Abstract
Efficient high-throughput expression of genes in mammalian cells can facilitate large-scale functional genomic studies. Towards this aim, we developed a simple yet powerful method to deliver genes into cells by cationic polymers on the surface of substrates. Transfection can be achieved by directly contacting nucleic acid–cell mixtures with the cationic substrates, e.g. polyethylenimine/collagen-coated wells. This single-step matrix-surface- mediated transfection method, termed ‘surfection’, can efficiently deliver multiple plasmids into cells and can successfully assay siRNA-mediated gene silencing. This technology represents the easiest method to transfer combinations of genes in large-scale arrays, and is a versatile tool for live-cell imaging and cell-based drug screening.
INTRODUCTION
Efficient and facile transfection of nucleic acids is conducive to biological research and clinical applications. Many gene transfer techniques have been developed including viral infection, cationic lipid-mediated and cationic polymer- mediated methods, electroporation, microinjection and biolistic bombardment. However, these methodologies usually entail complicated processes, involve delicate skills or require expensive instruments to achieve optimal results. In addition, because these methods are not readily scalable, there is a critical need for transfection methods that can be applied to large-scale cell arrays. Substrate-mediated transfection methods have been described for delivering DNA in a slow-release manner. These methods fall into three categories: (i) DNA has been embedded in poly (lactic-co-glycolic) acid (PLGA), gelatin or collagen (1–5); (ii) DNA/cationic polymer complexes have been embedded in PLGA (6,7) and collagen (6,8), or DNA/cationic polymer complexes have been immobilized on the surface of supports via biotin–neutravidin bridging (9,10); (iii) cationic polymers have also been immobilized on a solid support to create complexes with DNA (11). The cationic polymers employed for formation of DNA complexes include polylysine, dendrimer and polyethylenimine (12–14). Recently, Ziauddin and Sabatini (15) improved the solid phase transfection method and adapted it for the parallel transfection of hundreds of genes on a microarray slide. Subsequent addition of transfection reagents allowed cells growing on DNA/gelatin-printed microarray spots to take up plasmid DNA and express specific proteins in spatially distinct clusters for defining phenotypes of the cognate genes. Thus, high-throughput functional analysis of hundreds or thousands of genes is possible, for the first time, on a miniaturized glass slide. Using such ‘transfected cell arrays’, researchers have identified proteins involved in drug binding and apoptosis (15), as well as screened for effective small interfering RNA (siRNA) sequences (16,17). However, an automatic arrayer is required to produce satisfactory cell microarrays. We describe a simple, efficient and effective transfection method that can be accommodated in most laboratories for handling a few or hundreds of genes in an array format for functional characterization. This approach could potentially alleviate the bottleneck in functional genomics and has the potential for a broad range of high-throughput drug screening systems.
MATERIALS AND METHODS
Cell lines and medium
HEK293T, HeLa, CL1-0, MDCK, NIH3T3, HEP-2, H1299 and HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 mM l-glutamine and 10% fetal bovine serum. Cells were maintained in a subconfluent condition before use.
Preparation of polymer-coated slides
Polylysine-pretreated glass slides provide increased charges or roughness to allow a more even and stable coating. For this reason, the surfaces of glass slides or coverslips (Kimble Glass) were first treated with NaOH solution and washed, then soaked in poly-l-lysine solution (0.1 mg/ml; Sigma) for 1 h and washed five times with deionized water. After centrifugation at 500 r.p.m. for 5 min, the slides were dried at 40°C and exposed to UV light for sterilization. Three-millimeter holes were punched in a mini-array format in a silicone rubber sheet (Grace Bio-Labs) and placed onto the polylysine-pretreated glass slides or coverslips. The mini-wells were then coated with a mixture of gelatin (1 µg/cm2; Sigma) and different amounts of 25 kDa branched polyethylenimine (PEI) (Aldrich) ranging from 0.3 to 3 µg/cm2 with or without ligand-linked PEI (one-tenth the amount of PEI; PolyPlus-transfection), dried in a desiccator and used for transfection afterwards. The coated slides can be bulk prepared and stored in a dry cabinet for at least a month.
Preparation of polymer-coated cell culture plates
The stock solution of 25 kDa branched PEI (0.2 mg/ml) was prepared and sterilized. Equal amounts of PEI and collagen solutions were mixed together as the working solution and stored at 4°C before use. The 96-well plastic plates (Nunclon Delta Surface, NUNC) or borosilicate glass bottom plates (EZView glass bottom culture plates, IWAKI) were coated with a mixture of type I collagen (0.3–1 µg/cm2) and PEI (optimal concentration at 1–2 µg/cm2). These plates were dried in a desiccator or a vacuum oven at 40°C and used for transfection afterwards.
DNA preparation
Plasmids encoding enhanced GFP (pEGFP; Clontech), luciferase (pLuc; Clontech), nuclear fibroblast growth factor 3 binding protein (NoBP) fused with GFP (pNoBP-GFP, a gift from Dr J.J. Tsai-Wu at National Taiwan University Hospital), collapsin response mediator protein-1 (CRMP-1) fused with GFP (pCRMP-GFP) and coral fluorescent protein DsRed (pDsRed; Clontech), under the control of the cytomegalovirus promoter, were amplified in the DH5α strain of Escherichia coli and purified using the Plasmid Maxiprep Kit (Qiagen).
Transfection procedure
Cells (2 × 103–4 × 104) suspended in culture medium (20–100 µl) containing 10% fetal calf serum and 0.2–1 µg of plasmid DNA were added to each of the polymer-coated wells. After incubation at 37°C in 5% CO2 in air for 24–48 h, the relative fluorescent intensity and high resolution images were recorded with a fluorescent microscope. The relative fluorescent intensity was quantified by using MetaMorph Imaging System software (Universal Imaging Corporation). Data were normalized to a negative control such as pLuc-transfected cells.
shRNA design and gene silencing
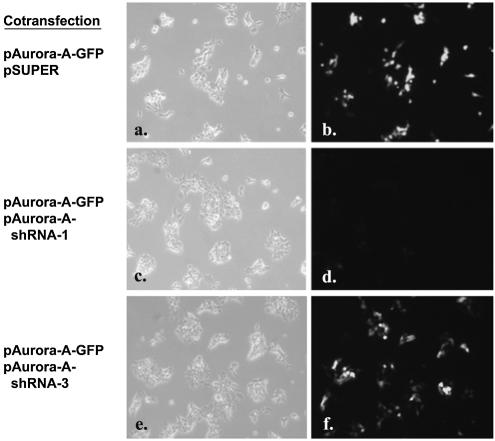
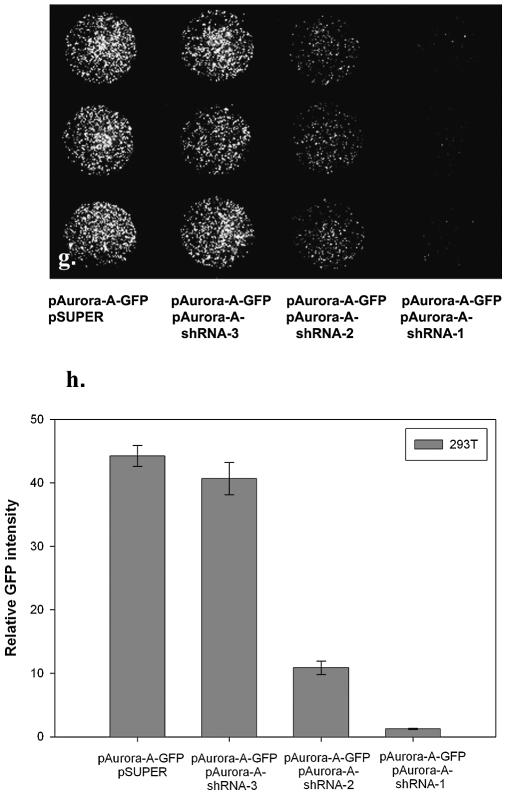
pSUPER plasmid (OligoEngine) was used to express the short hairpin RNA (shRNA) against the target gene. The effective pAurora-A-shRNA-1 was constructed based on a previously reported siRNA sequence against Aurora-A. The corresponding oligonucleotide sequence 5′GATCCCCATGCCCTGTCTTACTGTCATTCAAGAGATGACAGTAAGACAGGGCATTTTTTGGAAA3′ was synthesized, annealed, digested with BglII and HindIII and ligated into the pSUPER vector. Similarly, one ineffective pAurora-A-shRNA-3 and one partially active pAurora-A-shRNA-2 construct with corresponding oligonucleotide sequences 5′GATCCCCGCACATCAACTTGAGCCTCTTCAAGAGAGAGGCTCAAGTTGATGTGCTTTTTGGAAA3′ and 5′GATCCCCTTGAGCCT CCCTCCGTTTTTCAAGAGAAAACGGAGGGAGGCTCAAGTTTTTGGAAA3′ were used as controls. The 19 nucleotides of the Aurora-A target sequence are indicated in capitals. 293T cells were cotransfected with equal amounts (0.15 µg of each plasmid per well) of reporter plasmid containing Aurora-A gene fused to GFP (pAurora-A-GFP) and pSUPER vector alone, or pAurora-A-GFP and pAurora-A-shRNA-1, -2, -3, respectively. After transfection for 40 h, cells were subsequently fixed for 20 min in 3.7% paraformaldehyde solution. Fluorescent images were taken and the relative fluorescent intensities were quantified using MetaMorph Imaging System software.
RESULTS AND DISCUSSION
Substrate-mediated gene transfer methods have been described for delivering DNA in a slow-release manner (1–11). Owing to low transfection efficiency, they are not suitable for high-throughput transfection. Here, we developed a simple yet powerful method to deliver genes into cells by coating cationic polymers on the surface of substrates. The positively charged surface attracts negatively charged nucleic acids. As a result, the nucleic acids concentrate around the cells that adhere to the surface and are transfected into the cells with high efficiency. To enhance gene uptake, such cationic polymers can be mixed with ligand-linked cationic polymers before being coated onto the support. The cationic polymers can also be mixed with biocompatible biopolymers, such as collagen, gelatin and other extracellular matrix proteins. The biocompatible biopolymers can attenuate the toxicity of the cationic polymers, facilitate affixation of the cationic polymers to the support and provide a favorable environment for cell growth.
Using human HeLa and 293T cells, we optimized the single-step matrix-surface-mediated transfection method termed ‘surfection’ on polymer-coated mini-wells. Briefly, 3 mm holes were punched in a mini-array format in a silicone rubber sheet and placed onto polylysine-pretreated glass slides or coverslips. The mini-wells were then coated with a mixture of gelatin and PEI (optimal concentrations 1–2 µg/cm2, individually) (Fig. 1) with or without ligand-linked PEI (one-tenth the amount of PEI) and dried in a desiccator. Cells suspended in culture medium containing 10% fetal calf serum and plasmid DNA were added to the mini-wells. In the wells, DNA probably self-assembled with surface-exposed PEI and was picked up by cells during cell attachment. The noncovalent-bound PEI plays important roles in facilitating DNA condensation, cellular uptake, endosomal escape and nuclear transport (14). These four steps are all important for a transfection reagent, such as PEI, to achieve high transfection efficiency. Unexpectedly, serum in the culture medium did not interfere with the ‘surfection’ process. The cell–DNA mixture can be transferred to each well manually or by a robotic microdispenser. Alternatively, DNA solution can be transferred to the wells before the cells are added. This situation is especially suitable for automatic manipulation of multiple combinations of DNA, cells and drugs. The transfected cell array was monitored under a fluorescent microscope 24–48 h after seeding. Figure 2 shows that three concentrations of plasmid DNA encoding green fluorescent protein (pEGFP) were equally effective for the transfection of HeLa (Fig. 2a) and 293T (Fig. 2b) cells. The GFP-positive cells were evident on the entire surface of the wells on glass slides (Fig. 2a and b) and in 96-well plates (Fig. 2e) with a transfection frequency of up to 90%. The green fluorescence was not due to any artifacts from cells, reagents or the process, since cells treated the same way with luciferase-expressing vector did not display any fluorescence (Fig. 2g). Adding ligand-linked PEI or replacing gelatin with collagen I substantially benefited cell attachment and transfection efficiency (Fig. 2e). High transfection efficiency of several mammalian cell lines could be achieved by ‘surfection’, when these cells were seeded at 50–80% confluency (Fig. 2c). Furthermore, we were able to observe the subcellular distribution of proteins after transfecting human 293T cells with genes encoding CRMP-1 (18) fused with GFP (Fig. 3a, cytoplasm), NoBP (19) fused with GFP (Fig. 3a, nucleoli) and the coral fluorescent protein DsRed (20) (Fig. 3b). These three ectopically expressed fluorescent proteins, one in red and two in green but in distinct cellular locations, coexisted in at least 20% of the 293T (Fig. 3c, merged picture) and HeLa cells (data not shown). The efficiency of gene transfer and cell viability were usually much better than the traditional transfection methods using cationic lipids, cationic polymers or the combination [25–90% versus 5–50% as in Lee et al. (21)]. Since cell shape after ‘surfection’ was well maintained (Figs 2e and 3), it is feasible to perform live-cell imaging to analyze promoter function or protein–protein interactions during signal transduction. It is also easy to turn the transfected cell array into an expressed protein array for proteomic studies (22).
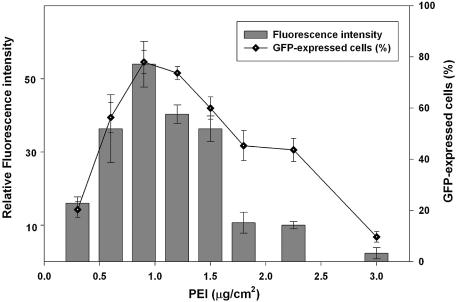
Figure 1.
Effect of PEI concentration on transfection efficiency in 293T cells. The mini-wells (3 mm in diameter) were coated with mixtures of PEI (0.3– 3.0 µg/cm2) and gelatin (1 µg/cm2). Six thousand 293T cells in culture medium containing 10% fetal calf serum and pEGFP plasmid DNA (0.3 µg/well) were aliquoted directly into each well. The fluorescent image was taken 40 h after seeding the cells. Data are expressed as a percentage of GFP-expressing cells in the total cell population, and relative fluorescent intensity using MetaMorph software (mean ± standard deviation, obtained from triplicate wells) with luciferase DNA (pLuc) transfected cells as the background control. The experiment was repeated three times with similar results.
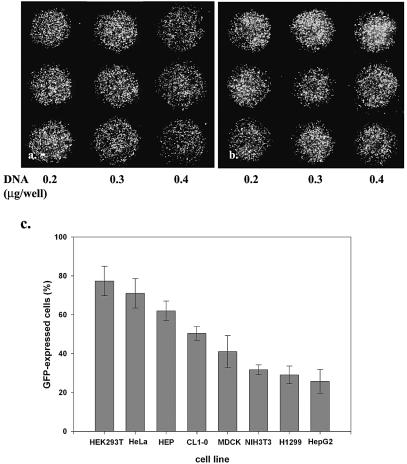
Figure 2.
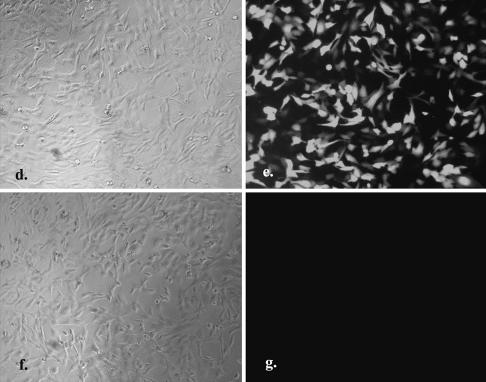
Genes delivered into cultured cells in polymer-coated wells. The mini-wells (3 mm in diameter) were coated with a mixture of PEI (1 µg/cm2) and gelatin (1 µg/cm2). Cells (2000–8000) in culture medium containing 10% fetal calf serum and plasmid DNA were aliquoted directly into each well. The fluorescent image was taken 40 h after seeding the cells. The dose effect of pEGFP DNA (0.2, 0.3 and 0.4 µg/well, respectively) on the transfection of (a) HeLa and (b) 293T cells is shown in triplicate. Magnification 40×. (c) The relative transfection efficiency of various cell lines was quantified by counting GFP-expressed cells in the total cell population (mean ± standard deviation, obtained from triplicate wells). A similar experiment was performed in a 96-well plate coated with PEI/RGD-PEI/collagen (1.8, 0.18 and 0.3 µg/cm2, individually) mixture using HeLa cells (2 × 104) and 1 µg of pEGFP (d and e) or pLuc (f and g). The fluorescent image was taken 24 h after seeding the cells. Magnification 100×. The experiments were repeated three times with similar results.
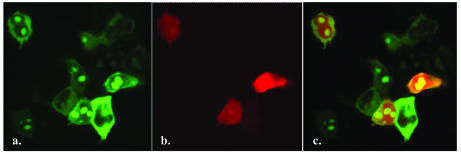
Figure 3.
High efficiency of multiple gene transfer in cultured cells. Five thousand 293T cells in culture medium containing 10% fetal calf serum and three plasmid DNAs (0.1 µg/well, individually) were aliquoted directly into the PEI/gelatin-coated wells (3 mm in diameter). The fluorescent image was taken 40 h after seeding the cells. Panels show the subcellular location of CRMP-1 protein fused with GFP (a, cytoplasm), NoBP protein fused with GFP (a, nucleoli), and DsRed fluorescent protein (b) in human 293T cells after triple gene transfer. (c) is the merged picture of (a) and (b). Magnification 400×. The cells were 80% confluent when the image was recorded.
Recently, short oligonucleotides, e.g. ribozymes (23,24) and siRNA (24), have demonstrated great potential for analyzing gene function in mammalian cells. Target validation, sequence prediction and large-scale efficacy screening are major barriers for the discovery and development of nucleic acid therapeutics. Further, delivering large amount of siRNA or small hairpin RNA (shRNA) has the potential to induce interferon response (25,26) that might affect the future use of siRNAs in the study of gene function and as therapeutic agents in vivo. One way to avoid this pitfall is to use the lowest effective dose of siRNA or shRNA vector (26). Therefore the efficacy of these double-stranded RNAs has to be carefully determined in vitro. To determine whether our transient transfection technology can be employed to assay shRNA-mediated gene silencing, we cotransfected a target-reporter vector containing full-length Aurora-A protein kinase (27) with enhanced GFP fused to the C-terminus (pAurora-A-GFP) and the pSUPER vector (28) expressing a previously reported siRNA sequence against Aurora-A (27) (pAurora-A-shRNA-1). Cotransfection of pAurora-A-GFP and pAurora-A-shRNA-1 (1:1 w/w) diminished cellular green fluorescence by >90% (Fig. 4d, g and h) compared with cotransfection of pAurora-A-GFP and control vectors (pSUPER or pAurora-A-shRNA-3) (Fig. 4b and f), demonstrating that ‘surfection’ of shRNA is effective. Thus, the ‘surfection’ platform can be tailored to facilitate the large-scale identification and functional validation of effective siRNA or ribozymes in mammalian systems.
Figure 4.
shRNA-mediated gene silencing. Five thousand 293T cells in culture medium containing 10% fetal calf serum and plasmid DNA were aliquoted directly into the PEI/gelatin-coated wells (3 mm in diameter). These cells were cotransfected with equal amounts (0.15 µg of each plasmid per well) of reporter plasmid containing Aurora-A gene fused to GFP (pAurora-A-GFP) and pSUPER vector alone (a and b), pAurora-A-GFP and pAurora-A-shRNA-1 (c and d) or pAurora-A-GFP and pAurora-A-shRNA-3 (e and f). The fluorescent image was taken 40 h after seeding the cells. Magnification 100×. In a separate experiment, pAurora-A-GFP was cotransfected with pSUPER vector or with pAurora-A-shRNA-1, 2, 3, respectively. After transfection of 293T cells for 40 h, cells were subsequently fixed for 20 minutes in 3.7% paraformaldehyde solution. The fluorescent image (magnification 40×) was taken (g) and the relative fluorescent intensity was quantified using MetaMorph Imaging System software (h) (mean ± standard deviation, obtained from triplicate wells) with luciferase DNA (pLuc)-transfected cells as the background control. The experiment was repeated three times with similar results.
Ziauddin and Sabatini (15) described a novel cell-based microarray system for identifying the cellular function of gene products. The advantage of their system is that it can handle hundreds or thousands of genes for functional analysis in a parallel format (15,16,29–31). But in addition to the need for an arrayer, there are technical problems in mixing and printing that must be solved before satisfactory cell microarrays can be produced. Here we have demonstrated an efficient and effective transfection method that can be accommodated to most laboratories for handling a few or hundreds of samples. With the availability of these polymer-coated devices that are bulk prepared, transfection can be achieved by simply contacting nucleic acids and cells with the cationic substrate without the need to prepare nucleic acid–lipid or nucleic acid–polymer complexes. The ‘surfection’ technology is also the easiest way to transfer multiple genes into cells. This less tedious method provides high-efficiency transfection in several mammalian cell lines and can be employed as a versatile and powerful tool for cell-based drug screening using the automated set-up (32–34). The transfected cell array described here has potential for a wide range of studies in the fields of functional genomics, high-throughput screening and tissue engineering.
REFERENCES
- 1.Shea L.D., Smiley,E., Bonadio,J. and Mooney,D.J. (1999) DNA delivery from polymer matrices for tissue engineering. Nat. Biotechnol., 17, 551–554. [DOI] [PubMed] [Google Scholar]
- 2.Klugherz B.D., Jones,P.L., Cui,X., Chen,W., Meneveau,N.F., DeFelice,S., Connolly,J., Wilensky,R.L. and Levy,R.J. (2000) Gene delivery from a DNA controlled-release stent in porcine coronary arteries. Nat. Biotechnol., 18, 1181–1184. [DOI] [PubMed] [Google Scholar]
- 3.Honma K., Ochiya,T., Nagahara,S., Sano,A., Yamamoto,H., Hirai,K., Aso,Y. and Terada,M. (2001) Atelocollagen-based gene transfer in cells allows high-throughput screening of gene functions. Biochem. Biophys. Res. Commun., 289, 1075–1081. [DOI] [PubMed] [Google Scholar]
- 4.Perlstein I., Connolly,J.M., Cui,X., Song,C., Li,Q., Jones,P.L., Lu,Z., DeFelice,S., Klugherz,B., Wilensky,R. et al. (2003) DNA delivery from an intravascular stent with a denatured collagen-polylactic-polyglycolic acid-controlled release coating: mechanisms of enhanced transfection. Gene Ther., 10, 1420–1428. [DOI] [PubMed] [Google Scholar]
- 5.Jang J.H. and Shea,L.D. (2003) Controllable delivery of non-viral DNA from porous scaffolds. J. Control. Release, 86, 157–168. [DOI] [PubMed] [Google Scholar]
- 6.Bielinska A.U., Yen,A., Wu,H.L., Zahos,K.M., Sun,R., Weiner,N.D., Baker,J.R.,Jr and Roessler,B.J. (2000) Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials, 21, 877–887. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y.C., Connell,M., Park,Y., Mooney,D.J. and Rice,K.G. (2003) Fabrication and in vitro testing of polymeric delivery system for condensed DNA. J. Biomed. Mater. Res., 67A, 1384–1392. [DOI] [PubMed] [Google Scholar]
- 8.Scherer F., Schillinger,U., Putz,U., Stemberger,A. and Plank,C. (2002) Nonviral vector loaded collagen sponges for sustained gene delivery in vitro and in vivo. J. Gene Med., 4, 634–643. [DOI] [PubMed] [Google Scholar]
- 9.Segura T. and Shea,L.D. (2002) Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug. Chem., 13, 621–629. [DOI] [PubMed] [Google Scholar]
- 10.Segura T., Volk,M.J. and Shea,L.D. (2003) Substrate-mediated DNA delivery: role of the cationic polymer structure and extent of modification. J. Control. Release, 93, 69–84. [DOI] [PubMed] [Google Scholar]
- 11.Zheng J., Manuel,W.S. and Hornsby,P.J. (2000) Transfection of cells mediated by biodegradable polymer materials with surface-bound polyethyleneimine. Biotechnol. Prog., 16, 254–257. [DOI] [PubMed] [Google Scholar]
- 12.Haensler J. and Szoka,F.C.,Jr (1993) Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug. Chem., 4, 372–379. [DOI] [PubMed] [Google Scholar]
- 13.Boussif O., Lezoualc’h,F., Zanta,M.A., Mergny,M.D., Scherman,D., Demeneix,B. and Behr,J.P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas M. and Klibanov,A.M. (2003) Non-viral gene therapy: polycation-mediated DNA delivery. Appl. Microbiol. Biotechnol., 62, 27–34. [DOI] [PubMed] [Google Scholar]
- 15.Ziauddin J. and Sabatini,D.M. (2001) Microarrays of cells expressing defined cDNAs. Nature, 411, 107–110. [DOI] [PubMed] [Google Scholar]
- 16.Kumar R., Conklin,D.S. and Mittal,V. (2003) High-throughput selection of effective RNAi probes for gene silencing. Genome Res., 13, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mousses S., Mousses,S., Caplen,N.J., Cornelison,R., Weaver,D., Basik,M., Hautaniemi,S., Elkahloun,A.G., Lotufo,R.A., Choudary,A. et al. (2003) RNAi microarray analysis in cultured mammalian cells. Genome Res., 13, 2341–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shih J.Y., Yang,S.C., Hong,T.M., Yuan,A., Chen,J.J., Yu,C.J., Chang,Y.L., Lee,Y.C., Peck,K., Wu,C.W. et al. (2001) Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. J. Natl Cancer Inst., 93, 1392–1400. [DOI] [PubMed] [Google Scholar]
- 19.Reimers K., Antoine,M., Zapatka,M., Blecken,V., Dickson,C. and Kiefer,P. (2001) NoBP, a nuclear fibroblast growth factor 3 binding protein, is cell cycle regulated and promotes cell growth. Mol. Cell. Biol., 21, 4996–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baird G.S., Zacharias,D.A. and Tsien,R.Y. (2000) Biochemistry, mutagenesis and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl Acad. Sci. USA, 97, 11984–11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C.-H., Ni,Y.-H., Chen,C.-C., Chou,C. and Chang,F.-H. (2003) Synergistic effect of polyethylenimine and cationic liposomes in nucleic acid delivery to human cancer cells. Biochim. Biophys. Acta, 1611, 55–62. [DOI] [PubMed] [Google Scholar]
- 22.Phizicky E., Bastiaens,P.I., Zhu,H., Snyder,M. and Fields,S. (2003) Protein analysis on a proteomic scale. Nature, 422, 208–215. [DOI] [PubMed] [Google Scholar]
- 23.Suyama E., Kawasaki,H., Nakajima,M. and Taira,K. (2003) Identification of genes involved in cell invasion by using a library of randomized hybrid ribozymes. Proc. Natl Acad. Sci. USA, 100, 5616–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherer L.J. and Rossi,J.J. (2003) Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol., 21, 1457–1465. [DOI] [PubMed] [Google Scholar]
- 25.Sledz C.A, Holko,M., de Veer,M.J., Silverman,R.H. and Williams,B.R. (2003) Activation of the interferon system by short-interfering RNAs. Nature Cell Biol., 5, 834–839. [DOI] [PubMed] [Google Scholar]
- 26.Bridge A.J., Pebernard,S., Ducraux,A., Nicoulaz,A.L. and Iggo,R. (2003) Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genet., 34, 263–264. [DOI] [PubMed] [Google Scholar]
- 27.Kufer T.A., Sillje,H.H., Korner,R., Gruss,O.J., Meraldi,P. and Nigg,E.A. (2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol., 158, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 29.Blagoev B. and Pandey,A. (2001) Microarrays go live–new prospects for proteomics. Trends Biochem. Sci., 26, 639–641. [DOI] [PubMed] [Google Scholar]
- 30.Bailey S.N., Wu,R.Z. and Sabatini,D.M. (2002) Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov. Today, 7, S113–S118. [DOI] [PubMed] [Google Scholar]
- 31.Wu R.Z., Bailey,S.N. and Sabatini,D.M. (2002) Cell-biological applications of transfected-cell microarrays. Trends Cell Biol., 12, 485–488. [DOI] [PubMed] [Google Scholar]
- 32.Regelin A.E., Fernholz,E., Krug,H.F. and Massing,U. (2001) High throughput screening method for identification of new lipofection reagents. J. Biomol. Screen., 6, 245–254. [DOI] [PubMed] [Google Scholar]
- 33.Liebel U., Starkuviene,V., Erfle,H., Simpson,J.C., Poustka,A., Wiemann,S. and Pepperkok,R. (2003) A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett., 554, 394–398. [DOI] [PubMed] [Google Scholar]
- 34.Chanda S.K. and Caldwell,J.S. (2003) Fulfilling the promise: drug discovery in the post-genomic era. Drug Discov. Today, 8, 168–174. [DOI] [PubMed] [Google Scholar]