Abstract
Reactive oxygen species cause damage to all of the major cellular constituents, including peroxidation of lipids. Previous studies have revealed that oxidative stress, including exposure to oxidation products, affects the progression of cells through the cell division cycle. This study examined the effect of linoleic acid hydroperoxide, a lipid peroxidation product, on the yeast cell cycle. Treatment with this peroxide led to accumulation of unbudded cells in asynchronous populations, together with a budding and replication delay in synchronous ones. This observed modulation of G1 progression could be distinguished from the lethal effects of the treatment and may have been due to a checkpoint mechanism, analogous to that known to be involved in effecting cell cycle arrest in response to DNA damage. By examining several mutants sensitive to linoleic acid hydroperoxide, the YNL099c open reading frame was found to be required for the arrest. This gene (designated OCA1) encodes a putative protein tyrosine phosphatase of previously unknown function. Cells lacking OCA1 did not accumulate in G1 on treatment with linoleic acid hydroperoxide, nor did they show a budding, replication, or Start delay in synchronous cultures. Although not essential for adaptation or immediate cellular survival, OCA1 was required for growth in the presence of linoleic acid hydroperoxide, thus indicating that it may function in linking growth, stress responses, and the cell cycle. Identification of OCA1 establishes cell cycle arrest as an actively regulated response to oxidative stress and will enable further elucidation of oxidative stress-responsive signaling pathways in yeast.
INTRODUCTION
Lipid peroxidation is a process that affects cellular lipid-containing structures, such as membranes, under prooxidative conditions. In addition to directly interfering with membrane integrity and function, peroxidative damage can also give rise to further toxic metabolites including lipid hydroperoxides, such as linoleic acid hydroperoxide (LoaOOH), and further breakdown products, such as malondialdehyde and 4-hydroxynonenal (Gutteridge and Halliwell, 1990; Girotti, 1998).
Oxidative stress occurs when the cellular redox balance is disturbed in favor of prooxidants. The yeast Saccharomyces cerevisiae, like other organisms, is capable of responding to oxidative stress. The response to sublethal doses of several oxidants, or oxidation products, leads to transient induction of increased resistance to normally lethal doses. Such adaptations occur in response to hydrogen peroxide, the superoxide-generating drug menadione, and products of lipid peroxidation, such as LoaOOH and malondialdehyde (Collinson and Dawes, 1992; Jamieson, 1992; Flattery-O'Brien et al., 1993; Turton et al., 1997; Evans et al., 1998). The responses include induction of several protective enzymes such as cytosolic catalase (Ctt1p; Marchler et al., 1993) and the superoxide dismutases (Sod1p and Sod2p; Galiazzo and Labbe-Bois, 1993), but they also appear to involve more general changes in cellular physiology. For example, yeast cells exposed to H2O2 redirect carbohydrate metabolism to the production of reducing power and increase protein degradation (Godon et al., 1998). In parallel with the general switch from biosynthetic to protective functions, cellular exposure to oxidants also results in impaired progression through the cell division cycle.
Cell cycle arrest forms a part of the general physiological response of cells to stress. It occurs when cells are exposed to DNA-damaging agents, heavy metals, oxidative stress, hyperthermia, and starvation, as well as other stresses (Rahman et al., 1988; Nunes and Siede, 1996; Weinert, 1998; Philipott et al., 1998; Flattery-O'Brien and Dawes, 1998). DNA-damage-induced arrest is under the control of genetically encoded checkpoint mechanisms (Weinert, 1998). In budding yeast, the elucidation of the mechanism controlling the response was initiated with the finding that the radiation sensitive rad9 mutant was deficient in the G2–arrest response (Weinert and Hartwell, 1988).
Oxidative stress-induced arrest has been well investigated in mammalian cells (for review see Shackelford et al., 2000); however, in S. cerevisiae, its role, extent, and mechanisms of occurrence are only now being elucidated. H2O2-induced G2 arrest is dependent on the RAD9 gene, whereas RAD9-independent G1 arrest occurs in response to a superoxide generator, menadione (Nunes and Siede, 1996; Flattery-O'Brien and Dawes, 1998). Treatment of wild-type cells with diethylmalate, a thiol-specific oxidant, or dioxygen stress in a sod1 mutant also result in G1 arrest (Lee et al., 1996; Wanke et al., 1999). Lipid peroxidation is known to have an effect on cell division of mammalian cells (Poot et al., 1988; Ji et al., 1998), and 4-hydroxynonenal delays the exit from G0/G1 in S. cerevisiae (Wonisch et al., 1998).
This work aims to examine the molecular mechanisms whereby oxidative stress, in particular lipid peroxidation, affects the cell cycle. With the use of LoaOOH as a model compound to investigate the effects of lipid peroxidation on yeast, we studied the effects of this peroxide on the cell division cycle and found that it caused a delay in G1. To further the analysis, we sought to define the molecular mechanism whereby this response occurs, and we report here the identification of a gene required for the LoaOOH-induced modulation of cell cycle progression.
MATERIALS AND METHODS
Strains and Media
The yeast strains used were from the European S. cerevisiae Archive for Functional Analysis strain collection (Frankfurt, Germany) and were in the FY1679 background. The strains are referred to by their accession codes (accession codes are catalogued online at http://www.rz.uni-frankfurt.de/FB/fb16/mikro/euroscarf/index.html). The wild-type reference strain C11#8 (MATa his3Δ200 ura3–52 G418s) was obtained by dissecting the FY10087D heterozygous diploid strain. The ynl099cΔ strain used was FY10459A (MATa his3Δ200 ura3–52 leu2Δ1 trp1Δ63 YNL099c(4714)::kanMX4). The plasmids used were pYCG YNL099c, containing the YNL099c clone and the vector control pRS416 (Sikorski and Hieter, 1989; Saiz et al., 1999). Standard yeast techniques were used for sporulation, spore dissection, and transformation.
Yeast was grown in YEPD containing 2% (wt/vol) glucose, 2% (wt/vol) bactopeptone, and 1% (wt/vol) yeast extract; in simple defined complete (SDC) medium containing 2% (wt/vol) glucose, 0.5% (wt/vol) ammonium sulfate, 0.17% (wt/vol) Yeast Nitrogen Base, supplemented with Ade (10 mg/l), Arg (50 mg/l), Asp (80 mg/l), His (20 mg/l), Iso (50 mg/l), Leu (100 mg/l), Lys (50 mg/l), Met (20 mg/l), Phe (50 mg/l), Thr (100 mg/l), Trp (100 mg/l), Tyr (50 mg/l), Ura (20 mg/l), Val (140 mg/l); or in the selective SD-ura medium (as SCD but lacking uracil). For solid media, 2% (wt/vol) agarose was added. Cultures were incubated at 30°C with aeration.
LoaOOH Synthesis, Oxidant Treatment, Viability Determination, and Tests of Sensitivity and Adaptation
LoaOOH was prepared and assayed according to the method of Evans et al. (1998). Oxidants were added directly from the stocks, aqueous for H2O2, in methanol for LoaOOH and in N,N-dimethylformamide for cumene hydroperoxide (CHP). For treatment of exponential cells, cultures were routinely grown overnight to an OD600 of 0.4 in SDC or SD-ura medium; in the screen of LoaOOH-sensitive deletion mutants, cells were grown for at least four doublings starting from an actively growing culture. Aliquots of a single culture were treated in culture medium with different oxidants or oxidant concentrations. For plate tests of sensitivity, cells were grown to exponential or stationary phase in the appropriate simple defined medium and then washed and diluted to the indicated OD600 in PBS; the suspensions (10 μl) were then spotted on plates containing the concentrations of oxidants indicated.
Colony-forming ability was determined by plating appropriate dilutions on either YEPD plates (for dose response) or simple defined medium plates (for experiments on synchronous populations). Total cell numbers were determined with the use of a hemocytometer. Cell integrity (vitality) was determined by examining their permeability to oxonol essentially as described by Deere et al. (1998): ∼106 cells were washed twice with 1 ml citrate buffer (50 mM Na citrate, 100 mM NaCl, pH 7.4) and stained with 0.5 μM oxonol (Molecular Probes, Eugene, OR) in the same buffer.
For growth determination and adaptation experiments, cells were grown to early exponential phase in SDC medium and diluted to an OD600 of 0.1 in the same medium. Cultures were then placed in the wells of a microtiter plate (200 μl per well) and pretreated for 1 h with the LoaOOH concentrations indicated, after which the indicated treatment was performed and growth was monitored as OD600 in a microtiter plate reader. Note that the OD values measured in microtiter plates are not the same as the conventionally determined ones.
Microscopy, Cell Counts, and Statistical Analysis
Cells were fixed in ice-cold, 70% (vol/vol) ethanol and were observed under phase-contrast with the 100x objective on a BH-2 Olympus microscope. The cells were classified into 3 groups: unbudded cells, cells with small buds (size <50% of the mother cell), and cells with large buds. The Chi-square test was used to compare the treated with the untreated populations for each strain and to calculate the p values presented. SDs for counts data were calculated based on a binomial distribution, as a percentage of the total number of cells counted, unless otherwise noted (Devore, 1995).
α-Factor Synchrony, Oxidant Treatment and Release, and Determination of α-Factor Resistant Cells and DNA Content
The cells were synchronized with the use of α-factor essentially according to the method of Breeden (1997). The appropriate simple defined medium was used in preference to YEPD due to the presence of antioxidants such as glutathione in YEPD medium. Synchronized cultures were split into several equal aliquots, corresponding to the nontreated control and the treated samples. The cells were removed from the medium and α-factor by centrifugation and washed once with 1 volume of PBS. The cells were then resuspended in 1 volume of PBS and left untreated or treated with the oxidant for 30 min at 30°C with shaking. Cells were removed from the treatments, washed twice with 1 volume of PBS, resuspended in fresh SDC or SD-ura medium and incubated. At intervals, samples were removed for microscopy, for determination of α-factor resistant cells, or for fluorescence analysis of DNA content, and the viability was determined.
To determine the numbers of α-factor resistant cells, the removed samples were incubated for a further 45 min in the presence of α-factor (8 mg/l). The cells were than fixed and the proportions of budded cells determined. For monitoring of cellular DNA content, the samples (∼106 cells) were resuspended in water and briefly sonicated to disperse clumps. Cells were then fixed in 70% (vol/vol) ethanol, 250 mM Tris, pH 7.5, for 1 h at room temperature and stored at 4°C. Cells were washed in 50 mM Tris, pH 7.8, and RNA was degraded by overnight incubation in 1 mg/ml RNase A in the same buffer at 37°C, followed by treatment with 5 mg/ml pepsin in 55 mM HCl for 30 min. Cells were washed in FACS buffer (180 mM Tris, pH 7.5, 180 mM NaCl, 70 mM MgCl2) and stained overnight at 4°C with 55 μg/ml propidium iodide in FACS buffer. Samples were analyzed on a FASCalibur flow cytometer as described by Haase and Lew (1997).
RESULTS
LoaOOH Treatment Causes Cell Division Cycle Delay in G1
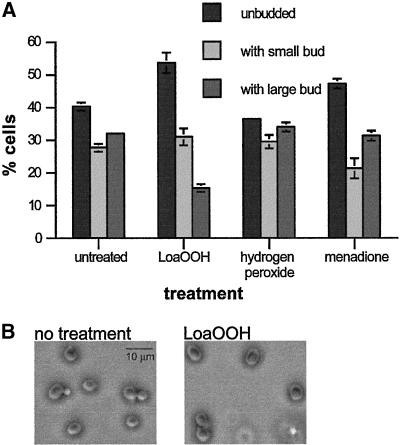
The effects of LoaOOH on cell cycle progression were initially examined by treating an asynchronous cell population with the compound and determining whether cells accumulated in a particular stage of the cell division cycle. A wild-type yeast (C11#8) population growing exponentially in SDC medium was divided into aliquots, and these were left untreated or treated with LoaOOH, menadione, or H2O2 for 2 h, which was approximately 1 doubling time for the untreated control. After treatment the cells were scored for the presence and the size of buds. LoaOOH treatment resulted in a marked increase in the proportion of unbudded cells (Figure 1). Since budding, together with DNA replication and resistance to the mating pheromone, occurs only once cells have passed Start (Pringle and Hartwell, 1981), this indicated that cells were accumulating in G1. Menadione also caused an increase in the proportion of unbudded cells (Figure 1), as shown previously (Flattery-O'Brien and Dawes, 1998), while in this strain, hydrogen peroxide treatment did not result in significant changes in the population.
Figure 1.
Treatment with LoaOOH leads to accumulation of unbudded cells. Wild-type cultures were grown to exponential phase in SDC medium, and aliquots were left untreated or treated with LoaOOH (0.04 mM), H2O2 (2 mM), or menadione (6 mM) for 2 h. Cells were fixed and scored for the presence and size of a bud. (A) Averages of 3 experiments are shown; error bars (± SD) are included if larger than 1%. (B) Photomicrographs of representative LoaOOH-treated or control cells.
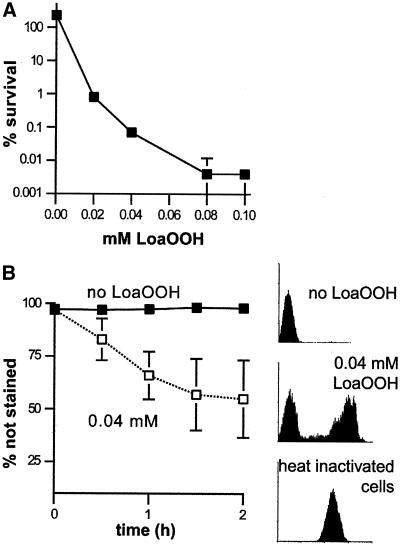
LoaOOH has been noted to be very toxic to yeast cells (Evans et al. 1998), and hence the effect of LoaOOH on cell viability under the above conditions was examined. Viability was found to be low — ∼0.1% of the initial number of colony-forming units (Figure 2A). Although not capable of forming macrocolonies after a 2-d incubation, many cells retained their immediate integrity, as shown by the use of the vital stain oxonol (Figure 2B). This may indicate that many cells were terminally arrested by the treatment. Since the treatment resulted in low viability, the observed accumulation of unbudded cells may have resulted either from heightened sensitivity of G1 cells to the treatment or from an active delay of the cell cycle in G1 in response to the treatment.
Figure 2.
Viability and vitality after treatment with LoaOOH. Wild-type cultures were grown to exponential phase, and aliquots were treated with the indicated concentrations of LoaOOH or left untreated. (A) Survival after a 2-h incubation is expressed as the colony-forming ability relative to that before treatment. Data are the averages ± SD of triplicate measurements from a representative 1 of 3 experiments. (B) Cell vitality was monitored during the 2-h incubation by staining with oxonol, as described in MATERIALS AND METHODS. Averages ± SD from 2 independent experiments are shown. Representative FACS analysis profiles of the oxonol-stained cells are included. Heat-inactivated cells illustrate the usual staining of dead cells. Note the change in axes from panels A to B. Some error bars are masked by the symbols.
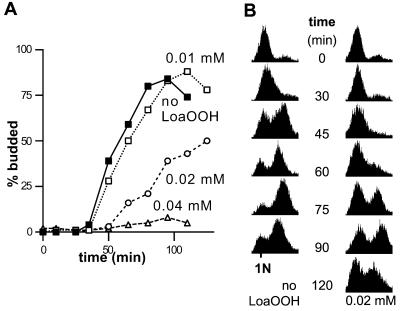
To discriminate between these 2 hypotheses and to determine if a G1 delay occurred in response to LoaOOH independent of viability loss, the effect of LoaOOH on G1 progression was examined in synchronous populations. Treatment with LoaOOH was found to lead to a budding delay that was correlated with the LoaOOH concentration and the resulting viability loss (Figure 3A). After treatment with 0.04 mM LoaOOH, the survival was only 5%, and very little budding could be seen after 2 h in fresh medium, indicating that cells died in G1 without progressing further. However, budding was also actively delayed after exposure to LoaOOH because, after treatment with 0.02 mM LoaOOH, a delay could be observed while cells mainly retained their viability, with 80% surviving. The delay was not due to LoaOOH retarding growth to the critical size necessary for Start, since no significant difference in cell volume between treated and untreated cells were found by microscopical measurements of unfixed cells during the 30 min period after resuspension in fresh medium (our unpublished results). To confirm that the delay occurred in G1, the progression through the cell cycle was also monitored by determining the DNA contents of individual cells by flow cytometry. As expected a similar replication delay was observed (Figure 3B). Thus, after LoaOOH treatment, cells delayed progression through G1.
Figure 3.
Treatment with LoaOOH leads to budding and replication delay. Wild-type cells were arrested with α-factor, and the culture was divided into aliquots, washed, resuspended in PBS, and treated with the indicated concentrations of LoaOOH for 30 min. Cells were then washed with PBS and resuspended in fresh SDC medium (time zero). Within the first 30 min, cells were plated on SDC medium for colony counts and total cell numbers were determined to give estimate of viability. (A) Progression through the cell division cycle was monitored by determining the proportion of budded cells. SDs were calculated for each sample as given in MATERIALS AND METHODS and were <3%. (B) Progression through the cell division cycle was monitored by determining the cellular DNA content as described in MATERIALS AND METHODS. FACS profiles show fluorescence on the X axis and event numbers on the Y axis. In both panels A and B, a representative of 2 independent experiments is shown.
Identification of a LoaOOH-Sensitive Mutant that Is Deficient in the Cell Cycle Response
The observed G1 delay could be due to the existence of a genetically encoded response mechanism. If so, it should be possible to obtain mutants in which this response is absent. We speculated that such mutants would also be hypersensitive to LoaOOH, but not to H2O2, because the cell cycle responses to the 2 oxidants appear different.
As part of the European Functional Analysis project, a large number of yeast deletion mutants was created. We recently screened 600 of these mutants for their hypersensitivity to 3 peroxides (H2O2, CHP, and LoaOOH) by examining the relative ability of strains to survive and grow in the presence of the oxidants (Higgins and Dawes, 1999). Eighty-three hypersensitive deletion mutants were identified including a set of 21 that were sensitive to LoaOOH but not to H2O2. Twelve of these mutants were examined for their ability to show a cell cycle response to LoaOOH by incubating exponentially growing cells with and without LoaOOH and determining the distributions of cells in different stages of budding. LoaOOH treatment resulted in a significant change in this distribution in the wild-type strain, for which the ratio of unbudded to large-budded cells nearly doubled (Table 1). A majority of the sensitive deletion mutants also showed significant population changes after addition of LoaOOH (p < 0.1), as illustrated by FY10349A (Table 1). However, the response was virtually abolished in at least 2 of the deletion strains (FY10459A and FY10023A, deleted for ORF YNL099c and PEX17, respectively) because for these the distributions of cells at different stages of budding did not change significantly after the addition of LoaOOH (Table 1).
Table 1.
Accumulation of unbudded cells in LoaOOH-sensitive deletion strains upon treatment with the hydroperoxide
| Strain | ORF/gene deleted | p | Unbudded/large
bud
|
||
|---|---|---|---|---|---|
| no LoaOOH | +LoaOOH | Change (times) | |||
| C11#8 | none | 1 × 10−4 | 1.6 | 3.1 | 1.9 |
| FY 10023A | PEX17 | 0.48 | 1.6 | 1.4 | 0.8 |
| FY 10349A | ECM33 | 0.02 | 1.6 | 2.9 | 1.8 |
| FY 10459A | YNL099c | 0.46 | 1.4 | 1.6 | 1.1 |
Wild-type and deletion strains were grown to exponential phase in SDC medium. The cultures were then split, and one part was treated with 0.04 mM LoaOOH for 2 h, while the other was left untreated. The cells at stages of budding were then counted (see Figure 1A). The Chi-square test was used to compare the treated with the untreated population for each strain, and the resulting p values are presented together with the ratios of unbudded to large-budded cells and their change upon LoaOOH treatment.
The YNL099c ORF, whose function was previously unknown, encodes a putative protein tyrosine phosphatase, indicating that it may be involved in a signal transduction cascade (Wishart and Dixon, 1998). Its sequence has homology to 2 other ORFs from S. cerevisiae, SIW14 and YNL056w, together with ORFs from several other organisms (Figure 4). The sequences show greatest homology around the putative active site containing the catalytically important CX5R motif (Figure 4; see Wishart and Dixon, 1998). Interestingly SIW14, which has 30% amino acid identity and 51% similarity with YNL099c, was isolated in a screen for mutations that were synthetic lethal with whi2Δ, a gene required for cell division cycle arrest after nutrient depletion (Binley et al., 1999). In the absence of SIW14, cells were reported to fail to arrest in G1 after nutrient depletion, to be unable to grow on nonfermentable carbon sources, and to show sensitivity to 1 M NaCl (Duffy et al., 1999). In contrast, the ynl099cΔ strain showed accumulation of unbudded cells in stationary phase, was capable of growth on glycerol, and was not sensitive to 1 M NaCl (our unpublished results). The ynl099cΔ mutant also showed no sensitivity to UV irradiation (254 nm), relative to the wild-type strain (our unpublished results). On the basis of its apparent involvement in oxidant-induced cell-cycle arrest, the YNL099c gene was designated OCA1. The function of the Oca1p was further examined by analyzing the phenotype of the available deletion mutant.
Figure 4.
Multiple sequence alignment of Ynl099cp and several of its homologues. Homology search was performed with the use of BLAST, and the sequences were aligned with CLUSTALW. Only a portion of the alignment is shown. Ynl099cp and its 2 homologues from S. cerevisiae (S.c.) Siw14p and Ynl056wp are included together with the closest homologues from Arabdidopsis thaliana (A.t., gene product of T7A14.14), Leishmania major (L.m., hypothetical protein L3291.05), and Schizosaccharomyces pombe (S.p., Pi043p). The CX5R motif is underlined. Consensus identity is indicated by black boxes, while consensus similarity is indicated with gray boxes. GeneBank numbers for the sequences are (top to bottom): CAA95895, AAC97999, CAB42360, CAA95975, CAB51762, and CAA95929.
G1 Arrest in Response to LoaOOH Requires OCA1
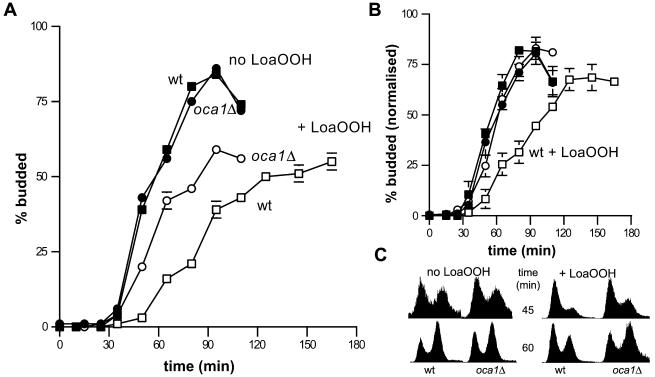
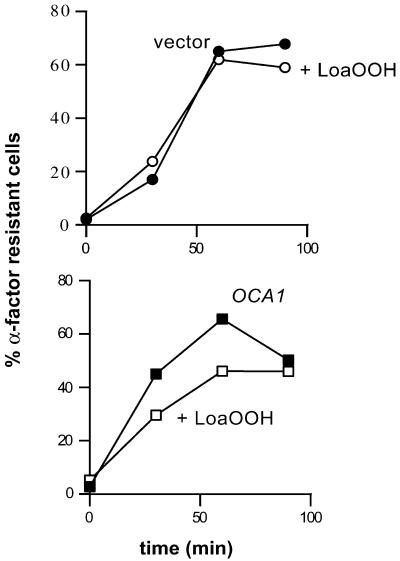
Although the oca1Δ strain showed little change in the proportion of unbudded cells after exposure to LoaOOH (Table 1), its loss of viability was equivalent to that of the wild-type strain under these conditions (see data below), indicating that it was not loss of cell survival that created the lack of response. To confirm that the oca1Δ strain was defective in the LoaOOH-induced G1 delay, α-factor synchronized populations were examined. When the mutant was exposed to 0.02 mM LoaOOH, the budding and replication delay was much shorter than in the wild-type strain (Figure 5A and 5C). To reduce any bias caused by differences in survival, the budding data were normalized for cell viability; this made the absence of a delay in the oca1Δ strain even more evident (Figure 5B), and it can be seen that all of the mutant cells that were viable after LoaOOH treatment budded with the same dynamics as the untreated control population. As expected, reintroduction of OCA1 into the oca1Δ strain on a single copy plasmid restored the budding delay in response to LoaOOH (our unpublished results). To further confirm that the lack of budding and replication delay in the oca1Δ strain reflected a true lack of cell-cycle delay, the timing of Start was monitored by determining the proportions of α-factor-sensitive cells at intervals after resuspending synchronized, LoaOOH-treated cells in fresh medium. In the absence of the Oca1p, cells did not delay crossing Start in response to LoaOOH, as indicated by the kinetics of disappearance of α-factor sensitive cells (Figure 6). Taken together, the evidence indicates that OCA1 is required for cell-cycle delay in G1 in response to LoaOOH. With this in mind, it was interesting to examine the effects of OCA1 deletion on cellular fitness with respect to the peroxide.
Figure 5.
Absence of a budding and replication delay in the oca1Δ strain. Wild-type and oca1Δ cells were treated with 0.02 mM LoaOOH as described in Figure 3A, and the timing of budding was determined. (A) Data from a representative of 2 independent experiments are presented. (B) The percentages of budded cells were normalized to the viabilities in the corresponding populations. Averages ± SE of 2 independent experiments are shown. In both panels A and B the error bars are shown if larger than 2.5%. (C) In a separate experiment, cells were treated as described for A, samples were taken at 45 and 60 min after resuspension in fresh medium and the DNA content of cells determined as described in MATERIALS AND METHODS. The data are presented as in Figure 3B, with LoaOOH treated populations shown on the right.
Figure 6.
Dependence of Start delay on OCA1. A single-copy plasmid carrying OCA1 or the vector alone as a control were separately introduced into the oca1Δ strain. Both resulting transformants were synchronized with α-factor, treated with 0.02 mM LoaOOH (open symbols), or left untreated (closed symbols), and released as described in Figure 3A, except that SD-ura replaced SDC medium. The proportions of α-factor resistant cells were determined as described in MATERIALS AND METHODS. SDs for all samples were <4%. A representative of 2 independent experiments is shown.
LoaOOH Sensitivity of the oca1Δ Strain
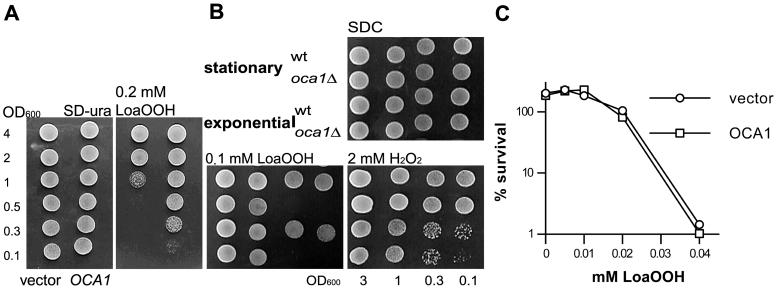
As expected from the initial screening results reintroduction of an OCA1 plasmid into the oca1Δ strain restored its resistance to LoaOOH (Figure 7A) and CHP but had no effect on its resistance to H2O2 (our unpublished results). The oca1Δ strain was more sensitive to LoaOOH than the wild-type when plated from either exponential or stationary growth phase (Figure 7B), although exponential-phase cells of both strains were more sensitive than those in stationary phase to H2O2 (Figure 7B; see also Steels et al., 1994).
Figure 7.
Absence of Oca1p results in LoaOOH sensitivity, irrespective of culture growth phase. (A) The oca1Δ strain carrying either the OCA1 clone (right colums) or the vector control (left columns) was grown in SD-ura medium to stationary phase. Cells were washed, diluted to the indicated OD600, and spotted on plates with or without LoaOOH (right and left panels respectively). Plates were photographed after 2-d growth. (B) Plate tests of sensitivity to LoaOOH and H2O2 were performed on wild-type (C11#8) and oca1Δ (FY10459A) strains grown to exponential (OD600 = 0.4) or stationary growth phase (OD600 ≈ 7), as described for A, except that SD-ura medium was replaced with SDC. In both A and B a representative of 2 experiments is shown. (C) Cell survival of the 2 transformants after 2-h incubation with different concentrations of LoaOOH was determined, as described in the legend to Figure 2A.
Both the initial screen and the plate sensitivity tests detect the ability of a strain to survive and grow in the presence of an oxidant. To determine whether the sensitivity of the oca1Δ strain to LoaOOH was due to impairment of its survival, of its growth, or both, in the presence of the peroxide, its ability to maintain viability during treatment with LoaOOH was examined. Survival did not depend on OCA1 (Figure 7C). This suggests that the sensitivity of the oca1Δ strain to LoaOOH was due to its inability to grow during exposure to the compound. To examine this possibility further, growth of the wild-type and oca1Δ strains was monitored for several hours in microtiter plates in the presence of a range of LoaOOH concentrations. Relative to the wild-type, the oca1Δ strain exhibited reduced ability to grow when exposed to 0.02 mM LoaOOH (Figure 8, no pretreatment). Hence, OCA1 was required for growth, but not cellular survival, in the presence of LoaOOH.
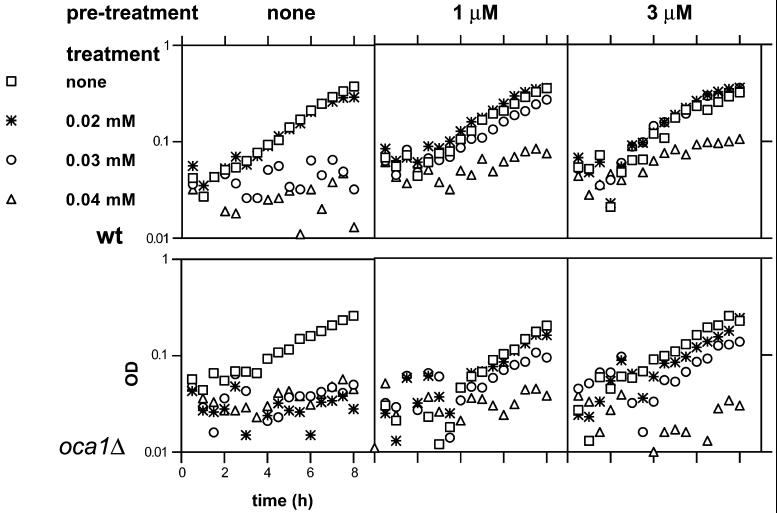
Figure 8.
Growth of wild-type and oca1Δ strains in the presence of LoaOOH and adaptation to the hydroperoxide. Wild-type (upper panels) and the oca1Δ (lower panels) strains were grown to early exponential phase and aliquoted into wells of a microtiter plate. Cells were then pretreated with the indicated concentrations of LoaOOH for 1 h. LoaOOH was then added to the indicated treatment concentrations at time zero, and growth was monitored as described in MATERIALS AND METHODS. Data from a representative of 4 experiments are shown.
The reduced growth of the oca1Δ strain during exposure to LoaOOH might have resulted from an absence of the adaptive response in the mutant strain. However, in the growth assay described above both the mutant and the wild-type strains showed better growth in the presence of LoaOOH after a pretreatment, indicating that both were capable of adaptation (Figure 8). It is interesting to note that both strains were capable of mounting an adaptive response to concentrations as low as 1 μM, which is considerably below those affecting growth or viability. The results also indicated that the oca1Δ strain may not be capable of adapting as well as the wild-type strain, because it showed less growth in the presence of 0.04 mM LoaOOH after pretreatment with 3 μM LoaOOH (Figure 8). However, it is not clear if this is a characteristic independent of its lack of ability to grow in the presence of the treatment.
DISCUSSION
Previous studies have shown that, among various stress conditions, oxidative stress results in modulation of cell cycle progression. In S. cerevisiae, the activity of the RAD9-dependent DNA-damage checkpoint pathway is essential for the response to H2O2 (Flattery-O'Brien and Dawes, 1998), whereas the response to diethylmalate requires a functional Ras-cAMP pathway (Wanke et al., 1999). The lipid peroxidation product 4-hydroxynonenal was found to cause a G1 delay, the basis of which was unclear (Wonisch et al., 1998). In this study, we demonstrated that LoaOOH treatment also causes a G1 delay, and we have identified a gene, OCA1, that is required for this delay.
What is the function of the G1 delay in response to LoaOOH? It is possible to deduce a benefit for the cell from the phenotype of the oca1Δ mutant. The G1 delay does not provide direct, immediate protection to the cells from LoaOOH, since in its absence cell survival is not impaired after an acute dose, nor is it absolutely required for adaptation to the peroxide. However, the oca1Δ strain cannot grow as well as the wild-type on plates containing LoaOOH. In this case LoaOOH is continuously present, and the G1 delay may therefore be needed to provide maximum fitness during chronic exposure. Peroxide treatment is known to result in reduction of growth and biosynthesis and an increase in cellular protective functions (Grant et al., 1998; Godon et al., 1998). G1 delay may act to strike a balance between these 2 modes of cellular operation and thus allow for net growth in the presence of a continuous low level of stress. This may be expected because a background level of lipid peroxidation would occur continuously in aerobically growing cells. At the same time, the deletion mutant may be somewhat deficient in its ability to adapt, indicating that G1 modulation may form a part of the adaptive response.
It is interesting that there were 2 levels of cellular responses that differed with respect to the concentrations at which they occurred. An adaptive response was elicited by concentrations (as low as 1 μM) that caused no net effect on either cellular survival or growth. Modulation of cell cycle progression, on the other hand, occurred at higher concentrations that did affect survival to some extent. It is possible that the G1 delay is activated in response to a different signal —one that is formed when cellular damage becomes significant. The absence of the cell division cycle response in the pex17Δ strain may shed some light on this question. Pex17p is required for the normal biogenesis of peroxisomes (Huhse et al., 1998), and hence the absence of a G1 delay in the deletion mutant may indicate that peroxisomal processing of LoaOOH is required for the compound to affect cell cycle progression.
How does Oca1p effect its function? Oca1p is a putative protein tyrosine phosphatase and may form part of a stress-activated signaling cascade. A role for Pyp1p, another protein tyrosine phosphatase, has been described in stress responses of Schizosaccharomyces pombe. The phosphatase affects the activity of a mitogen-activated protein kinase cascade homologous to the S. cerevisiae Hog1p pathway, and its involvement in the activation of this pathway in response to oxidative stress and heat shock has been reported (Samejima et al., 1997; Nguyen and Shiozati, 1999). However, the Hog1p mitogen-activated protein kinase cascade in S. cerevisiae was shown not to be activated in response to oxidative stress (Schuller et al., 1994). The cellular signaling pathways responsive to oxidative stress have not yet been thoroughly investigated in S. cerevisiae (Dawes, 1998), and further analysis of the role of Oca1p may allow a better understanding of oxidative stress signaling in this organism. Interestingly, there is growing awareness of the involvement of protein tyrosine phosphatases in oxidative stress responses in mammals and of the regulation of these enzymes by oxidants (Keyse and Emslie, 1992; Carballo et al., 1999; Barrett et al., 1999).
Oca1p is 1 of 3 homologous proteins in S. cerevisiae (Wishart and Dixon, 1998). 2 of the 3, namely Oca1p and Siw14p, encode putative protein tyrosine phosphatases. Both appear necessary for cell cycle arrest after a stress condition. It is therefore possible that both Siw14p and Oca1p may be involved in integrating different forms of stress signaling and cell cycle progression. The third protein, Ynl056wp, appears to be an inactive phosphatase, having the catalytically important cysteine replaced by a serine residue (Wishart and Dixon, 1998). Such inactive phosphatases retain the ability to bind to the phosphorylated substrates (Wishart et al., 1995; Wang et al., 2000) and can thus still modulate their activity (Wishart and Dixon, 1998). Indeed, a ynl056wΔ mutant exhibits sensitivity to caffeine (Rieger et al., 1999), an indication that it is not an inactive ORF. The functions of the 3 proteins in the cell have not been clearly identified. Their activities and the identity of their substrates may allow further insight into the molecular mechanism of the stress responses in S. cerevisiae and other organisms.
ACKNOWLEDGMENTS
We thank P. Attfield and D. Veal for their assistance with flow cytometry.
Abbreviations used:
- CHP
cumene hydroperoxide
- LoaOOH
linoleic acid hydroperoxide
- SDC
simple defined complete (medium)
Footnotes
This work was supported by Australian Research Council Grant AO-9917158 and by an Australian Postgraduate Award to N.A.
REFERENCES
- Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of protein-tyrosine phosphatase 1B. J Biol Chem. 1999;274:34543–34546. doi: 10.1074/jbc.274.49.34543. [DOI] [PubMed] [Google Scholar]
- Binley KM, Radcliffe PA, Trevethick J, Duffy KA, Sudbery PE. The yeast PRS3 gene is required for cell integrity, cell cycle arrest upon nutrient deprivation, ion homeostasis, and the proper organization of the actin cytoskeleton. Yeast. 1999;15:1459–1469. doi: 10.1002/(SICI)1097-0061(199910)15:14<1459::AID-YEA472>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Breeden LL. Alpha-factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–342. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- Carballo M, Conde M, El Bekay R, Martin-Nieto J, Camacho MJ, Monteseirin J, Conde J, Bedoya FJ, Sobrino F. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem. 1999;274:17580–17586. doi: 10.1074/jbc.274.25.17580. [DOI] [PubMed] [Google Scholar]
- Collinson LP, Dawes IW. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- Dawes IW. Stress responses. In: Dickinson JR, Schweizer M, editors. The Metabolism and Molecular Physiology of Saccharomyces cerevisiae. London: Taylor and Francis; 1998. pp. 277–326. [Google Scholar]
- Deere D, Shen J, Vesey G, Bell P, Bissinger P, Veal D. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast. 1998;14:147–160. doi: 10.1002/(SICI)1097-0061(19980130)14:2<147::AID-YEA207>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Devore JL. Probablity and Statistics for Engineering and the Sciences. 4th ed. Belmon: Wadsworth Publising Company; 1995. [Google Scholar]
- Duffy KA, Binley KM, Radcliffe PA, Sudbery PE. S/W14, a tyrosine phosphatase involved in cell integrity, the co-ordination of the cell cycle arrest upon nutrient limitation and organisation of the actin cytoskeleton. Curr Genet. 1999;35:383. doi: 10.1002/(SICI)1097-0061(199910)15:14<1459::AID-YEA472>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Evans MV, Turton HE, Grant CM, Dawes IW. Toxicity of linoleic acid hydroperoxide to Saccharomyces cerevisiae: involvement of a respiration related process for maximal sensitivity and adaptive response. J Bacteriol. 1998;180:483–490. doi: 10.1128/jb.180.3.483-490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flattery-O'Brien J, Collinson LP, Dawes IW. Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J Gen Microbiol. 1993;139:501–507. doi: 10.1099/00221287-139-3-501. [DOI] [PubMed] [Google Scholar]
- Flattery-O'Brien JA, Dawes IW. Hydrogen peroxide causes RAD9-dependent cell cycle arrest in G2 in Saccharomyces cerevisiae whereas menadione causes G1 arrest independent of RAD9 function. J Biol Chem. 1998;273:8564–8571. doi: 10.1074/jbc.273.15.8564. [DOI] [PubMed] [Google Scholar]
- Galiazzo F, Labbe-Bois R. Regulation of Cu, Zn- and Mn-superoxide dismutase transcription in Saccharomyces cerevisiae. FEBS Lett. 1993;315:197–200. doi: 10.1016/0014-5793(93)81162-s. [DOI] [PubMed] [Google Scholar]
- Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- Grant CM, Perrone G, Dawes IW. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;253:893–898. doi: 10.1006/bbrc.1998.9864. [DOI] [PubMed] [Google Scholar]
- Gutteridge JMC, Halliwell B. The measurement and mechanism of lipid peroxidation in biological systems. Trends Biochem Sci. 1990;15:129–135. doi: 10.1016/0968-0004(90)90206-q. [DOI] [PubMed] [Google Scholar]
- Haase SB, Lew DJ. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 1997;283:322–332. doi: 10.1016/s0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- Higgins VJ, Dawes IW. Phenotype screening of gene deletant strains for sensitivity to oxidative stress. Curr Genet. 1999;35:391. doi: 10.1002/yea.811. [DOI] [PubMed] [Google Scholar]
- Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau WH. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Rouzer CA, Marnett LJ, Pietenpol JA. Induction of cell cycle arrest by endogenous product of lipid peroxidation, malondialdehyde. Carcinogenesis. 1998;19:1275–1283. doi: 10.1093/carcin/19.7.1275. [DOI] [PubMed] [Google Scholar]
- Keyse SM, Emslie E. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359:664–667. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- Lee J, Romeo A, Kosman DJ. Transcriptional remodelling and G1 arrest in dioxygen stress in Saccharomyces cerevisiae. J Biol Chem. 1996;271:24885–24893. doi: 10.1074/jbc.271.40.24885. [DOI] [PubMed] [Google Scholar]
- Marchler G, Schuller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AN, Shiozaki K. Heat shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 1999;13:1653–1663. doi: 10.1101/gad.13.13.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes E, Siede W. Hyperthermia and paraquat-induced G1 arrest in the yeast Saccharomyces cerevisiae is independent of the RAD9 gene. Radiat Environ Biophys. 1996;35:55–57. doi: 10.1007/BF01211243. [DOI] [PubMed] [Google Scholar]
- Philipott CC, Rashford J, Yamaguchi IY, Rouault TA, Dancis A, Klausner RD. Cell-cycle arrest and inhibition of G1 cyclin translation by iron in AFT-1up yeast. EMBO J. 1998;17:5026–5036. doi: 10.1093/emboj/17.17.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Esterbauer H, Rabinovitch PS, Hoehn H. Disturbance of cell proliferation by 2 model compounds of lipid proliferation contradicts causative role in proliferative senescence. J Cell Physiol. 1988;137:421–429. doi: 10.1002/jcp.1041370305. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1981. pp. 97–143. [Google Scholar]
- Rahman DJR, Sudbery PE, Kelly S, Marison IW. The effects of dissolved oxygen concentration on the growth physiology of Saccharomyces cerevisiae whi2 mutants. J Gen Microbiol. 1988;134:2241–2248. doi: 10.1099/00221287-134-8-2241. [DOI] [PubMed] [Google Scholar]
- Rieger KJ, El-Alama M, Stein G, Bradshaw C, Slonimski PP, Kinsley M. Chemotyping of yeast mutants using robotics. Yeast. 1999;15:973–986. doi: 10.1002/(SICI)1097-0061(199907)15:10B<973::AID-YEA402>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Saiz JE, Santos MA, Vazquez de Aldana CR, Revuelta JL. Disruption of six unknown open reading frames from Saccharomyces cerevisiae reveals 2 genes involved in vacuolar morphogenesis and one gene required for sporulation. Yeast. 1999;15:155–164. doi: 10.1002/(SICI)1097-0061(19990130)15:2<155::AID-YEA342>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Samejima I, Mackie S, Fantes PA. Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 1997;16:6162–6170. doi: 10.1093/emboj/16.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller C, Brewster JL, Alexander MR, Gustin MC, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford RE, Kaufmann WK, Paules RS. Oxidative stress and cell cycle checkpoint function. Free Radical Biol Med. 2000;28:1387–1404. doi: 10.1016/s0891-5849(00)00224-0. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steels EL, Learmonth RP, Watson K. Stress tolerance and membrane lipid unsaturation in Saccharomyces cerevisiae grown aerobically or anaerobically. Microbiology. 1994;140:569–576. doi: 10.1099/00221287-140-3-569. [DOI] [PubMed] [Google Scholar]
- Turton HE, Dawes IW, Grant CM. Saccharomyces cerevisiae exhibits a yAP-1 mediated adaptive response to malondialdehyde. J Bacteriol. 1997;179:1096–1101. doi: 10.1128/jb.179.4.1096-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tabernero L, Zhang M, Harms E, Van Etten RL, Stauffacher CV. Crystal structure of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate. Biochemistry. 2000;39:1903–1904. doi: 10.1021/bi991348d. [DOI] [PubMed] [Google Scholar]
- Wanke V, Accorsi K, Porro D, Esposito F, Russo T, Vanoni M. In budding yeast, reactive oxygen species induce both RAS-dependent and RAS-independent cell cycle-specific arrest. Mol Microbiol. 1999;32:753–764. doi: 10.1046/j.1365-2958.1999.01391.x. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–341. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Wishart JM, Denu DM, Williams JA, Dixon JE. A single mutation converts a novel phosphotyrosine binding domain into a dual-specificity phosphatase. J Biol Chem. 1995;270:26782–26785. doi: 10.1074/jbc.270.45.26782. [DOI] [PubMed] [Google Scholar]
- Wishart MJ, Dixon JE. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem Sci. 1998;23:301–306. doi: 10.1016/s0968-0004(98)01241-9. [DOI] [PubMed] [Google Scholar]
- Wonisch W, Kohlwein SD, Schaur J, Tatzber F, Guttenberger H, Zarkovic N, Winkler R, Esterbauer H. Treatment of budding yeast Saccharomyces cerevisiae with the lipid peroxidation product 4-HNE provokes a temporary cell cycle arrest in G1 phase. Free Radical Biol Med. 1998;25:682–687. doi: 10.1016/s0891-5849(98)00110-5. [DOI] [PubMed] [Google Scholar]