (SNP) allele frequencies in pooled DNA samples using real time PCR is a promising approach for large-scale diagnostics and genotyping. The limits of detection (LOD) and limits of quantification (LOQ) for mutant SNP alleles are of particular importance for determination of the working range, which, in the case of allele-specific real time PCR, can be limited by the variance of calibration data from serially diluted mutant allele samples as well as by the variance of the 100% wild-type allele samples (blank values). In this study, 3σ and 10σ criteria were applied for the calculation of LOD and LOQ values. Alternatively, LOQ was derived from a 20% threshold for the relative standard deviation (%RSD) of measurements by fitting a curve for the relationship between %RSD and copy numbers of the mutant alleles. We found that detection and quantification of mutant alleles were exclusively limited by the variance of calibration data since the estimated LODcalibration (696 in 30 000 000 copies, 0.0023%), LOQ20%RSD (1470, 0.0049%) and LOQcalibration (2319, 0.0077) values were significantly higher than the LODblank (130, 0.0004%) and LOQblank (265, 0.0009%) values derived from measurements of wild-type allele samples. No significant matrix effects of the genomic background DNA on the estimation of LOD and LOQ were found. Furthermore, the impact of large genome sizes and the general application of the procedure for the estimation of LOD and LOQ in quantitative real time PCR diagnostics are discussed.
INTRODUCTION
Single nucleotide polymorphisms (SNP) provide a powerful tool for genetic marker analyses due to their abundance and high potential for automation. However, SNP genotyping surveys in a large number of individuals are still costly and time consuming. Therefore, a promising approach is the application of current high throughput genotyping platforms for pooled DNA samples. DNA pooling reduces the genotyping effort, but the methods used have to provide precise estimates of allele frequencies in a wide range of wild-type/mutant allele ratios in DNA pools. Several genotyping methods were found to be suitable for measuring SNP allele frequencies in DNA pools, including cleaved amplified polymorphic DNA (1–3), pyrosequencing (1,2,4), primer extension chemistry (2,5–9), single strand conformation polymorphism (10) and denaturing high performance liquid chromatography (3,9,11). Quantitative real time PCR assays especially meet all the requirements for highly sensitive and accurate estimation of very low SNP allele frequencies in DNA pools, due to enabling the quantification of gene copy number over a wide range of linearity (1,2,12). The discrimination of wild-type versus mutant sequences can be improved in fluorogenic 5′→3′ exonuclease PCR assays by using allele-specific primers with artificially mismatched or single locked nucleic acid bases in the 3′-terminal regions (13–15). However, prior to the estimation of very low SNP frequencies, the assay-specific limits of quantification (LOQ) and limits of detection (LOD) should be determined to utilize the full working range of quantification and to avoid the occurrence of false positive results. The LOQ can be defined as the lowest ratio of wild-type to mutant alleles above which quantitative results may be obtained, whereas the LOD is the lowest ratio of wild-type to mutant alleles that can be determined to be different from the wild-type allele with a specific degree of confidence. LOD and LOQ values of an analytical method can be derived either from the standard deviations of calibration data and/or from the standard deviations of blank measurements. This is the first report in which LOD and LOQ values for a molecular SNP quantification assay were exactly determined by exploiting statistical parameters. Besides SNP allele frequency estimation, the procedure can be applied generally for validation of the lower working range limits in quantitative real time PCR diagnostics.
MATERIALS AND METHODS
Plasmid construction
Primers Pv_f (5′-ATTATCAACGGCGAATCCACCC-3′) and Pv_r (5′-ACACCCCGCATATTGATTTAGCAT-3′) were used to amplify a 528 bp fragment of the cytochrome b gene of single spore Plasmopara viticola isolates, which were sensitive (wild-type) and resistant (mutant) to QoI fungicides. The sequence spans a single point mutation, which leads to a change in the amino acid at position 143 from glycine to alanine, conferring resistance to QoI respiration inhibitors. Cloning of PCR products was performed using TA cloning of the pCR®4-TOPO® vector (Invitrogen, Karlsruhe, Germany). The ‘wild-type’ and ‘mutant’ inserts were confirmed by DNA sequencing on an ABI Prism® 377 platform (Applied Biosystems, Foster City, USA) using the DYEnamic ET Terminator Cycle Sequencing Kit™ (Amersham Pharmacia Biotech, Uppsala, Sweden).
Plasmid DNA extraction, restriction and quantification
Plasmid DNA was extracted using the High Pure™ Plasmid Isolation Kit (Boehringer, Mannheim, Germany) and linearized with ScaI (MBI Fermentas, St Leon-Rot, Germany) according to the manufacturer’s instructions. The concentrations of plasmid DNA samples were determined by fluorescence using PicoGreen dsDNA Quantification Reagent® (Molecular Probes, Eugene, OR) according to the manufacturer’s protocol on a Storm™ fluorescent scanner (Molecular Dynamics, Sunnyvale, CA). All samples were measured in duplicate. If duplicates differed by >5%, the sample was remeasured. Copy numbers of the cloned cytochrome b sequences were derived from the molecular weights of the cloning vector and insert.
Genomic DNA extraction and quantification
Total genomic DNA from infected grapevine leaf segments was isolated using the NucleoSpin® Plant DNA Isolation Kit (Macherey Nagel) according to manufacturer’s protocol. For complete removal of PCR inhibitors, the genomic DNA was treated three times with wash buffer CW. The concentration of genomic DNA was determined by UV spectrophotometry.
Construction of DNA pools
Plasmid DNA stocks were diluted with 10 mM Tris buffer to working concentrations of 50 000 copies/µl. DNA pools with a total number of 30 000 000 copies and defined ‘mutant’ allele concentrations of 10.0%, 1.0%, 0.1%, 0.01% and 0.001% were generated by serial dilution of ‘mutant’ plasmid DNA in 1:10 ratios with ‘wild-type’ plasmid DNA. Pooled DNA samples were allowed to equilibrate at room temperature for 1 h.
Oligonucleotide primers and probes
PCR primers and fluorescent 3′ minor groove binding DNA probes were purchased from Applied Biosystems. The real time PCR oligonucleotides were designed using Primer Express™ version 1.5 (Applied Biosystems). The probes of the mutation-specific and endogenous reference systems were labelled at their 5′-ends with the fluorescent dyes FAM (6-carboxyfluorescein) and VIC, respectively.
The 3′ terminal sequence of forward primer Pv-M-f was selected for specific amplification of the ‘mutant’ allele. Allelic discrimination was enhanced by introducing an amplification refractory mutation system with an artificial mismatch (T:T) in the 3′ subterminal region of Pv-M-f (16). Sequences and specifications of real time PCR primers and probes used are compiled in Table 1.
Table 1. Real time PCR primers and probes.
| PCR system | Name | Sequence (5′→3′) | Amplicon (bp) |
|---|---|---|---|
| Reference system |
Pv-C-f |
ATCGAATTAACCAACCGTTATTTACATC |
75 bp |
| |
Pv-C-r |
CCCCGCATATTGATTTAGCATTTA |
|
| |
Pv-C-p |
VIC-CGCATAATATGTTCAACACT-MGB |
|
| Mutant-specific system |
Pv-M-f |
CGAGAATAAATTTGTAATAACTGTTGCTG |
129 bp |
| |
Pv-M-r |
AGAGAAGCTTTATGGTGTTCAGGG |
|
| Pv-M-p | FAM-ATTTGTCCCCAAGGCA-MGB |
VIC and FAM, fluorescent dyes; MGB, minor groove binder; f, forward primer; r, reverse primer; p, probe; bp, base pair; the introduced artificial mismatch base in the 3′-terminal region of primer Pv-M-f for allele discrimination is underlined.
Real time PCR assays
The quantitative PCR reactions were performed in an ABI Prism™ 7700 sequence detection system using the TaqMan® PCR Master Mix (Applied Biosystems). Reactions contained 900 nmol primers, 200 nmol probes, 400 µM each dATP, dCTP and dGTP, 800 µM dUTP, 1 U Amplitaq Gold DNA polymerase, 0.2 U AmpErase uracil N-glycosylase (UNG) and 1× TaqMan buffer in a total volume of 25 µl. After a decontamination step at 50°C, a two step protocol was followed for 50 cycles: 95°C for 15 s and 61°C for 1 min.
Data analyses
Relative standard deviations and confidence intervals. The pooled DNA samples were measured 5-fold (m) at each concentration level of the mutant allele on one plate in order to circumvent plate-to-plate variations. For the estimation of copy number, calibration curves were produced by plotting the threshold cycle values (Ct values) versus the logarithm of the copy number. The relative standard deviations %RSD and the 95% relative confidence intervals (95%CI) were calculated by:
%RSD = (σ/![]() ) × 100
) × 100
95%CI = (RSD × tdf;α)/√m
where ![]() is the mean value, σ is the standard deviation, m is the number of replicas at each concentration level, tdf;α is the student factor at α = 0.05 and df = m – 1 degrees of freedom.
is the mean value, σ is the standard deviation, m is the number of replicas at each concentration level, tdf;α is the student factor at α = 0.05 and df = m – 1 degrees of freedom.
PCR efficiencies (E) were calculated by:
E = 10–1/b – 1
where b is the slope of the linear regression equation.
Limits of detection (LOD) and quantification (LOQ). The LOQ can be approximately estimated from the concentration of mutant alleles at which an acceptable degree of performance, in terms of %RSD, is obtained. For determination of the LOQRSD for analytical chemistry, a commonly used threshold of 20% was set.
Assuming a normal distribution of measured Ct values, LODcalibration and LOQcalibration were calculated from the residual standard deviation of the regression data according to the criteria 3 × Sxy/b and 10 × Sxy/b (17):
LODcalibration = 3 × (Sxy/b)
LOQcalibration = 10 × (Sxy/b)
where Sxy is the residual standard deviation and b is the slope of the linear regression equation.
‘Wild-type allele’ samples (blanks) were measured 5-fold on one plate and LODblank and LOQblank were calculated according to the 3σ and 10σ criteria of the IUPAC recommendations for analytical nomenclature (18):
LODblank = ![]() wild-type + 3σwild-type
wild-type + 3σwild-type
LOQblank = ![]() wild-type + 10σwild-type
wild-type + 10σwild-type
where ![]() wild-type is the mean Ct value and σwild-type is the standard deviation of the wild-type allele Ct measurements.
wild-type is the mean Ct value and σwild-type is the standard deviation of the wild-type allele Ct measurements.
RESULTS
Establishment of real time PCR systems for SNP quantification
Primers and probes for the reference and mutant-specific real time PCR assays were designed to be used at the same hybridization temperature and MgCl2 concentration. Thus, it was possible to use the same reaction master mix and to run the PCR reactions on the same plate. Concentrations of forward and reverse primers were varied independently in steps of 50, 300 and 900 nM to evaluate the optimal reaction conditions for the primers. In both systems the steepest amplification curves were achieved for a fixed copy number of the target template with primer concentrations of 900 nM. The specificity was confirmed by agarose gel electrophoresis and direct sequencing of amplified PCR products (data not shown). The annealing temperature in the mutant-specific real time PCR was increased in 1°C increments to achieve unambiguous discrimination of mutant and wild-type SNP alleles with allele-specific ARMS primers. An annealing temperature of 61°C resulted in the maximum ΔCt values for Ct mutant and Ct wild-type.
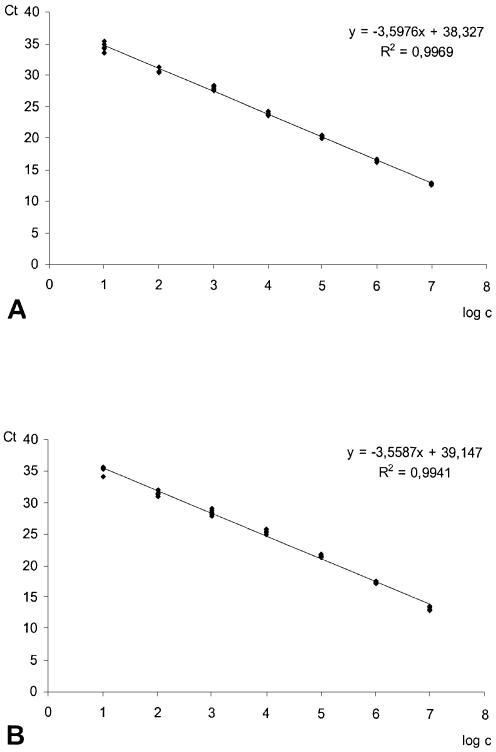
To ensure correct normalization of copy numbers of the mutant allele with the endogenous reference sequence in real time PCR the amplification efficiencies should be similar. Both PCR systems were tested with plasmid DNA samples from the mutant allele, which were serially diluted in 1:10 ratios with 10 mM Tris. Almost identical PCR efficiencies of 0.90 and 0.91 for the reference and mutant-specific PCR assays, respectively, were derived from the slopes of the standard curves (Fig. 1).
Figure 1.
Reference (A) and mutant allele (B) PCR specific calibration curves for plasmid DNA samples, which were serially diluted in 1:10 ratios with 10 mM Tris.
LOD and LOQ values derived from the calibration curve
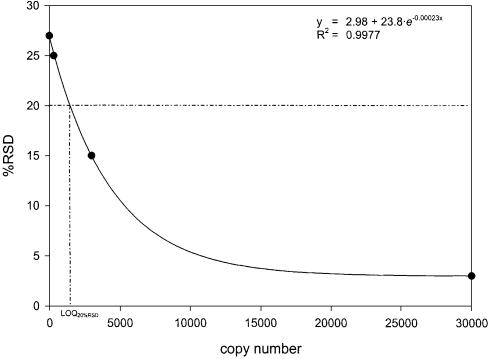
Plasmid DNA of the mutant allele was serially diluted in 1:10 ratios with plasmid DNA of the wild-type allele with 30 000 000 copies in total at each concentration level. The Ct values of the reference and mutant-specific PCR systems were measured in five replicates and the precision parameters %RSD and 95%CI for ΔCt values (mean of Ct reference – Ct mutant) were calculated (Table 2). The critical %RSD threshold value of 20% (equivalent to a 95%CI of 24.8%) was exceeded in the range of 3.000–300 copies of the mutant allele. Assuming an exponential decrease in %RSD, a curve was created using SigmaPlot version 6.0 for Windows (SPSS, Chicago, IL). Linear least squares regression for the equation y = y0 + a·e–bx resulted in the best fit (R2 = 0.9977) for the variables y0 = 2.98, a = 23.84 and b = 0.000229. Figure 2 displays the fitted curve and the derivation of LOQ20%RSD. Based upon the derived equation the absolute LOQ20%RSD can be estimated as 1470 copies (0.0490%) of the mutant allele (Fig. 3).
Table 2. Validation parameters of calibration curve and blank measurements.
| Copy number of the mutant allele | m | ΔCt | Estimated copy number | %RSD | 95%CI |
|---|---|---|---|---|---|
| 30 000 000 |
5 |
–1.48 |
27 359 424 |
16.3 |
20.2 |
| 3 000 000 |
5 |
1.37 |
3 817 996 |
17.0 |
21.2 |
| 300 000 |
5 |
5.32 |
296 521 |
12.0 |
14.9 |
| 30 000 |
5 |
8.91 |
27 164 |
3.3 |
4.1 |
| 3000 |
5 |
12.44 |
2 619 |
15.5 |
22.0 |
| 300 |
5 |
15.63 |
339 |
24.8 |
30.8 |
| 0 |
5 |
17.88 |
77 |
26.5 |
33.0 |
a = 24.40
b = –3.49
R2 = 0.997
Sxy = 808.3
m, number of replicates minus outliers; ΔCt, difference of threshold cycle means (Ct mutant – Ct reference); %RSD, relative standard deviation; 95%CI, relative confidence interval of measured copies at a 95% probability level; a, intercept of the regression; b, slope of regression; R2, correlation coefficient; Sxy, residual standard deviation.
Figure 2.
Linear least squares curve fit for relationship between residual standard deviation and copy number at low concentration levels of the mutant allele. The derivation of LOQ20%RSD is illustrated by dotted lines.
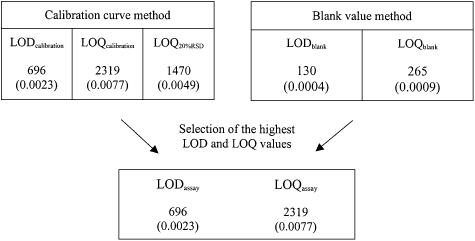
Figure 3.
Estimated detection and quantification limits; The absolute LOD and LOQ values are given in copy numbers. The relative LOD and LOQ values in brackets are given as percentages.
To test the linearity of SNP allele quantification, ΔCt values were used for linear regression analysis. The regression coefficient of the calibration curve presented is R2 = 0.997 (slope b = –3.49; intercept a = 24.40), demonstrating the high correlation of the original number of mutant allele copies and the ΔCt values obtained by amplification of mutant alleles over five magnitudes of concentration (Table 2). The absolute LODcalibration and LOQcalibration were calculated from the residual standard deviation of the calibration curve Sxy = 803.3, resulting in 696 and 2319 copies of the mutant allele, respectively. These copy numbers represent relative amounts of 0.0023 and 0.0077% mutant alleles in the pooled DNA samples (Fig. 3).
LOD and LOQ values derived from the blank values
Although allele-specific ARMS primers and real time PCR conditions (particularly the annealing temperature) were optimized for discrimination of wild-type and mutant SNP alleles, there was still significant amplification of the wild-type templates. For the determination of LOD and LOQ values these blank signals also have to be considered. Assuming that the Ct values belong to a normal distribution, a ‘practical’ LODblank can be set at a concentration that gives a signal equal to three times the standard deviation of the wild-type sample measures. This 3σ criterion represents a 99.73% probability that the value is above the background level. The LOQ was set at 10 times the standard deviation of the wild-type sample measurements. Ct values for the reference and mutant-specific PCR systems were measured for wild-type allele samples in five replicates containing a total of 30 000 000 plasmid copies. The mean ΔCt value for the wild-type allele was 17.88 (Table 2). Based on the standard deviation of estimated copy numbers of the wild-type allele (σwild-type = 19.24), the absolute LODblank and LOQblank were estimated as 130 copies (0.0004%) and 265 copies (0.0009%) of the mutant allele, respectively (Fig. 3).
Assay-specific LOD and LOQ
The relevant LOD and LOQ values limiting the detection and quantification of the SNP real time PCR assay (LODassay and LOQassay) were derived from LODcalibration (696 copies, 0.0023%) and LOQcalibration (2319 copies, 0.0077%) because they significantly exceed the values of LOD20%RSD, LODblank and LOQblank (Fig. 3).
Influence of sample background matrix DNA on LOD and LOQ
Genomic DNA was extracted from grapevine leaves which were fully infected with a strobilurin-sensitive (wild-type allele) P.viticola isolate. PCR inhibitors were removed completely by thorough washing during extraction. The Ct values of the reference PCR system were used to adjust the unknown copy number of P.viticola cytochrome b wild-type alleles in the total genomic DNA to 30 000 000 wild-type allele copies in plasmid DNA. Reference and mutant PCR-specific Ct values of genomic wild-type allele samples were measured in five replicates. The mean ΔCt value for the wild-type allele was 17.28. Based on the standard deviation of estimated copy numbers of the wild-type allele (σwild-type = 35.41), LODblank and LOQblank of 186 (0.0006%) and 434 copies (0.0014%) of the mutant allele were estimated. These values are slightly higher than LODblank (130 copies, 0.0004%) and LOQblank (265 copies, 0.0009%) values derived from plasmid wild-type samples without genomic DNA. However, the differences can rather be explained by the large deviations of Ct values (σwild-type) in this range of measurements than by background matrix effects. For the estimations of LODcalibration and LOQcalibration, plasmid DNA containing the mutant allele was serially diluted with genomic background DNA. Ct values for the reference and mutant-specific PCR systems were remeasured in five replicates. The regression coefficient of the calibration curve was R2 = 0.995 (slope b = –3.40; intercept a = 23.64). The estimated LODcalibration, LOQcalibration and LOQ20%RSD values of 526 (0.0018%), 1753 (0.0058%) and 1946 copies (0.0065%), respectively, are similar to LODcalibration (696 copies, 0.0023%), LOQcalibration (2319 copies, 0.0077%) and LOQ20%RSD (1470 copies, 0.0049%) values derived from plasmid wild-type samples without genomic DNA. Thus it can be assumed that the estimations of LOD and LOQ values were only slightly influenced by inhibitory background matrix effects provided that PCR inhibitors were completely removed and the input of total genomic DNA to the PCR does not exceed 200 ng (data not shown).
DISCUSSION
SNP quantification in DNA pools by real time PCR is a powerful tool for cost-efficient large-scale diagnostics and genotyping. This approach is particularly suitable for the estimation of very low SNP allele frequencies in large populations of genotypes. To use the full capacity of the quantification system, the lower limits of the working range have to be determined exactly. In this study the following approach was used to determine the assay-specific LOD and LOQ values for SNP quantification using real time PCR. First, two real time PCR assays were established; one for the estimation of total copy numbers of the target sequence and one for the estimation of copy numbers of the mutant allele. The mutant allele-specific PCR assay was optimized regarding the discrimination of mutant and wild-type alleles (the difference between the Ct values of mutant and wild-type allele samples should be as large as possible). According to guidelines for validation of analytical methods, LOD and LOQ were determined not only from the variances of interpolated regression line data (LODRSD and LOD/LOQcalibration) but also from the variance of the blank measurements (LOD/LOQblank). Both approaches were applied because the detection and quantification of SNP allele frequencies can be limited by the uncertainty of the calibration curve as well as by the signals of wild-type allele samples (blanks). In order to achieve the highest statistical certainty, the assay-specific LODassay and LOQassay values were derived from the highest LODcalibration/LODRSD/LODblank and LOQcalibration/LODblank values. Detection and quantification of the mutant alleles were limited exclusively by the uncertainty of the calibration curve since the blank value approach yields considerably lower LOD and LOQ than the calibration curve approach. The low LODblank and LOQblank values in SNP quantification were achieved by improving the specificity of the mutant-specific PCR assay. Allele-specific SNP discrimination was enhanced by using minor groove binding probes, which allow the optimization of the assay in a wide range of annealing temperatures without loss of hybridisation sensitivity (19). The power of the oligonucleotide for allelic discrimination was also increased by introducing an artificial mismatch at the 3′-subterminal base. Compared to primers with a G:G mismatch only at the 3′ residue, primers with an additional T:T mismatch resulted in considerably lower LODblank and LOQblank values (data not shown). In addition to optimization of the allele discrimination, the gap between the blank and the calibration curve should be bridged. The distance between Ct values of calibration samples at the lowest concentration level and Ct values of wild-type samples should be as small as possible since the actual LOD and LOQ are located near or in this range of mutant allele concentrations with the utmost probability.
The value of residual standard deviation used for the calculation of LODcalibration and LOQcalibration has widespread acceptance as a measure of the error associated with calibration. However, the residual standard deviation is derived from the least squares of all the calibration data and fails to account for the variability of measurements at a specific level of concentration. This is of particular importance for accurate estimations of LOD and LOQ, since decreasing amounts of the mutant allele at low concentration levels cause the variance of calibration data to rise. Hence, in addition to LOQcalibration, the relative standard deviations of measures at low concentration levels were used for more precise estimation of the calibration curve derived LOQ. Using the 20%RSD threshold, LODRSD was estimated to be in the range 3.000–300 copies of the mutant allele. This result provides only a rough estimate for LODRSD. The relationship between %RSD values and copy numbers of the mutant allele at low concentration levels should follow the Horwitz curve (%RSD = 2(1 – 0.5 log(concentration))). It has been shown to be more or less independent of analyte, matrix and method by the analysis of many method validation studies (20). The simple, empirically derived equation is explained by the decreasing influence of the analyte on the measured value and the increasing effects of blanks or the matrix near zero concentration (21). In this study the %RSD values always exceeded the values calculated by the Horwitz equation for low concentrations of the mutant allele (Table 3). The SNP quantification assay does not appear to completely fulfil the criteria of the equation, therefore, instead of the generalized Horwitz function, we used a fitted model based on the individual analytical uncertainty of the assay. The calculated LOD20%RSD of 1470 copies (0.0049%) was below LODcalibration. Regarding the alternatively derived LODRSD values of 546 (0.0018%) and 5332 (0.0178%) copies for thresholds of 10 and 30%, respectively, the 20% RSD limit seems to be suitable for the estimation of LOD in real time PCR systems.
Table 3. Calculated and Horwitz %RSD values for different concentrations of the mutant allele.
| Concentration | 10–7 | 10–6 | 10–5 | 10–4 | 10–3 |
|---|---|---|---|---|---|
| %RSDHorwitz |
23 |
16 |
11 |
8 |
5.6 |
| %RSDcalculated | 24.8 | 15.5 | 3.3 |
All LOD and LOQ values in this study were derived from Ct values of plasmid copies containing the target sequence. In practice, the LOD values for SNP allele frequency estimation also depends on the genome size of the investigated species. The DNA content of the haploid complement of an organism is defined as the 1C value. For example, up to 29 026 copies of the haploid human genome are present in PCR, given the 1C value of 3.476 pg and a typical 100 ng genomic DNA sample (too much genomic DNA can attenuate or even inhibit PCR). Therefore, a single copy of the haploid genome is present at a level of 0.003445% (Table 4). Calculated LODcalibration and LODblank values below these genome specific thresholds are invalid since the target sequence simply cannot be reliably detected in samples of this size. As shown in Table 4, the genome size is not critical for the estimation of LOD and LOQ in small genomes. However, for species with large genomes such as maize (Zea mays), mouse (Mus musculus) and human the LODgenome can reach or even exceed the estimated LODblank and LODcalibration values.
Table 4. Genome specific limits of detection.
| Species | Genome size (×106 bp)a | Haploid C value (pg)b | Genome copies (per 100 ng) | LODgenome (%) |
|---|---|---|---|---|
|
Zea mays |
5000 |
5.112 |
19.560 |
0.005112 |
|
Mus musculus |
3454 |
3.532 |
28 571 |
0.003500 |
|
Homo sapiens |
3400 |
3.476 |
29 026 |
0.003445 |
|
Arabidopsis thaliana |
100 |
0.102 |
986 900 |
0.000101 |
|
Saccharomyces cerevisae |
12 |
0.012 |
8 178 313 |
0.000012 |
| Escherichia coli | 5 | 0.005 | 21 272 968 | 0.000005 |
aGenome sizes were taken from database of genome sizes DOGS (http://www.cbs.dtu.dk/databases/DOGS).
bMass in pg = number of base pairs/(0.9869 × 109).
The estimation of LOD and LOQ values as described in this study can be applied not only to SNP quantification in pooled DNA samples but also for many other real time quantitative PCR applications, like the quantification of genetically modified organsims (GMO) in food or feed. The LOD and LOQ for GMO quantification have to be derived only from the variance of calibration data because the blank measurements (GMO-free samples) should produce no signal at all. In several validation studies, the LOD and LOQ values for quantification of GMO were only roughly estimated by the variation of relative 95%CI, which should not exceed 30% for quantification and 100% for detection, respectively (22,23). The LOQ threshold of 30% of the 95%CI corresponds approximately to the LOD20%RSD used in this study. The application of a fitted curve for the relationship of relative standard deviations and low concentrations of the target sequence would enable specification of an exact number of copies for LOD and LOQ.
REFERENCES
- 1.Breen G., Harold,D., Ralston,S., Shaw,D. and St Clair,D. (2000) Determining SNP allele frequencies in DNA pools. Biotechniques, 2, 464–470. [DOI] [PubMed] [Google Scholar]
- 2.Shifman S., Pisante-Shalom,A., Yakir,B. and Darvasi,A. (2002) Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol. Cell. Probes, 16, 429–434. [DOI] [PubMed] [Google Scholar]
- 3.Bäumler S., Sierotzki,H., Hall,A., Gisi,U, Mohler,V., Felsenstein,F.G. and Schwarz,G. (2002) Evaluation of Erysiphe graminis f. sp. tritici field isolates for resistance to strobilurin fungicides with different SNP detection systems. Pest. Manag. Sci., 59, 310–314. [DOI] [PubMed] [Google Scholar]
- 4.Gruber J.D., Colligan,P.B. and Wolford,J.K. (2002) Estimation of single nucleotide polymorphism allele frequency in DNA pools by using pyrosequencing. Hum. Genet., 110, 395–401. [DOI] [PubMed] [Google Scholar]
- 5.Mohlke K.L., Erdos,M.R., Scott,L.J., Fingerlin,T.E., Jackson,A.U., Silander,K., Hollstein,P., Boehnke,M. and Collins,F.S. (2002) High-throughput screening for evidence of association by using mass spectrometry genotyping on DNA pools. Proc. Natl Acad. Sci. USA, 99, 16928–16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norton N., Williams,N.M., Williams,H.J., Spurlock,G., Kirov,G., Morris,D.W., Hoogendoorn,B., Owen,M.J. and O’Donovan,M.C. (2002) Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum. Genet., 110, 471–478. [DOI] [PubMed] [Google Scholar]
- 7.Le Hellard S., Ballereau,S.J., Visscher,P.M., Torrance,H.S., Pinson,J., Morris,S.W., Thomson,M.L., Semple,C.A.M., Muir,W.J., Blackwood,D.H.R. et al. (2002) SNP genotyping on pooled DNAs: comparison of genotyping technologies and a semi-automated method for data storage and analysis. Nucleic Acids Res., 30, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou G.H., Kamahori,M., Okano,K., Chuan,G., Harada,K. and Kambara,H. (2001) Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER). Nucleic Acids Res., 29, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoogendoorn B., Norton,N., Kirov,G., Williams,N., Hamshere,M.L., Spurlock,G., Austin,J., Stephens,M.K., Buckland,P.R., Owen,M.J. et al. (2000) Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Hum. Genet., 107, 488–493. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki T., Tahira,T., Suzuki,A., Higasa,K., Kukita,Y., Baba,S. and Hayashi,K. (2001) Precise estimation of allele frequencies of single-nucleotide polymorphisms by a quantitative SSCP analysis of pooled DNA. Am. J. Hum. Genet., 68, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolford J.K., Blunt,D., Ballecer,C. and Prochazka,M. (2000) High-throughput SNP detection by using DNA pooling and denaturing high performance liquid chromatography (DHPLC). Hum. Genet., 107, 483–487. [DOI] [PubMed] [Google Scholar]
- 12.Germer S., Holland,M.J. and Higuchi,R. (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res., 10, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaab W.E. and Skopek,T.R. (1999) A novel assay for allele discrimination that combines the fluorogenic 5′ nuclease polymerase chain reaction (TaqMan®) and mismatch amplification mutation assay. Mutat. Res., 430, 1–12. [DOI] [PubMed] [Google Scholar]
- 14.Latorra D., Campbell,K., Wolter,A. and Hurley,J.M. (2003) Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum. Mutat., 22, 79–85. [DOI] [PubMed] [Google Scholar]
- 15.Bates J.A. and Taylor,E.J.A. (2001) Scorpion ARMS primers for SNP real-time PCR detection and quantification. Mol. Plant Pathol., 2, 275–280. [DOI] [PubMed] [Google Scholar]
- 16.Newton C.R., Graham,A., Heptinstall,L.E., Powell,S.J., Summer,C., Kalsheker,N., Smith,J.C. and Markham,A.F. (1989) Analysis of any point mutation in DNA. The amplification retractory mutation system (ARMS). Nucleic Acids Res., 17, 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J.C. and Miller,J.N. (1993) Statistics for Analytical Chemistry, 3rd Edn. Ellis Horwood Ltd, Chichester, UK. [Google Scholar]
- 18.Inczédy J., Lengyel,T. and Ure,A.M. (1998) Compendium on Analytical Nomenclature. Definitive Rules 1997, 3rd Edn. Blackwell Science, New York, NY. [Google Scholar]
- 19.Kutyavin I.V., Afonina,I.A., Mills,A., Gorn,V.V., Lukhatanov,E.A., Belousov,E.S., Singer,M.J., Walburger,D.K., Lokhov,S.G., Gall,A.A. et al. (2000) 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res., 28, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz W. (1982) Evaluation of analytical methods for regulation of foods and drugs. Anal. Chem., 54, 67A–76A. [Google Scholar]
- 21.Albert R. and Horwitz,W. (1997) A heuristic derivation of the Horwitz curve. Anal. Chem., 69, 789–790. [Google Scholar]
- 22.Hübner P., Waiblinger,H.U., Pietsch,K. and Brodmann,P. (2001) Validation of PCR methods for the quantification of genetically modified plants in food. J. AOAC Int., 84, 1855–1864. [PubMed] [Google Scholar]
- 23.Block A. and Schwarz,G. (2003) Validation of different genomic and cloned DNA calibration standards for construct-specific quantification of LibertyLink in rapeseed by real-time PCR. Eur. Food Res. Technol., 216, 421–427. [Google Scholar]