Abstract
Recent studies show that Arf, a bona fide tumor suppressor, also plays an essential role during mouse eye development. Tgfβ is required for Arf promoter activation in developing mouse eyes, and its capacity to induce Arf depends on Smads 2/3 as well as p38 Mapk. Substantial delay between activation of these pathways and increased Arf transcription imply that changes in the binding of additional transcription factors help orchestrate changes in Arf expression. Focusing on proteins with putative DNA binding elements near the mouse Arf transcription start, we now show that Tgfβ induction of this gene correlated with decreased expression and DNA binding of C/ebpβ to the proximal Arf promoter. Ectopic expression of C/ebpβ in mouse embryo fibroblasts (MEFs) blocked Arf induction by Tgfβ. Although basal levels of Arf mRNA were increased by C/ebpβ loss in MEFs and in the developing eye, Tgfβ was still able to increase Arf, indicating that derepression was not the sole factor. Chromatin immunoprecipitation (ChIP) assay showed increased Sp1 binding to the Arf promotor at 24 and 48 hours after Tgfβ treatment, at which time points Arf expression was significantly induced by Tgfβ. Chemical inhibition of Sp1 and its knockdown by RNA interference blocked Arf induction by Tgfβ in MEFs. In summary, our results indicate that C/ebpβ and Sp1 are negative and positive Arf regulators that are influenced by Tgfβ.
Introduction
Arf, a bona fide mammalian tumor suppressor gene transcribed from the Cdkn2a locus, encodes p19Arf in an alternative reading frame when compared to p16Ink4a, the first gene found at this chromosomal locus [1]. Mouse p19Arf is primarily known to physically interact with and block Mdm2, thereby stabilizing p53 and contributing to cancer surveillance [2]. Genetically engineered mice that lack the first coding exon for Arf, but retaining the Ink4a coding sequence, develop spontaneous tumors from as early as two months of age [3]. Although Arf coding sequence can be deleted in mouse and human tumors, in a substantial number the gene is intact but silenced alone or together with INK4A [4], [5]. Therefore, understanding how Arf expression is controlled is relevant to understanding a fundamental mechanism that cancer cells utilize to evade its tumor suppressive activity.
A number of findings indicate that transcriptional control of Arf is the major determinant of p19Arf protein level and function. Throughout most of the developing mouse embryo, Arf expression is essentially silenced [6]. Indeed, our studies reveal that Arf expression is detectable only in the developing eye and internal umbilical vessels [7]. Global silencing of its expression is mediated by chromatin remodeling proteins such as Bmi1 since the expression of both Arf and Ink4a increase when Bmi1 is deleted in mouse models [8]. In this regard, a long non-coding RNA (ANRIL), transcribed anti-parallel to human ARF and INK4a (and the INK4b gene lying further 5′ of ARF/INK4a) [9] acts in cis to foster CBX7 binding to this region in cultured human PC3 cells [10]. Despite evidence for global repression of the Cdkn2a locus, it is also clear that transcription activators contribute to the selective induction or repression of the Arf promoter. Examples include E2Fs 1 and 3 [11], [12], [13], [14], Dmp1 [15], [16], AP1 [17], and Pokemon [18]. FoxO proteins are also implicated as Arf regulators and they appear to act by binding an element in the first Arf intron, far from the transcription start site [19]. It is important to note that many of these conclusions stem from highly tractable cell culture models, but the in vivo relevance is less clear in most cases.
Adding to the concept that Arf must have tissue-specific control is the fact that the gene plays an essential role in eye development [20]. Arf-deficient mice develop persistent hyperplastic primary vitreous (PHPV) that is evident at embryonic day (E) 13.5 and persists in the postnatal period [20]. In this setting, p19Arf blocks the expression of Pdgfrβ, a growth factor receptor that is essential for hyperplastic accumulation of cells in the primary vitreous in the absence of Arf [21]. Tgfβ2 is essential for Arf expression in the developing mouse [7]; and in cultured MEFs, Arf induction by Tgfβ depends on activation of TbrII, Smad 2/3, and p38 Mapk [22]. Interestingly, RNA polymerase II binding to the Arf promoter and increased Arf mRNA lag substantially behind activation of these pathways and the binding of Smad 2/3 to the Arf gene [22]. Moreover, Tgfβ2 has numerous effects during mouse embryo development whereas Arf expression is principally localized to the primary vitreous [7]. Both findings indicate that other proteins must cooperate with Smad 2/3 to control Arf. Taking advantage of mouse and cell culture-based models, we identify two such cooperating events: de-repression of Arf by C/ebpβ down-regulation and loss of promoter binding, and transcriptional activation by Sp1.
Materials and Methods
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center, Dallas, Texas. Methods such as the use of isoflurane for anesthetization of animals were used to minimize suffering during surgeries.
Mice, Cells and Reagents
Arf lacZ/+ [7] mice were maintained in a mixed C57BL/6 × 129/Sv genetic background. Tgfβ2+/− mice [23] and C/ebpβ +/− mice [24], also in a mixed C57BL/6 × 129/Sv genetic backgrounds, were purchased from Jackson Laboratories.
Primary MEFs from wild type (WT), Arf lacZ/lacZ, and C/ebpβ −/− mice were obtained and cultivated as previously described [6]. MSCV-based retrovirus vectors encoding mouse C/ebpβ [Liver Activating Protein (LAP) isoform] were produced in our laboratory using vectors from Addgene (Cambridge, MA). The following chemical agents were used in some analyses: HLM006474 (HLM), from EMD Millipore Chemicals Inc (Billerica, MA); and Mithramycin A, from Sigma (St. Louis, MO). Tgfβ1 (Tgfβ), obtained from R&D Systems, Inc (Minneapolis, MN), was added to cell culture medium at a dose of 5 ng/ml; an equivalent volume of vehicle (4 mM HCl) was added into the medium as a control.
Real Time RT PCR
Cell pellets were dissolved in 800 µl Trizol (Invitrogen); total RNA was extracted from Trizol solution after addition of chloroform, precipitated with isopropanol, and dissolved in water. Two µg total RNA was use to synthesize cDNA with Superscript III RT kits (Invitrogen) according to the manufacturer’s recommendations. Then, quantitative RT-PCR (qRT-PCR) was performed using Fast SYBR Green Master mix and a model 7900 HT Fast Cycler instrument (both from Applied Biosystems). The primers were as follows: Arf: 5′-TTCTTGGTGAAGTTCGTGCGATCC-3′ (forward) and 5′-CGTGAACGTTGCCCATCAT CATCA-3′ (reverse); C/ebpβ: 5′-GTTTCGGGACTTGATGCAAT-3′ (forward) and 5′- CCCCGCAGGAACATCTTTA-3′ (reverse); Sp1: 5′-TCATGGATCTGGTGGTGATGGG-3′ (forward) and 5′-GCTCTTCCCTCACTGTCTTTGC-3′ (reverse); Gapdh: 5′-TCAACAGCAACTCCCACTCTTCCA-3′ (forward) and 5′-ACCCTGTTGCTGTAGCCGTAT TCA-3′ (reverse). Results are pooled from three separate experiments.
Western Blotting and β-Gal Assay
Cells were collected, lysed, separated by SDS-PAGE and transferred to PVDF membrane with 50–100 µg total protein per sample. The membrane was incubated with primary antibody for two hours, washed trice in Tris-Buffered Saline Tween-20 (TBST) for 15 minutes each time; and then incubated with horseradish peroxidase (HRP)-labeled secondary antibody for one hour. After washing in TBST, the membrane was incubated with 2 ml ECL (GE Healthcare Life Sciences) for 5 minutes and visualized by exposure to film. β-galactosidase assays were performed in Arf lacZ/lacZ MEFs as previously described [7] using a commercial kit (Applied Biosystems; Foster City, CA). For western blotting, antibodies directed against the following proteins were utilized: C/ebpβ, and Hsc70 (Santa Cruz Biotechnology, Inc; Santa Cruz, CA); phospho-p38 Mapk, and phospho-Smad2 (Cell Signaling Technology; Danvers, MA); and p19Arf (Abcam Inc; Cambridge, MA). Experimental findings were confirmed in at least two independent experiments, with quantitative data from β-galactosidase assays pooled from all representative experiments.
Laser Capture Microdissection (LCM)
LCM was done as previously described [25]. Briefly, mouse embryos ware harvested at E13.5 for LCM. Embryo heads were immediately embedded in OCT freezing medium without fixation. Fourteen µm thick sections were cut on a CryoStar NX70 cryostat, which were mounted on PEN Membrane Metal Slides (Applied Biosystems) and stained with hematoxylin and eosin (H&E) (Molecular Machines & Industries AG; Glattbrugg, Switzerland). LCM was carried out using an Arcturus Veritas Microdissection System. Cells in the vitreous, lens, and retina were dissected from each eye and collected separately. Samples were pooled from at least 5 microdissected sections from the same embryo. Total RNA was extracted using an Arcturus PicoPure LCM RNA isolation kit (Applied Biosystems) and the expression of specific genes was analyzed with real time RT-PCR as described above.
ChIP Assay
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described [22]. Briefly, wild type MEFs (3×106/ChIP) were treated with Tgfβ (5 ng/ml) or vehicle for 1.5, 24 or 48 hours. Cells were cross-linked and sonicated, and then subjected to immunoprecipitation using antibodies against C/ebpβ (sc150, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), or Sp1 (sc59, Santa Cruz). Rabbit IgG (sc2027, Santa Cruz) was used as a negative control. Protein A/G sepharose beads (sc2003, Santa Cruz) were used to collect the antibody-chromatin complexes. The beads were washed sequentially with low salt, high salt, LiCl and TE buffers (Upstate ChIP Kit, Millipore) and eluted in 0.1 M NaHCO3, 1% SDS. Cross-linking was reversed by incubation at 67°C overnight, and the genomic DNA was extracted using Qiagen PCR Purification Kit. Quantitative analysis of the precipitated and input DNA was carried out using specific primer sets and Fast SYBR green master mix on a model 7900 HT Fast Cycler instrument (both from Applied Biosystems). The primer sets for proximal promoter regions of Arf were as follows: 5′-AGATGGGCGTGGAGCAAAGAT-3′ (forward) and 5′- ACTGTGACAAGCGAGGTGAGAA (reverse).
siRNA
We purchased siRNA against mouse SP1 (catalog # 74195; Life Technologies, Grand Island, NY). The siRNA was dissolved in 1× siRNA buffer (Dharmacon) and used for transfection (100 nM final concentration). Scrambled siRNA (siGENOME Non-Targeting siRNA #3, Dharmacon) was used as control. 24 hours after the initial transfection, the cells were treated with either Tgfβ or vehicle, and they were harvested 48 hours later for western blotting or RT-PCR.
Statistical Analysis
Quantitative data are presented as the mean±S.D. from three or more representative experiments. Statistical significance (p value <0.05) was calculated using Student’s t test.
Results
Recognizing the substantial delay between Smad binding to the Arf promoter and increased synthesis of Arf primary transcript [22], we considered potential roles for other transcription factors whose function might be influenced by Tgfβ. Among those, C/ebpβ was an attractive candidate because previous work had implicated it as an Arf repressor in primary epidermal keratinocytes [26], and putative consensus DNA binding elements are found within 500 bp 5′ to the Arf translation initiation codon (Figure 1A). Utilizing chromatin immunoprecipitation (ChIP), we demonstrated that C/ebpβ was bound to this region in cultured mouse embryo fibroblasts (MEFs) at passage 3 (YZ and SXS, unpublished data).
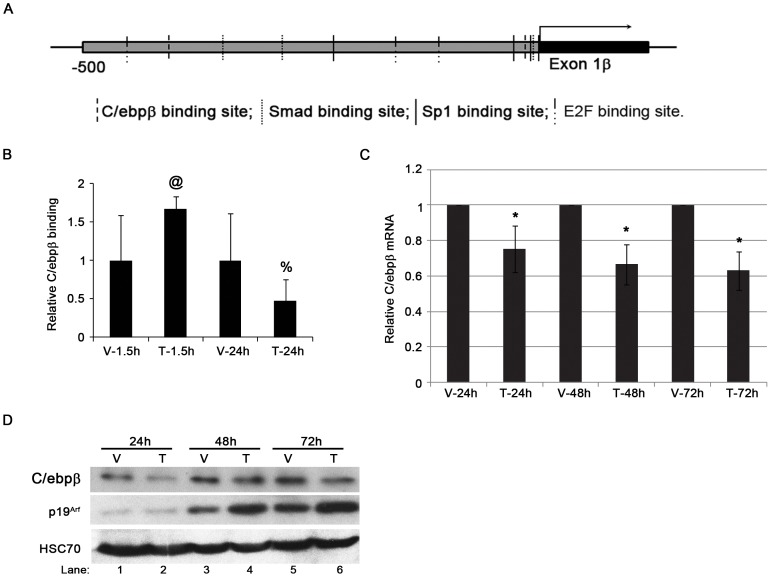
Figure 1. Inverse correlation of C/ebpβ and Arf expression during Tgfβ treatment.
(A). Schematic diagram showing potential C/ebpβ, Smad, Sp1 and E2F binding sites at the Arf promoter. (B). Tgfβ decreases C/ebpβ binding to the Arf locus in MEFs. Quantitative analysis of representative chromatin immunoprecipitation (ChIP) assays of using wild type MEFs exposed to vehicle (V) or Tgfβ (T) for 1.5 hours or 24 hours. ChIP assay was carried out using antibodies specific to C/ebpβ and IgG. Immunoprecipitated DNA and input DNA were amplified with primers for proximal regions genomic Arf promoter. p-values as follows: 0.1 (@) and 0.2 (%) for Tgfβ versus corresponding vehicle. (C). Quantitative analysis of real time, RT-PCR using total RNA isolated from WT MEFs shows the expression of C/ebpβ mRNA changes during Tgfβ treatment up to 72 hours. The data is plotted as the fold changes of target genes from cells treated with Tgfβ (T) (5 ng/ml) versus the same cells treated with vehicle (V) (4 mM HCl). The significant changes between Tgfβ treatment and vehicle treatment was marked as * (p<0.05). (D) Representative western blot of lysates from wild type MEFs treated with Tgfβ (T) and vehicle (V) at different time points showing the inverse correlation of C/ebpβ and Arf protein expression.
We next investigated whether Tgfβ influenced the binding of endogenous C/ebpβ to the Arf promoter. We previously established that Smad 2/3 binding to elements in the proximal Arf promoter (Figure 1A) is enhanced within 1.5 hours following the addition of Tgfβ2 to the culture medium, whereas RNA polymerase II (RPolII) binding is not increased until 24 hours, after which Arf mRNA increases [22]. Paralleling the delayed RPolII binding, C/ebpβ localization to a proximal promoter element in the Arf promoter was diminished at 24 hours following an initial increase at 1.5 hours (Figure 1B). Interestingly, Tgfβ stimulation diminished C/ebpβ mRNA and protein between 24 and 72 hours (Figures 1C and D). The effect on C/ebpβ protein expression was evident when it was ectopically expressed (Figures 2B, lane 3 versus 4), implying that the decreased repression was not simply due to decreased transcription of the native mRNA. Of note, the fact that p19Arf level did not strictly inversely correlate with C/ebpβ (Figure 1D, lane 3 versus 1) indicates that other factors, such as cell “culture shock” that has been described for cultured mouse fibroblasts [27], must play a role in expression of this tumor suppressor and these other factors maybe be independent of Tgfβ signaling (see more below).
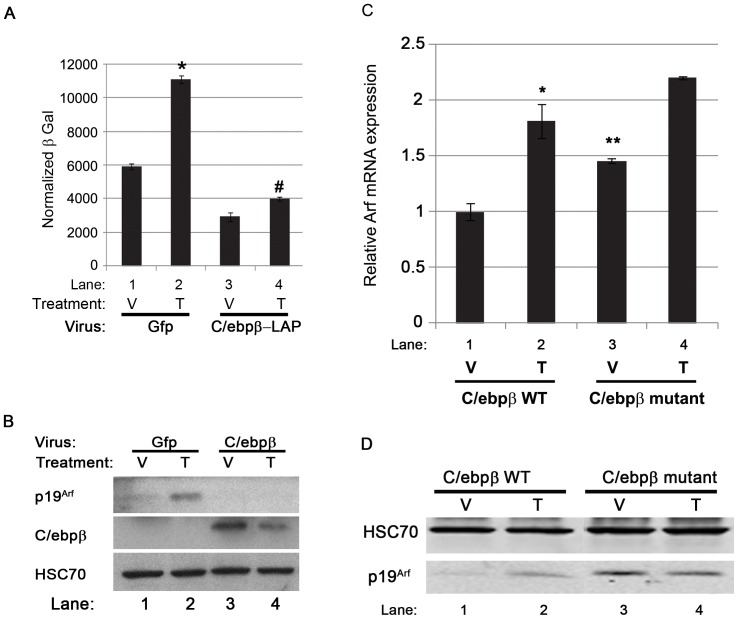
Figure 2. The effects of overexpression or absence of C/ebpβ on Arf induction by Tgfβ.
(A). β-galactosidase activity in Arf lacZ/lacZ MEFs showing the effects of ectopically-expressed C/ebpβ (LAP form) on Arf induction following 48 hour exposure to Tgfβ. Significant increase (*) and decrease (#) of ArflacZ expression is represented in the figure. *, #, p<0.05. (B) Representative western blot for the indicated proteins using lysates from wild type MEFs, exposed to 48 hours of Tgfβ (T) and vehicle (V) after transduction using Gfp- or C/ebpβ (LAP form)-expressing retrovirus. (C) qRT-PCR using total RNA isolated from C/ebpβ +/+ and C/ebpβ −/− MEFs exposed to vehicle (V) or Tgfβ (T) for 48 hours. Differences in transcript level between Tgfβ- and vehicle-treated C/ebpβ +/+ MEFs are significant [p<0.05 (*)]. Differences in transcript level between vehicle-treated C/ebpβ +/+ and C/ebpβ −/− MEFs are significant, too [p<0.05 (**)]. (D) Representative western blot for the indicated proteins using lysates from C/ebpβ +/+ and C/ebpβ −/− MEFs exposed to vehicle (V) or Tgfβ (T) for 48 hours.
We confirmed that ectopically expressed C/ebpβ blunted Arf transcription by showing that β-galactosidase activity was repressed in cultured Arf lacZ/lacZ MEFs infected with retrovirus encoding the liver-enriched activator protein (LAP) isoform of C/ebpβ, which includes a transactivation domain [28], [29] (Figure 2A, lane 3 versus 1). Consistent with the concept that p19Arf expression is primarily controlled by Arf transcription, Western blotting showed that ectopic C/ebpβ also diminished the low basal p19Arf evident in wild type MEFs at passage 3 (Figure 2B, lane 3 versus 1). Further, ectopic expression of C/ebpβ also blunted Tgfβ-dependent induction of Arf transcription and p19Arf expression in cultured MEFs (Figures 2A and B, lane 2 versus 4). These data indicate that C/ebpβ can repress Arf expression in MEFs in a manner that is dominant over Tgfβ-dependent induction of p19Arf.
We next took advantage of C/ebpβ −/− mice to begin to address whether de-repression by C/ebpβ down-regulation contributes to Arf induction by Tgfβ. C/ebpβ −/− mice have been previously shown to exhibit increased postnatal lethality, abnormal hematopoiesis, abnormal glucose homeostasis and immune system defects, among their abnormalities [24], [30]. The mice were generated by introducing a MCI-Neo poly(A)+ mutation at the 3′ terminus of C/ebpβ to abolish translation of the LAP and LIP isoforms [24]. As previously described [26], analysis of cultured MEFs derived from wild type and C/ebpβ −/− embryos demonstrated that basal Arf mRNA and p19Arf protein were increased upon C/ebpβ loss (Figure 2C and D, lane 3 versus 1). Despite the increased baseline Arf expression, though, absence of C/ebpβ only minimally influenced the further induction of Arf mRNA by Tgfβ (Figure 2C, compare lane 4 versus 3 with 2 versus 1). This further increase in p19Arf was not as evident by western blotting (Figure 2D, compare lane 4 versus 3 with 2 versus 1), suggesting that additional factors may act by post-transcriptional mechanisms to control p19Arf protein level. Taken together, these findings indicate that loss of C/ebpβ binding to the Arf promoter cannot fully account for the increased Arf mRNA in response to Tgfβ stimulation.
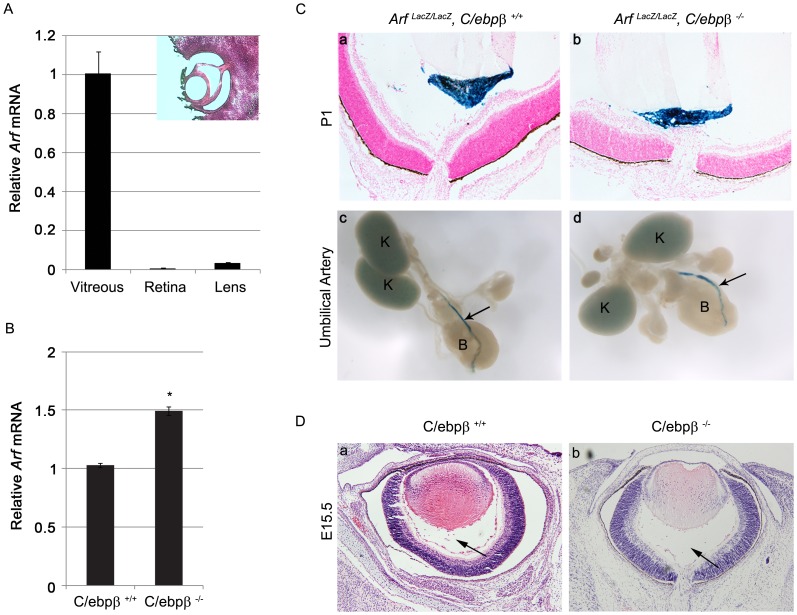
We extended our studies to the in vivo setting by examining how the presence or absence of C/ebpβ influences Arf expression and Tgfβ2 effects in the developing vitreous, the only well-characterized site of p19Arf activity in the developing mouse embryo [7], [21]. At E13.5, Arf mRNA is principally detected in the primary vitreous (Figure 3A), where p19Arf represses Pdgfrβ expression to block vascular mural cell hyperplasia [21], [25]. Consistent with its role as a bona fide repressor, Arf mRNA was elevated in the primary vitreous of C/ebpβ −/− embryos as compared to wild type (Figure 3B). In addition to de-repressing Arf expression in a tissue known to express the transcript, we investigated whether loss of C/ebpβ was sufficient to drive ectopic Arf expression beyond its normal expression pattern. Utilizing Arf lacZ/lacZ animals in which the β-galactosidase reporter reflects Arf mRNA [7], we did not find enhanced Arf expression in ocular tissues that do not normally express Arf, nor did its expression in genitourinary structures extend beyond the internal umbilical artery (Figure 3C). Finally, we found no apparent ocular abnormalities at E15.5 or in the postnatal period (Figure 3D and additional data not shown), indicating that the increased Arf mRNA was not obviously detrimental.
Figure 3. Loss of C/ebpβ increases Arf mRNA expression in vitreous of developing eye.
(A). qRT-PCR analysis using total RNA isolated from the vitreous (V), lens (L) and retina (R) from E13.5 WT mouse embryos. Expression was normalized to that of Gapdh. (B) qRT-PCR analysis using total RNA isolated from the vitreous from E13.5 C/ebpβ +/+ and C/ebpβ −/− mouse embryos. Expression was normalized to that of Gapdh. (C) Arf expression is limited to previously identified sites in C/ebpβ −/− mice during development. (a, b) Representative photomicrographs of hematoxylin- and eosin-stained and X-Gal stained slides of P1 mouse eye of the indicated genotype. Note that Arf-expressing cells are limited to the vitreous (blue staining) in the Arf lacZ/lacZ, C/ebpβ −/− embryo, similar to the littermate Arf lacZ/lacZ, C/ebpβ +/+ control embryo. (c,d) Representative whole-mount, E13.5 embryo from mice of the indicated genotype, following X-gal staining. Note that Arf-expressing cells are limited to the umbilical artery (arrow) in the Arf lacZ/lacZ, C/ebpβ −/− embryo, similar to its littermate Arf lacZ/lacZ, C/ebpβ +/+ control embryo. K, kidney; B, bladder. (D). Representative photomicrographs of hematoxylin- and eosin-stained slides of E15.5 embryos showing there is no primary vitreous hyperplasia in C/ebpβ −/− embryos. Arrows denote the cellular area of the primary vitreous.
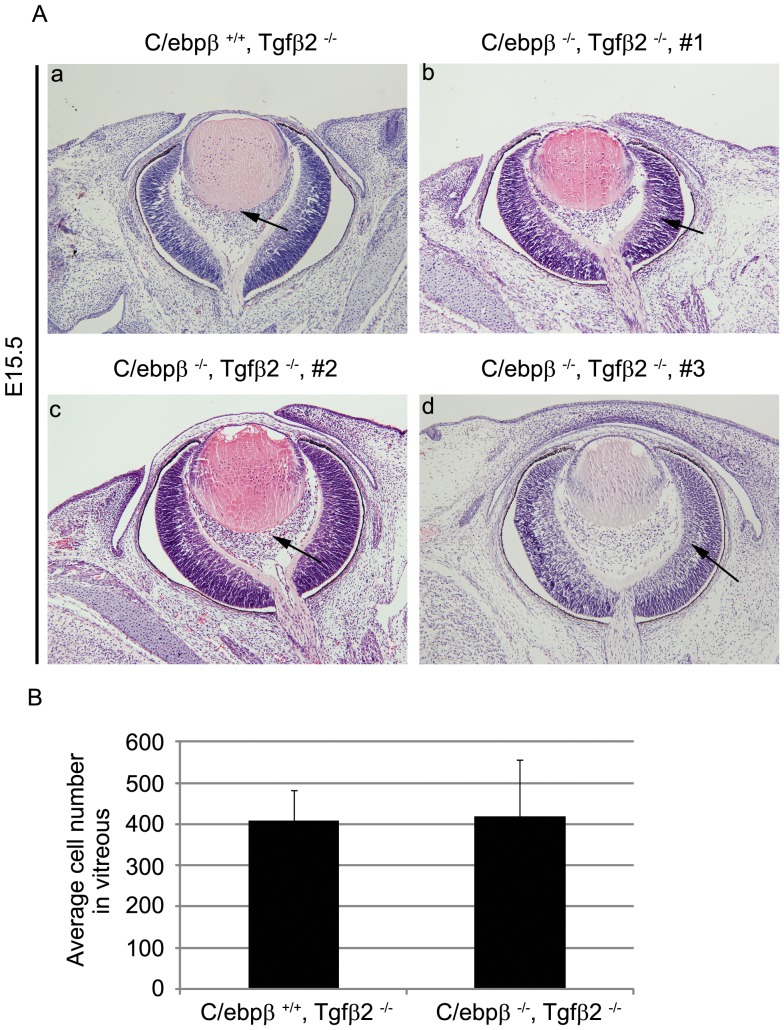
We previously established that p19Arf expression is diminished in the primary vitreous of Tgfβ2−/− embryo eyes and this results in primary vitreous hyperplasia, mimicking that observed in Arf −/− embryos [7]. That exogenous Tgfβ1 reverses this phenotype in Tgfβ2−/− embryos – but not in Arf −/− embryos – demonstrates that p19Arf is the key Tgfβ-dependent target that prevents primary vitreous hyperplasia [22]. If Tgfβ2 solely acts to reverse C/ebpβ-driven Arf repression, the primary vitreous hyperplasia in Tgfβ2−/− embryos should be rescued in C/ebpβ −/− embryos. We investigated this by analyzing the ocular phenotype in Tgfβ2−/− embryos that had or lacked C/ebpβ. Our analyses demonstrated that the eyes of Tgfβ2−/− embryos were indistinguishable from those lacking both genes (Figure 4A and B). That the absence of an Arf repressor cannot reverse the developmental abnormality illustrates that Tgfβ2 likely also influences a positively acting factor to drive p19Arf expression in the primary vitreous.
Figure 4. Loss of C/ebpβ is insufficient to rescue PHPV like eye phenotype of Tgfβ2 KO mouse.
(A) Representative photomicrographs of hematoxylin- and eosin-stained slides of E15.5 embryos showing the primary vitreous hyperplasia in C/ebpβ+/+, Tgfβ2−/− embryos (a) is NOT corrected by additional loss of expression of C/ebpβ in C/ebpβ −/−, Tgfβ2−/− embryos (b-d). Arrows denote the cellular area of the primary vitreous. (B) Quantitative analyses show that the average cell numbers in the vitreous have little change in C/ebpβ −/−, Tgfβ2−/− embryos at E13.5 as compared with C/ebpβ +/+, Tgfβ2−/− littermates.
Considering potential positive regulators of Arf, E2Fs and Sp1 are reasonable candidates based, in part, on DNA binding elements near the Arf transcription start site (Figure 1A). E2Fs have been proven to participate in Arf regulation in various cell contexts [11], [14], [31], [32]. Sp1 has been implied to be important in Arf regulation because deletion of potential Sp1 binding sites diminishes Arf promoter expression, and because Sp1 can bind to the Arf promoter [11], [33].
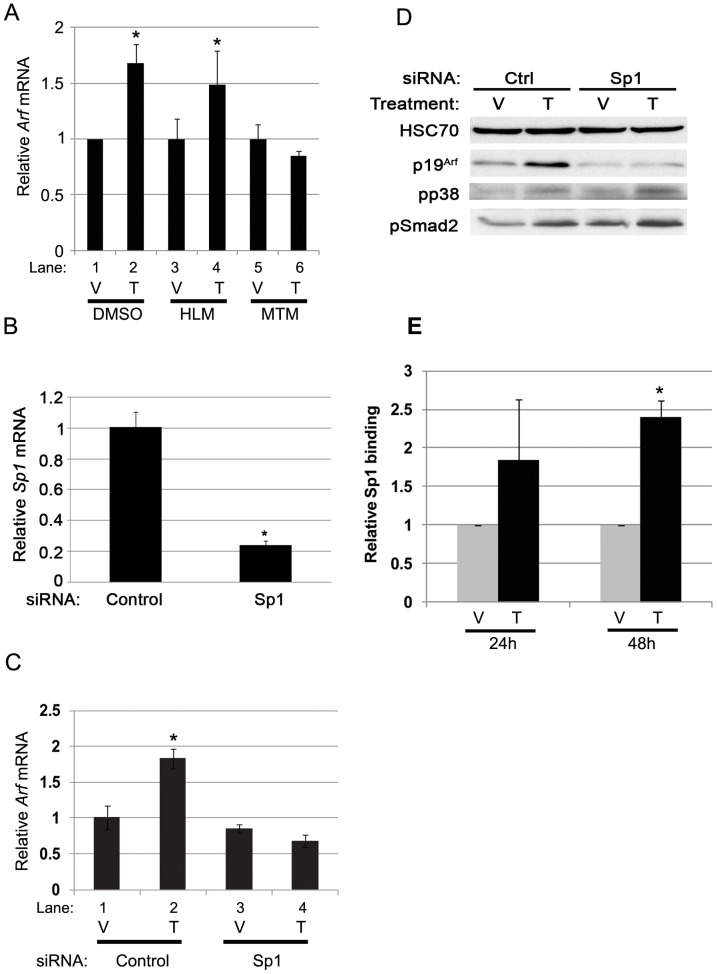
To begin to test whether these candidates act in response to Tgfβ, we first investigated whether chemical inhibition of either pathway interfered with Arf induction by Tgfβ. We utilized HLM006474 (HLM), which inhibits the DNA-binding activity of E2Fs [34], and mithramycin A (MTM) which, among other things, interferes with Sp1 binding to GC-rich DNA [35]. Induction of Arf mRNA by Tgfβ proceeded unabated in the absence or presence of HLM (Figure 5A, lane 3 and 4 versus lane 1 and 2), even though it restored the repression of other E2F-dependent genes like PAI-1 [36](YZ and SXS, unpublished data). In contrast, MTM blocked Arf mRNA induction (Figure 5A, land 5 and 6 versus lane 1 and 2), but MTM did not significantly block Smad 2/3 binding to the proximal region of Arf promoter (YZ and SXS, negative data not shown). To exclude potential off-target effects of MTM, we showed that transient Sp1 knockdown by siRNA transfection (Figure 5B) also blocked Arf mRNA and protein induction by Tgfβ (Figures 5C and D). Of note, Sp1 knockdown did not block phosphorylation of Smad 2/3 or p38 Mapk (Figure 5D), two events that are required downstream of Tgfβ2 [22]. Finally, ChIP demonstrated that the minimal Sp1 binding to the proximal Arf promoter at baseline was significantly increased by Tgfβ at 24 and 48 hours (Figure 5E and additional data not shown), paralleling the time course for Arf mRNA increase we previously described [22]. These findings suggest that direct binding of Sp1 to the Arf promoter is required for Tgfβ to augment p19Arf expression.
Figure 5. Inhibition or knockdown of Sp1 blocks Arf mRNA induced by Tgfβ.
(A) qRT-PCR analysis using total RNA isolated from WT MEFs treated with Sp1 inhibitor, mithramycin A (MTM), E2F inhibitor, HLM006474 (HLM) and control DMSO, following 48 hour exposure to Tgfβ (T) or vehicle (V). The significant changes between Tgfβ treatment and vehicle treatment is marked as * (p<0.05). (B) qRT-PCR analysis of Sp1 using total RNA isolated from WT MEFs treated with either siRNA control (Scram), or siRNA targeting mouse Sp1 as indicated for 48 hours. *, p<0.05. (C) qRT-PCR analysis using total RNA isolated from WT MEFs treated with Tgfβ (T) or vehicle (V) for 48 hours following 24 hours transfection with either siRNA control (Scram), or siRNA targeting mouse Sp1 as indicated. Sp1 knockdown significantly dampens the induction of Arf mRNA by Tgfβ (*, p<0.05). (D) Representative western blot for the indicated proteins using lysates from wild type MEFs treated with Tgfβ (T) or vehicle (V) for 48 hours following 24 hours transfection with either siRNA control (Scram), or siRNA targeting mouse Sp1 as indicated. (E) Tgfβ promotes Sp1 binding to the Arf locus in MEFs. Quantitative analysis of representative ChIP assays using wild type MEFs exposed to vehicle (V) or Tgfβ (T) for 24 hours or 48 hours. ChIP assay was carried out using antibodies specific to Sp1 and IgG as control. Immunoprecipitated DNA and input DNA were amplified with primers for proximal region of Arf promoter. *, p<0.05 for Tgfβ versus corresponding vehicle.
Discussion
We recently demonstrated that Tgfβ is an essential regulator of Arf during eye development [7], [22]. However, Arf expression is limited given the protean effects of Tgfβs during mouse embryo development [7], and Arf mRNA induction is delayed following immediate Smad 2/3 binding to the promoter [22]. Both suggest that Arf expression is orchestrated by Tgfβ-dependent changes in transcriptional regulators beyond the Smad proteins. Our new data indicate that Sp1 and C/ebpβ represent such cooperating factors, influencing Arf induction in opposing ways. We have the following evidence: First, ectopic expression of C/ebpβ blocked Arf induction by Tgfβ. Second, C/ebpβ binding to the Arf promoter is diminished by Tgfβ treatment in a time frame coincident with Arf mRNA induction. The concept that Tgfβ orchestrates de-repression of Arf by C/ebpβ down-regulation in vivo is supported by the fact that Arf expression in the vitreous is elevated in C/ebpβ −/− animals. However, absent the essential Arf inducer – Tgfβ2– loss of C/ebpβ is not sufficient to correct the PHPV-like eye phenotype in Tgfβ2−/− mice; hence, removing C/ebpβ repression is not the whole story. Searching for a positive trans-acting factor induced by Tgfβ, we found chemical and genetic evidence supporting a role for Sp1. In summary, our data provide new insight into the molecular basis underlying Arf control by Tgfβ during eye development, and this may inform our understanding of certain disease processes.
Our work extends previous reports implicating both C/ebpβ and Sp1 as potential regulators of p19Arf expression. That C/ebpβ can repress Arf was previously suggested primarily by the elevated Arf mRNA and protein observed in C/ebpβ −/− keratinocytes in culture and in the adult mouse [26]. Sp1 is well known to bind to GC-rich promoter elements [37], [38], and the mouse and human Arf promoters contain numerous Sp1 binding sites within CpG islands [15], [33]. Several previous studies showed the potential importance of Sp1 binding to the human ARF promoter in cultured cells [11], [39]. However, the potential physiological importance of either in Arf regulation is not yet clear. For example, C/ebpβ −/− mice are completely refractory to chemically induced skin cancer [40], which concept is consistent with higher p19Arf expression as a tumor suppressor. However, Arf does not seem to play a role in tumor resistance in this model [26]. Nonetheless, our findings demonstrating increased Arf mRNA in the vitreous of C/ebpβ −/− embryos indicates that C/ebpβ can repress Arf in a normal developmental context. The lack of widespread Arf promoter activation in these embryos and newborn Arf lacZ/lacZ, C/ebpβ −/− mice, though, still highlights the importance of tissue-specific positive transcriptional regulators of Arf.
The fact that the phenotype due to blunted Arf expression in Tgfβ2-deficent embryo eyes was not reversed in animals also lacking C/ebpβ provides additional in vivo evidence for the importance of positively-acting factors. That Sp1 cooperates with Smad signaling is consistent with previous findings that Tgfβ2 regulates p15Ink4b through direct Sp1 binding to the promoter [41], [42]. Sp1 also collaborates with Smad proteins to induce the expression of vimentin in cultured cells undergoing the epithelial-mesenchymal transition in response to Tgfβ [43]. Our preliminary work shows that decreased expression of C/ebpβ in response to Tgfβ depends on TbrII and Smad 2/3 activation (YZ and SXS, unpublished data), but we do not yet know whether Sp1 binding to the Arf promoter similarly depends on the activation of that pathway. Sp1 is also known to work cooperatively with E2Fs [44], which are also implicated as both positive and negative regulators of Arf [11], [31], [32], [45]. Our finding that HLM does not significantly block Arf induction by Tgfβ suggests that Sp1 seems to act independently of E2Fs in this context. It will obviously be important to demonstrate the functional importance of Sp1 in vivo using our mouse model for PHPV. Regrettably, Sp1−/− mice display an embryonic lethal phenotype at E11.5, before primary vitreous development [46]. Tissue specific Sp1 knockout using a Wnt1-Cre driver would be very informative.
Finally, we have carried out this line of investigation in the mouse to gain insight into human diseases, like cancer and PHPV. Repression of human ARF expression is a relatively common mechanism by which cancers can evade this tumor suppressor activity [47]; presumably, restoring ARF expression could represent a novel therapeutic approach, especially for that subset of cancers also retaining wild type p53. As a human disease, PHPV is typically sporadic, but several reports of familial disease suggest that it could have an underlying genetic basis [48], [49], [50]. C/ebpβ is frequently expressed in human cancer and has been implicated as an oncogenic factor (as in the keratinocyte model noted above) [26], [40] or tumor suppressor with the capacity to foster senescence [51], [52]. These disparate effects may be due, in part, to the capacity of C/ebpβ to form homo- and heterodimeric complexes with either activating or transcriptional repressive activity [28]. Sp1, too, could act as a Tgfβ-dependent tumor suppressor, by controlling Ink4b [41], [42] or Arf (this work), or as an oncogene by facilitating EMT [43]. Again, one could envision that the net effect of Sp1 could depend on the underlying cellular or genetic context. As more sophisticated, “next-generation” genome sequencing and analytical tools are applied – particularly to diseases like PHPV – the role for these genes might be revealed.
Acknowledgments
We gratefully acknowledge Syann Lee and Joel Elmquist (both at UTSW) for assistance with LCM. We thank other members of the Skapek lab for technical assistance and helpful discussion.
Funding Statement
This research was funded by the National Eye Institute (EY 014368 and EY 019942), http://www.nei.nih.gov/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Quelle DE, Zindy F, Ashmun RA, Sherr CJ (1995) Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83: 993–1000. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y, Xiong Y, Yarbrough WG (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734. [DOI] [PubMed] [Google Scholar]
- 3. Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, et al. (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649–659. [DOI] [PubMed] [Google Scholar]
- 4. Esteller M, Cordon-Cardo C, Corn PG, Meltzer SJ, Pohar KS, et al. (2001) p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res 61: 2816–2821. [PubMed] [Google Scholar]
- 5. Xing EP, Nie Y, Song Y, Yang GY, Cai YC, et al. (1999) Mechanisms of inactivation of p14ARF, p15INK4b, and p16INK4a genes in human esophageal squamous cell carcinoma. Clin Cancer Res 5: 2704–2713. [PubMed] [Google Scholar]
- 6. Zindy F, Quelle DE, Roussel MF, Sherr CJ (1997) Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15: 203–211. [DOI] [PubMed] [Google Scholar]
- 7. Freeman-Anderson NE, Zheng Y, McCalla-Martin AC, Treanor LM, Zhao YD, et al. (2009) Expression of the Arf tumor suppressor gene is controlled by Tgfbeta2 during development. Development 136: 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168. [DOI] [PubMed] [Google Scholar]
- 9. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, et al. (2007) Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67: 3963–3969. [DOI] [PubMed] [Google Scholar]
- 10. Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, et al. (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parisi T, Pollice A, Di Cristofano A, Calabro V, La Mantia G (2002) Transcriptional regulation of the human tumor suppressor p14(ARF) by E2F1, E2F2, E2F3, and Sp1-like factors. Biochem Biophys Res Commun 291: 1138–1145. [DOI] [PubMed] [Google Scholar]
- 12. Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K (2005) Distinct E2F-mediated transcriptional program regulates p14ARF gene expression. EMBO J 24: 3724–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elliott MJ, Dong YB, Yang H, McMasters KM (2001) E2F-1 up-regulates c-Myc and p14(ARF) and induces apoptosis in colon cancer cells. Clin Cancer Res 7: 3590–3597. [PubMed] [Google Scholar]
- 14. del Arroyo AG, El Messaoudi S, Clark PA, James M, Stott F, et al. (2007) E2F-dependent induction of p14ARF during cell cycle re-entry in human T cells. Cell Cycle 6: 2697–2705. [DOI] [PubMed] [Google Scholar]
- 15. Inoue K, Roussel MF, Sherr CJ (1999) Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci U S A 96: 3993–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K (2005) Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol 25: 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ameyar-Zazoua M, Wisniewska MB, Bakiri L, Wagner EF, Yaniv M, et al. (2005) AP-1 dimers regulate transcription of the p14/p19ARF tumor suppressor gene. Oncogene 24: 2298–2306. [DOI] [PubMed] [Google Scholar]
- 18. Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, et al. (2005) Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 433: 278–285. [DOI] [PubMed] [Google Scholar]
- 19. Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, et al. (2007) FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev 21: 2775–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McKeller RN, Fowler JL, Cunningham JJ, Warner N, Smeyne RJ, et al. (2002) The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc Natl Acad Sci U S A 99: 3848–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silva RL, Thornton JD, Martin AC, Rehg JE, Bertwistle D, et al. (2005) Arf-dependent regulation of Pdgf signaling in perivascular cells in the developing mouse eye. EMBO J 24: 2803–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng Y, Zhao YD, Gibbons M, Abramova T, Chu PY, et al. (2010) Tgfbeta signaling directly induces Arf promoter remodeling by a mechanism involving Smads 2/3 and p38 MAPK. J Biol Chem 285: 35654–35664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, et al. (1997) TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 124: 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, et al. (1995) Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J 14: 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Widau RC, Zheng Y, Sung CY, Zelivianskaia A, Roach LE, et al. (2012) p19Arf represses platelet-derived growth factor receptor beta by transcriptional and posttranscriptional mechanisms. Mol Cell Biol 32: 4270–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ewing SJ, Zhu S, Zhu F, House JS, Smart RC (2008) C/EBPbeta represses p53 to promote cell survival downstream of DNA damage independent of oncogenic Ras and p19(Arf). Cell Death Differ 15: 1734–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sherr CJ, DePinho RA (2000) Cellular senescence: mitotic clock or culture shock? Cell 102: 407–410. [DOI] [PubMed] [Google Scholar]
- 28. Ramji DP, Foka P (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J 365: 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Descombes P, Schibler U (1991) A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67: 569–579. [DOI] [PubMed] [Google Scholar]
- 30. Liu S, Croniger C, Arizmendi C, Harada-Shiba M, Ren J, et al. (1999) Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPbeta gene. J Clin Invest 103: 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR (1997) Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A 94: 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bates S, Phillips AC, Clark PA, Stott F, Peters G, et al. (1998) p14ARF links the tumour suppressors RB and p53. Nature 395: 124–125. [DOI] [PubMed] [Google Scholar]
- 33. Robertson KD, Jones PA (1998) The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol 18: 6457–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma Y, Kurtyka CA, Boyapalle S, Sung SS, Lawrence H, et al. (2008) A small-molecule E2F inhibitor blocks growth in a melanoma culture model. Cancer Res 68: 6292–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, et al. (1991) Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest 88: 1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koziczak M, Krek W, Nagamine Y (2000) Pocket protein-independent repression of urokinase-type plasminogen activator and plasminogen activator inhibitor 1 gene expression by E2F1. Mol Cell Biol 20: 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Briggs MR, Kadonaga JT, Bell SP, Tjian R (1986) Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234: 47–52. [DOI] [PubMed] [Google Scholar]
- 38. Dynan WS, Tjian R (1983) The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35: 79–87. [DOI] [PubMed] [Google Scholar]
- 39. Zhang HJ, Li WJ, Yang SY, Li SY, Ni JH, et al. (2009) 8-Chloro-adenosine-induced E2F1 promotes p14ARF gene activation in H1299 cells through displacing Sp1 from multiple overlapping E2F1/Sp1 sites. J Cell Biochem 106: 464–472. [DOI] [PubMed] [Google Scholar]
- 40. Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC (2002) CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A 99: 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feng XH, Lin X, Derynck R (2000) Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J 19: 5178–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li JM, Nichols MA, Chandrasekharan S, Xiong Y, Wang XF (1995) Transforming growth factor beta activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem 270: 26750–26753. [DOI] [PubMed] [Google Scholar]
- 43. Jungert K, Buck A, von Wichert G, Adler G, Konig A, et al. (2007) Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res 67: 1563–1570. [DOI] [PubMed] [Google Scholar]
- 44. Karlseder J, Rotheneder H, Wintersberger E (1996) Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol 16: 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aslanian A, Iaquinta PJ, Verona R, Lees JA (2004) Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev 18: 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marin M, Karis A, Visser P, Grosveld F, Philipsen S (1997) Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89: 619–628. [DOI] [PubMed] [Google Scholar]
- 47. Sherr CJ (2012) Ink4-Arf Locus in Cancer and Aging. Wiley Interdiscip Rev Dev Biol 1: 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu YS, Chang BL (1997) Persistent hyperplastic primary vitreous in male twins. Korean J Ophthalmol 11: 123–125. [DOI] [PubMed] [Google Scholar]
- 49. Lin AE, Biglan AW, Garver KL (1990) Persistent hyperplastic primary vitreous with vertical transmission. Ophthalmic Paediatr Genet 11: 121–122. [DOI] [PubMed] [Google Scholar]
- 50. Wang MK, Phillips CI (1973) Persistent hyperplastic primary vitreous in non-identical twins. Acta Ophthalmol (Copenh) 51: 434–437. [DOI] [PubMed] [Google Scholar]
- 51. Sebastian T, Johnson PF (2009) RasV12-mediated down-regulation of CCAAT/enhancer binding protein beta in immortalized fibroblasts requires loss of p19Arf and facilitates bypass of oncogene-induced senescence. Cancer Res 69: 2588–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sebastian T, Malik R, Thomas S, Sage J, Johnson PF (2005) C/EBPbeta cooperates with RB:E2F to implement Ras(V12)-induced cellular senescence. EMBO J 24: 3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]