Abstract
The use of primary somatic cells in nuclear transfer procedure has opened a new opportunity to manipulate domestic animal genomes via homologous recombination. To date, while a few loci have been targeted in somatic cells using similar enrichment strategies as those used in mouse ES cells, there have been problems of low efficiency, mixed targeted and non-targeted cells within a colony and difficulties in cloning the cell after targeting. Utilizing the hypoxanthine guanine phosphoribosyl transferase (HPRT) as a test locus, it was determined that while no targeted colonies were identified using a conventional targeting construct, an average of 1 per million targeted cells were identified when a nuclear localization signal (nls) was added to the construct. When the nls was combined with cell synchronization using a thymidine block, targeting efficiency increased 7-fold. Moreover, the number of random integrants decreased by over 54-fold resulting in a 1:3 targeted to random integration ratio. This method should facilitate the application of homologous recombination to primary somatic cells.
INTRODUCTION
Targeted gene inactivation by homologous recombination (HR) has been very successful in mouse ES cells. However, the lack of functional ES cells in other animal species has been a major barrier in the development of similarly genetically modified animals. As a result, progress in the development of transgenic non-murine animals of importance in biomedical applications has been severely hampered. The use of primary somatic cells in nuclear transfer procedure (1,2) has opened a new opportunity to manipulate non-murine animal genomes via homologous recombination. Unfortunately, targeting in primary somatic cells is hampered by cell senescence, lower overall rates of HR and higher rates of non-homologous end joining (NHEJ), than ES cells (3–6). In expressed loci, allowing the highly efficient promoter trap enrichment approaches, targeting efficiency in somatic cells is still low compared to that seen in mouse ES cells (7–9). Our own experience using both systems (10–13) supports the observation that targeting in somatic cells is more difficult than in ES cells. In primary somatic cells, the meager available data on successfully targeted loci, and the sporadic reports of failed attempts at targeting (14), reinforce the difficulties associated with working with these cells. Moreover, in all cases reported to date, there have been problems of low efficiency, and/or mixed targeted and non-targeted cells within a colony, and difficulties in cloning the cell after targeting (15,16).
In various gene therapy protocols, it has been demonstrated that a nuclear localization signal (nls) enhances both the nuclear transport and the expression of transfected plasmid DNA (17,18). At present, however, there are no reports showing a link between the use of nls and the frequency of HR. Here, we present evidence that the use of a nls and S-phase cell cycle synchronization by thymidine block, enhances targeting efficiency at the hypoxanthine guanine phosphoribosyl transferase (HPRT) locus in primary fetal bovine fibroblasts.
MATERIALS AND METHODS
Cell culture and selection
Male primary fetal bovine fibroblasts were isolated from a 35-day-old male fetus. The head and viscera were removed and the remaining tissue was minced with a sterile razor blade. The tissue was added to 10 ml of 0.05% trypsin (Gibco Laboratories, Grand Island, NY) supplemented with 0.9 mM potassium chloride, 0.9 mM dextrose, 0.7 mM sodium bicarbonate, 0.1 mM EDTA (Sigma–Aldrich, St Louis, MO) and 20 mM sodium chloride. The tissue/trypsin solution was shaken at 37°C for 15 min, supernatant removed and the process repeated three times. After incubation, the supernatant was collected, pooled and pelleted. The cell pellet was resuspended in DMEM/F12 media (Gibco) supplemented with 10% fetal bovine serum (FBS) and 5% calf serum (CS) (Hyclone), 30 mM sodium bicarbonate, 0.5 mM pyruvic acid and 2 mM N-acetyl-l-cysteine (Sigma). In addition, 100 U penicillin and 250 ng amphotericin (Gibco) were added to inhibit microbial growth. The cells were plated in 10 cm tissue culture plates, placed in a 5% CO2 incubator at 37°C, allowed to attach, grown to confluency and passaged 1:2 or 1:3. The cells were then trypsinized and frozen in 50% FBS, 40% media and 10% DMSO (Sigma), for long time storage and future use.
When needed, cells were thawed and grown in DMEM/F12 (Gibco) media supplemented with 15% fetal bovine serum and incubated at 37°C in a 5% CO2 atmosphere. At 70–80% confluency, cells were harvested by trypsinization and 9–10 million cells were resuspended in 0.8 ml F10 nutrient mixture (Gibco). Asc1 linearized targeting construct at 2 µg/million cells were mixed with the cells, placed in a 4 mm gap cuvette, and electroporated at 450 V, four pulses of 1 ms duration using the BTX Electro-cell manipulator ECM2001. Electroporated cells were mixed with fresh medium and plated in 10 cm plates at 5 × 105 cells per plate. Five days later, cells were placed on selection media containing 1.25 µg/ml puromycin (Clontech) for 4 days followed by 50 µg/ml 8-azaguanine (8-AG, Sigma) for 8 additional days. For each replication, 3–5 plates were maintained on puromycin selection for the entire period to obtain both random and targeted frequencies. For the thymidine block, media was replaced with media containing 2 mM thymidine at 60% confluency. Twenty-four hours later, cells were harvested and electroporated as above. There was no significant difference in cell survival between untreated and thymidine-synchronized cells before or after electroporation as determined by viability counting (data not shown).
Statistical analysis
All analyses were done in triplicates once (Fig. 1C), or twice (Fig. 2B). Comparison of the effect of addition of nls on targeting efficiency was determined using one-way ANOVA, with presence or absence of nls as the main factor. For comparison of the effects of nls and thymidine, data were analyzed by two-way ANOVA with treatment and replication (set) as the main factors. In both cases, mean separation was accomplished using the Games-Howell test using the Statview/SAS statistical package. Significance was set at P < 0.05 unless otherwise stated.
Figure 1.
Targeted disruption of HPRT locus. (A) Maps of the targeting constructs, bovine HPRT locus and structure of targeted locus. Locations of HPRT exons, pGK-puro cassette, nls, PCR primer sites, PCR product size and end sequencing regions are shown. (B) Sequence of nls, two 72 bp tandem repeats from SV40 enhancer. (C) Number of random and targeted insertions obtained with each targeting vector per 9.5 × 106 cells. (D) Diagnostic PCR of four 8-AG-resistant colonies. (E) End sequence of PCR product. PCR primer sites are shown in italics, exon 4 and exon 6 sequences are shown in bold.
Figure 2.
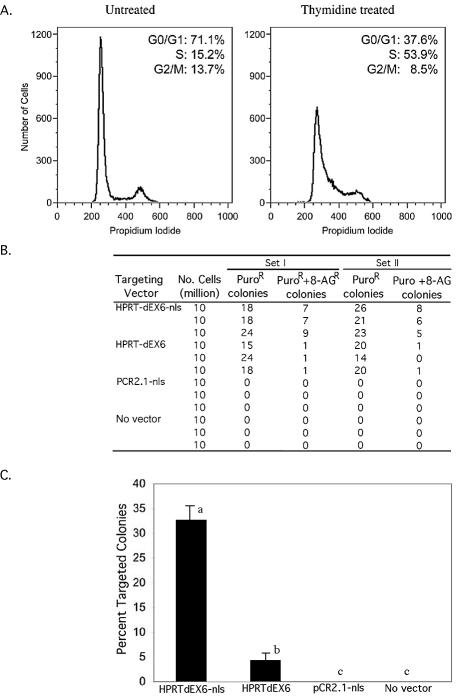
Effect of cell synchronization on targeting efficiency. (A) Syn chronization of cells in S-phase with 2 mM thymidine for 24 h. (B) Number of random and targeted insertions obtained with each targeting construct per 107 cells in S-phase synchronized cells. No colony was obtained either with a control vector containing only nls or with no vector thereby indicating that 8-AG resistance does not occur in response to the nls or cell synchronization process. (C) Percent targeted colonies in thymidine-synchronized cells with different targeting constructs. Columns with different superscripts are different at P < 0.001. Means ± SE.
Cell cycle analysis
Cells were grown to 60% confluency and treated with 2 mM thymidine (Sigma) for times ranging from 12 to 36 h. Following thymidine treatment, 1–2 × 106 cells were harvested, washed in PBS, fixed in 70% ethanol in PBS and stored overnight at –20°C. For cell cycle analysis, cells were centrifuged, rinsed with PBS, resuspended in 1 ml of 0.05 mg/ml propidium iodide, 0.1% Triton X-100 and 0.2 mg/ml Rnase-A solution in PBS and incubated at 4°C for 1 h. Cells were then analyzed by FACSCalibur™ flow cytometer (Becton Dickinson) to determine DNA content. Sync Wizard Model was used to model the cell cycle distribution within each sample.
Construction of targeting vectors
The targeting vectors were designed to allow targeting of the HPRT locus, but equally important to determine the number of random integrations. Selected regions of the HPRT gene were amplified from genomic DNA isolated from male bovine fibroblasts by LR-PCR. A forward primer from exon 4 (5′-CAACAGGGGACATAAAAGTAATTGGTGG-3′) and a modified reverse primer from exon 6 containing a non-priming EcoR1 site (5′-GAATTCATTTGCCAGTGTCAATTATATC-3′) were used to amplify a ∼6.5 kb fragment. This fragment was cloned into PCR2.1 Topo vector (Invitrogen) and a HindIII 2.5 kb fragment containing exon 4 and part of intron 4 was deleted. A HindIII–FseI–AscI–HindIII linker was cloned into the HindIII site. For the right arm of the targeting vector, a 4 kb fragment from exon 6 to intron 8 was amplified and cloned into PCR2.1 Topo vector (Invitrogen). Primers used were a forward primer from exon 6 containing a non-priming NotI site (5′-GCGGCCGCATAAACCAAAGATGGTCAAG-3′) and a reverse primer from intron 8 (5′-GCATTCTCCAGATTCCTGCC-3′). The targeting vector HPRT-ΔEx6 was constructed by digesting the PCR2.1-exon 4–6 plasmid with EcoRI and NotI and an EcoRI–NotI pGK-Puro cassette inserted into it. This was followed by addition of the 4 kb exon 6–intron 8 fragment into the NotI site in the proper orientation. Each of these steps was confirmed by sequencing. For construction of the HPRT-ΔEx6-nls vector, a 180 bp nls cassette containing FseI sites at both ends was cloned into the above vector at the FSI site present in the HindIII–FseI–AscI–HindIII linker introduced previously.
PCR analysis of targeted colonies
The colonies were expanded and genomic DNA was extracted with wizard genomic purification kit (Promega) as per the instructions of the manufacturer. To increase yield, 1 µl glycogen (20 mg/ml) was added to DNA before isopropanol precipitation. A diagnostic LR-PCR was performed with a forward primer BovHPRT46F from exon4 (5′-CAACAGGGGACATAAAAGTAATTGGTGG-3′) and reverse primer pGK-P1 from pGK-Puro (5′-TCTGCACGAGACTAGTGAGACGTGCTAC-3′) using 50 ng of genomic DNA in a 25 µl reaction using Hi-fidelity PCR kit (Roche) as per the instructions of the manufacturer. The PCR conditions were 94°C, 2 ms (94°C, 10 s; 57°C, 30 s; 68°C, 8 ms) × 23 cycles (94°C, 10 s; 57°C, 30 s; 68°C, 8 ms + 20 s/cycle increment) × 10 cycles, 86°C, 7 ms.
Southern blot analysis
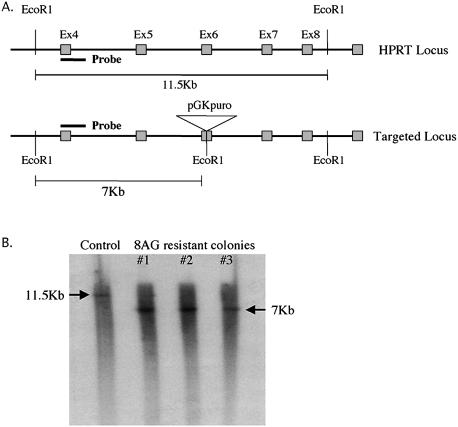
8AG-resistant colonies were grown in a 10 cm plate until they reached confluency. The DNA was extracted with Wizard genomic DNA purification kit (Promega) as per the instructions of the manufacturer; 5 µg genomic DNA was digested overnight with EcoR1 (New England Biolabs), separated on 1% agarose gel and transferred to Hybond-XL (Amersham). A 500 bp DNA probe corresponding to exon4–intron4 of the HPRT gene 2 kb outside the targeting construct (Fig. 3) was labeled using [α-32P]dCTP (ICN) by random oligopriming using High Prime kit (Roche). Membranes were subsequently hybridized overnight at 65°C with the radiolabeled probe in Rapid-hyb buffer (Amersham), washed with increasing levels of stringency and exposed to Kodak BioMax MS film.
Figure 3.
Southern analysis of 8-AG-resistant colonies. (A) Bovine HPRT locus and structure of targeted locus. Location of 500 bp exon4–intron4 probe 2 kb outside the targeting construct, EcoR1 sites and the size of EcoR1 genomic fragments are shown. The targeted locus has an additional EcoR1 site within exon6 that results in polymorphism between the targeted and untargeted locus. (B) Lanes 1–4 show EcoR1 digested genomic DNA samples hybridized with the probe shown in (A). Arrows indicate 11.5 kb EcoR1 fragment from the non-targeted bovine HPRT locus and the 7 kb fragment from the HPRT targeted locus.
RESULTS
Effect of nls on targeting efficiency
To test the effect of nls and to monitor frequencies of random and targeted insertions, we designed two HPRT targeting constructs, HPRT-ΔEx6 and HPRT-ΔEx6-nls (Fig. 1A). Both constructs have a 31 bp deletion and PGK-puro insertion in exon 6 to ensure inactivation of the HPRT locus. Additionally, HPRT-ΔEx6-nls construct contains a 180 bp cassette comprised of two 72 bp tandem repeats from SV40 enhancer known to act as nuclear localization signal (19) (Fig. 1B). Diploid male cells that undergo targeted gene disruption at the single copy X-linked HPRT locus can be selected with 8-AG as HPRT– cells are resistant to 8-AG, while all transformants, random and targeted, can be selected in puromycin.
Male primary bovine fibroblasts were electroporated with linearized targeting constructs, and plated in media containing puromycin or puromycin plus 8-AG. While the total number of insertions obtained with both these constructs are equivalent, HPRT-ΔEx6-nls construct produced 1–2 8-AG-resistant colonies per 9.5 × 106 cells in contrast to HPRT-ΔEx6 where no targeted events were seen (P < 0.05; Fig. 1C). To confirm targeting, the structure of HPRT locus in all the four 8AG-resistant colonies was determined by Long Range PCR (LR-PCR) and sequencing. In all cases, the expected product size and sequence was obtained (Fig. 1D and E). For sequencing, 600 bp from each end of the predicted insertion point were analyzed.
Effect of cell synchronization on targeting efficiency
To determine the relationship between cell cycle and the use of the nls, an additional set of experiments were carried out. Cells were synchronized in the S-phase of the cell cycle by a 2 mM thymidine treatment for 24 h (Fig. 2A) and electroporated with the linearized targeting constructs. As compared to non-synchronous cells, the total number of insertions (random plus targeted) were reduced by 54-fold with constructs with or without nls, while targeted insertions increased 6-fold with HPRT-ΔEx6-nls construct (Fig. 2B and C). Percent of targeted colonies per 8-AG-resistant colony increased from 4.4% in the absence of nls to 32.7% with nls (P < 0.001). No colony was obtained either with a control vector containing only nls, or with no vector, thereby showing that 8-AG resistance does not occur in response to the nls or cell synchronization process. As previously, all 8-AG-resistant colonies were verified by LR-PCR, and PCR products confirmed by end sequencing.
Genomic Southerns of 8AG-resistant colonies
To ensure that our PCR and selection data were accurate, we amplified three representative 8-AG-resistant colonies, extracted DNA and determined the structure of the HPRT loci. Targeted colonies should have a diagnostic fragment of 7 kb due to the presence of an additional EcoR1 site in the targeting loci. In contrast, the single endogenous HPRT allele present in a male cell line should have the expected 11.5 kb diagnostic fragment when probed with exon 4 and part of intron 4. As shown in Figure 3, all three 8-AG-resistant colonies showed a correctly targeted HPRT locus confirming the validity of the selection protocols as well as the PCR assays.
DISCUSSION
Enhancement of gene targeting by delivering targeting construct during S-phase of the cell cycle is consistent with the earlier observations that HR occurs mainly at late S/G2 phase (20) and thymidine synchronizes cells in S-phase and enhances HR (21). A potential explanation for this enhancement is that targeting constructs that lack nls cannot enter the nucleus when the nuclear membrane is intact but must wait until the first mitosis (M-phase) when the nuclear membrane breaks down. This restricts their availability during S-phase when HR is optimal and increases their availability during G1 phase when NHEJ is dominant (20). This eventually results in an increased number of random insertions. In contrast, targeting constructs containing nls are available predominantly during S-phase in S-phase synchronized cells, which results in enhancement of targeted events and suppression of random insertions.
Our results are striking; no targeted event was seen with non-nls targeting construct after screening 3306 colonies. This increased to four targeting events per 3534 insertions using nls targeting construct (Fig. 1C). Moreover, the combination of nls and S-phase cell synchronization had a dramatic effect on both suppressing NHEJ and increasing HR; 43 targeted events were obtained out of a total of 130 insertions, i.e. one targeted event per 3.0 colonies (Fig. 2B and C). Additionally, control plasmids containing an nls signal but no HPRT targeting homology, or cells thymidine blocked and electroporated resulted in no 8-AG colonies, demonstrating that the appearance of the 8-AG colonies is not due to increased mutation rates. Moreover, PCR and genomic Southern validated that the 8-AG colonies were correctly targeted.
Overall, these finding have important implications for targeting in somatic cells, as with a drastic reduction in the number of random insertions, identification of a targeted colony is greatly facilitated even in cases where no enrichment protocols are available. However, these numbers may not be absolute as targeting efficiency varies from locus to locus (12,22).
Polyadenylation signal-less, promoter-less constructs, and positive–negative selection (PNS) has been used to enrich for targeting events (23–25). Using these enrichment schemes in parallel with nls should greatly facilitate targeting of somatic cells by HR. This approach will have broader applications in gene targeting in all cells in general, and somatic cells in particular, and should speed up functional genomic analysis and creation of knockout animals for biomedical and agricultural purposes.
Acknowledgments
ACKNOWLEDGEMENT
This study was funded by NIH grant HL51587 to J.A.P.
REFERENCES
- 1.Campbell K.H., McWhir,J., Ritchie,W.A. and Wilmut,I. (1996) Sheep cloned by nuclear transfer from a cultured cell line. Nature, 380, 64–66. [DOI] [PubMed] [Google Scholar]
- 2.Wilmut I., Schnieke,A.E., McWhir,J., Kind,A.J. and Campbell,K.H. (1997) Viable offspring derived from fetal and adult mammalian cells. Nature, 385, 810–813. [DOI] [PubMed] [Google Scholar]
- 3.Brown J.P., Wei,W. and Sedivy,J.M. (1997) Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science, 277, 831–834. [DOI] [PubMed] [Google Scholar]
- 4.Arbones M.L., Austin,H.A., Capon,D.J. and Greenburg,G. (1994) Gene targeting in normal somatic cells: inactivation of the interferon-gamma receptor in myoblasts. Nature Genet., 6, 90–97. [DOI] [PubMed] [Google Scholar]
- 5.Hanson K.D. and Sedivy,J.M. (1995) Analysis of biological selections for high-efficiency gene targeting. Mol. Cell. Biol., 15, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waldman A.S. (1992) Targeted homologous recombination in mammalian cells. Crit. Rev. Oncol. Hematol., 12, 49–64. [DOI] [PubMed] [Google Scholar]
- 7.Denning C., Dickinson,P., Burl,S., Wylie,D., Fletcher,J. and Clark,A.J. (2001) Gene targeting in primary fetal fibroblasts from sheep and pig. Cloning Stem Cells, 3, 221–231. [DOI] [PubMed] [Google Scholar]
- 8.Denning C. and Priddle,H. (2003) New frontiers in gene targeting and cloning: success, application and challenges in domestic animals and human embryonic stem cells. Reproduction, 126, 1–11. [DOI] [PubMed] [Google Scholar]
- 9.Templeton N.S., Roberts,D.D. and Safer,B. (1997) Efficient gene targeting in mouse embryonic stem cells. Gene Ther., 4, 700–709. [DOI] [PubMed] [Google Scholar]
- 10.Piedrahita J.A. (2000) Targeted modification of the domestic animal genome. Theriogenology, 53, 105–116. [DOI] [PubMed] [Google Scholar]
- 11.Piedrahita J.A., Oetama,B., Bennett,G.D., van Waes,J., Kamen,B.A., Richardson,J., Lacey,S.W., Anderson,R.G. and Finnell,R.H. (1999) Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nature Genet., 23, 228–232. [DOI] [PubMed] [Google Scholar]
- 12.Piedrahita J.A., Zhang,S.H., Hagaman,J.R., Oliver,P.M. and Maeda,N. (1992) Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc. Natl Acad. Sci. USA, 89, 4471–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vazquez J.C., Nogues,C., Rucker,E.B. and Piedrahita,J.A. (1998) Factors affecting the efficiency of introducing precise genetic changes in ES cells by homologous recombination: tag-and-exchange versus the Cre-loxp system. Transgenic Res., 7, 181–193. [DOI] [PubMed] [Google Scholar]
- 14.Williams S., Sahota,V., Palmai-Pallag,T., Tebbutt,S., Walker,J. and Harris,A. (2003) Evaluation of gene targeting by homologous recombination in ovine somatic cells. Mol. Reprod. Dev., 66, 115–125. [DOI] [PubMed] [Google Scholar]
- 15.McCreath K.J., Howcroft,J., Campbell,K.H., Colman,A., Schnieke,A.E. and Kind,A.J. (2000) Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature, 405, 1066–1069. [DOI] [PubMed] [Google Scholar]
- 16.Denning C., Burl,S., Ainslie,A., Bracken,J., Dinnyes,A., Fletcher,J., King,T., Ritchie,M., Ritchie,W.A., Rollo,M., de Sousa,P., Travers,A., Wilmut,I. and Clark,A.J. (2001) Deletion of the alpha(1,3)galactosyl transferase (GGTA1) gene and the prion protein (PrP) gene in sheep. Nat. Biotechnol., 19, 559–562. [DOI] [PubMed] [Google Scholar]
- 17.Wilson G.L., Dean,B.S., Wang,G. and Dean,D.A. (1999) Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J. Biol. Chem., 274, 22025–22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludtke J.J., Zhang,G., Sebestyen,M.G. and Wolff,J.A. (1999) A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J. Cell Sci., 112, 2033–2041. [DOI] [PubMed] [Google Scholar]
- 19.Dean D.A., Dean,B.S., Muller,S. and Smith,L.C. (1999) Sequence requirements for plasmid nuclear import. Exp. Cell Res., 253, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takata M., Sasaki,M.S., Sonoda,E., Morrison,C., Hashimoto,M., Utsumi,H., Yamaguchi-Iwai,Y., Shinohara,A. and Takeda,S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundin C., Erixon,K., Arnaudeau,C., Schultz,N., Jenssen,D., Meuth,M. and Helleday,T. (2002) Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol., 22, 5869–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smithies O., Gregg,R.G., Boggs,S.S., Koralewski,M.A. and Kucherlapati,R.S. (1985) Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature, 317, 230–234. [DOI] [PubMed] [Google Scholar]
- 23.Mansour S.L., Thomas,K.R. and Capecchi,M.R. (1988) Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature, 336, 348–352. [DOI] [PubMed] [Google Scholar]
- 24.Joyner A.L., Skarnes,W.C. and Rossant,J. (1989) Production of a mutation in mouse En-2 gene by homologous recombination in embryonic stem cells. Nature, 338, 153–156. [DOI] [PubMed] [Google Scholar]
- 25.Charron J., Malynn,B.A., Robertson,E.J., Goff,S.P. and Alt,F.W. (1990) High-frequency disruption of the N-myc gene in embryonic stem and pre-B cell lines by homologous recombination. Mol. Cell. Biol., 10, 1799–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]