Abstract
Strawberry (Fragaria spp) is an emerging model for the development of basic genomics and recombinant DNA studies among rosaceous crops. Functional genomic and molecular studies involve relative quantification of gene expression under experimental conditions of interest. Accuracy and reliability are dependent upon the choice of an optimal reference control transcript. There is no information available on validated endogenous reference genes for use in studies testing strawberry-pathogen interactions. Thirteen potential pre-selected strawberry reference genes were tested against different tissues, strawberry cultivars, biotic stresses, ripening and senescent conditions, and SA/JA treatments. Evaluation of reference candidate’s suitability was analyzed by five different methodologies, and information was merged to identify best reference transcripts. A combination of all five methods was used for selective classification of reference genes. The resulting superior reference genes, FaRIB413, FaACTIN, FaEF1α and FaGAPDH2 are strongly recommended as control genes for relative quantification of gene expression in strawberry. This report constitutes the first systematic study to identify and validate optimal reference genes for accurate normalization of gene expression in strawberry plant defense response studies.
Introduction
Transcriptomic analyses are essential in understanding complex molecular processes occurring in plants. Although global evaluation techniques such as microarrays or RNAseq provide a representative snapshot of a transcriptome, these techniques can only be practically applied to a limited number of tissues, treatments or time points. The data found by global expression techniques need to be then considered carefully, typically using relative quantification of gene expression by quantitative reverse transcription (RTqPCR). This method is used as a primary source of in-depth molecular expression information for a smaller set of gene candidates due to its wide range of quantification, reproducibility, and higher precision and accuracy [1], [2], [3]. However, this approach requires knowledge of stably expressed reference genes for data normalization of target genes under specific experimental conditions. Failure to use an appropriate reference or internal control gene may result in biased gene expression profiles, as well as low reproducibility. Consequently, either only gross changes in gene expression level are declared statistically significant, or the pattern of gene expression is inaccurately characterized [4], [5].
To date, some of the best known and most frequently used reference gene transcripts for RTqPCR in plants and animals include those coding for 18S rRNA, glyceraldehyde-3-phosphate dehydrogenase, elongation factor-1α, actin, and α- and β-tubulin [6], [7], [8], [9], [10], [11], [12], [13]. These genes have been recognized as stably expressed housekeeping genes, and they have been historically used as reference genes in many plants when normalizing RNA-gel blots and semi-quantitative PCR. However, numerous reports have indicated that transcript accumulation is not always consistent under some experimental conditions or across tissues. Such variation, may introduce a significant level of error in interpreting the actual expression pattern of a target gene [14], [15]. Identification of most appropriate and highly-stable internal reference genes for normalization in any given experimental plant system is a prerequisite and compulsory step to obtain reliable and reproducible results from RTqPCR. A strong reference is the foundation of accurate RTqPCR analyses following the golden rules which have been detailed recently in Udvardi et al. [16].
Over the last few years efforts have been made to identify suitable reference genes for quantification of gene expression in model plant species such as Arabidopsis [17]. Efforts have been extended to crop plants such as pea [18], banana [19], [20], sulla [21], zucchini [22], and citrus [23]. However, reference genes still have yet been identified and tested in other species of high agricultural interest including strawberry (Fragaria spp), a small fruit crop of great value throughout the world (FAOSTAT Agriculture Data [http://faostat.fao.org/, updated 7 aug 2012]).
Due to its broad horticultural importance and relatively close relationship to other valuable rosaceous crops, strawberry has been proposed as a model for functional genomics and transgenic studies within the Rosaceae [24], [25]. Strawberry’s rapid cycling, fast growth and relative transformability make it an attractive system for functional evaluation of genes associated with plant traits not testable in Arabidopsis. An increasing number of molecular studies are being reported in this species. Many of these studies have performed RTqPCR analysis using traditional reference genes to describe a wide variety of molecular events occurring in strawberry. The technique has been used to query gene expression during plant development, fruit ripening, aroma production, and responses to many biotic and abiotic stresses [26], [27], [28], [29], [30]. However, the suite of strawberry reference genes has not been carefully vetted to determine their optimal suitability for comparative expression analyses across a range of conditions, tissues or treatments.
It is necessary to identify candidate genes specifically chosen for transcript normalization for the conditions under study [31], [32]. Also, when using only one reference gene, its stability cannot be properly evaluated. The use of multiple reference genes does not only produce more reliable data, it permits an internal evaluation of the stability of these reference transcripts as well.
In the present study a subset of candidate reference genes for strawberry RTqPCR normalization in plant defense studies were identified and tested. Candidates were evaluated across a range of forty-eight situations distributed over seven experimental conditions including fruit ripening stages, biotic stress after Colletotrichum acutatum infection, and treatments with plant hormones such as SA and MeJA. Also, different cultivars of strawberry (Fragaria × ananassa), and growth conditions were tested. Recommendations for the use of these candidate genes are provided to ensure an accurate normalization of transcript level under a given condition in strawberry gene expression studies by RTqPCR.
Results
Selection of Candidate Reference Genes in Strawberry for Gene Expression Analysis
Candidate genes were selected for further analysis based on in-house data and information obtained from a range of microarrays experiments ([33], Amil-Ruiz et al., unpublished). Specific strawberry transcripts have been identified that exhibit a high degree of stability among biological replicates and in varying experimental conditions. Due to the fact that low abundance transcripts generally show high variation in their basal expression [34] they were not considered further. The analysis was performed only with candidates whose primers match prescribed conditions described below. In addition, the analysis sought to examine transcripts representing a cross-section of functional diversity to avoid a putative co-regulation effect among genes that may respond in parallel in response to a particular experimental assay. Such precautions are a prerequisite for one of the statistical procedures (the geNORM algorithm) reported to identify stably expressed genes [4].
Under all of these restrictive conditions, thirteen preselected candidate genes were identified (Table 1). These genes encode molecular components associated with a wide variety of biological functions in plant cell physiology such as 18S rRNA (gene FaRIB413), a ribosome component; GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (genes FaGAPDH1 and FaGAPDH2), an essential enzyme for carbohydrate metabolism in cytoplasm; ELONGATION FACTOR-1α (gene FaEF1α), a component of the protein synthesis machinery; ACTIN (gene FaACTIN), α-TUBULIN (gene FaTUBα) and β-TUBULIN (gene FaTUBβ), major components of microfilament and microtubule of the cytoskeleton, respectively; the UBIQUTIN CONJUGATING ENZYME E2 (gene FaUBQ1), a basic component of the ubiquitin-mediated protein tagging system; chromatin remodeling protein CHC1 (gene FaCHC1), an essential part of the chromodomain remodeling complex; an S-adenosyl-L-methionine-dependent methyltransferase (gene FaMT1), an enzyme implicated in secondary metabolism; a strawberry ortholog of the Arabidopsis AtBZIP61 regulatory transcription factor (gene FaBZIP1); a mitochondrial import inner membrane translocase (gene FaTIM1); a protein with a forkhead-associated domain and unknown molecular function (gene FaFHA1). In addition, the FaWRKY1 gene, a previously reported strawberry gene known to respond to all the different biological conditions used in this study [30], was chosen as a target gene to test the validity of these strawberry candidate genes as good reference genes in RTqPCR analyses.
Table 1. Information of selected genes after evaluation, and characteristics of PCR products and primers used in this analysis.
| Fragaria x ananassagene ID | Fragaria vesca orthologe (a) | Gene description | Oligo orientation | Sequence (5′-3′) | Primer melting temp(°C) (b) | Product size (bp) | Optimal anealing (°C) (c) | PCR product melting temp (°C) (d) | PCR efficiency ± SD (e) | Ref (f) |
| FaGAPDH2 | gene07104 | Glyceraldehyde-3-phosphate dehydrogenase | sense chain | CCCAAGTAAGGATGCCCCCATGTTCG | 82,1 | 117 | 65 | 85 | 1,769±0,028 | [26] |
| anti-sense chain | TTGGCAAGGGGAGCAAGACAGTTGGTAG | 81,2 | ||||||||
| FaUBQ1 | gene08438 | Ubiquitin E2 | sense chain | CCCATCTCCGACAACCGCCACATCTAAA | 83,1 | 130 | 66 | 89,5 | 1,727±0,028 | |
| anti-sense chain | CCGCCGCCCCAATCTTCTGTACTCC | 82,7 | ||||||||
| FaGAPDH1 | gene18492 | Glyceraldehyde-3-phosphate dehydrogenase | sense chain | GGCTTCTATCTCAACCGGCTCGTCTT | 77,7 | 121 | 65 | 85 | 1,925±0,025 | [51] |
| anti-sense chain | CTTCCCACTGCTCCCTGATCTCTGATAC | 77,3 | ||||||||
| FaEF1α | gene28639, gene28622, gene23217 | Elongation factor 1-alpha | sense chain | TGGATTTGAGGGTGACAACATGA | 73,1 | 145 | 65 | 87 | 1,798±0,028 | [27] |
| anti-sense chain | GTATACATCCTGAAGTGGTAGACGGAGG | 73,3 | ||||||||
| FaTUBα | gene01798, gene05604, gene03851, gene26908 | Tubulin alpha | sense chain | CATGGCTTGCTGTTTGATGTACCGTG | 78,5 | 156 | 65 | 87 | 1,765±0,027 | |
| anti-sense chain | GGGACAACAGTGGGTGGCTGGTAGTT | 79,1 | ||||||||
| FaTUBβ | gene07781, gene13266, gene20192, gene08531, gene18775 | Tubulin beta | sense chain | ACACTGTTGTGGAGCCTTACAATGCTAC | 74,7 | 172 | 65 | 85,5 | 1,773±0,024 | |
| anti-sense chain | GACATTGTTGCGGAGATCAAGTGATT | 74,8 | ||||||||
| FaACTIN | gene26612, gene18390, gene22626, gene18570, gene14112, gene01836 | Actin | sense chain | GGGCCAGAAAGATGCTTATGTCGG | 77 | 152 | 65 | 85 | 1,801±0,029 | [28] |
| anti-sense chain | GGGCAACACGAAGCTCATTGTAGAAG | 76,2 | ||||||||
| FaCHC1 | gene25887 | SWI/SNF complex component | sense chain | CATCTGTTTCCGCCACAACCTATACAT | 75,1 | 156 | 65 | 83,5 | 1,762±0,03 | |
| anti-sense chain | TTTGTTTTTCTCTGAGTTGGCCATTAGA | 74,1 | ||||||||
| FaMT1 | gene10517 | S-adenosyl-L-methionine-dependent methyltransferase | sense chain | AGGAGATAAGATAGCATTCGAAGTACCC | 71,5 | 153 | 65 | 83,5 | 1,782±0,028 | |
| anti-sense chain | CTGTACTTAGGATCACAAGGCTTGAAC | 70,9 | ||||||||
| FaBZIP1 | gene17796 | Basic leucine zipper transcription factor | sense chain | AGGGTCAACAAAACCAGAATGGGGATAA | 77,7 | 151 | 65 | 85,5 | 1,712±0,027 | |
| anti-sense chain | CTGCGTTCCAGCTCTGAAATGTATTGC | 77,8 | ||||||||
| FaTIM1 | gene17570 | Mitochondrial import inner membrane translocase | sense chain | GCTCCGCCACTTACGCCGCTAATTT | 80,2 | 100 | 65 | 86,5 | 1,811±0,028 | |
| anti-sense chain | AGATCATCAGGCCCCGTCTTTCTCGTTA | 80 | ||||||||
| FaFHA1 | gene17571 | SMAD/FHA domain-containing protein | sense chain | ATTGCATGCTAAGTTGGTGGAACAGTAT | 73,9 | 179 | 65 | 85,5 | 1,744±0,025 | |
| anti-sense chain | GACCCTTAGACCTTGTGTTGATGACAAA | 74,6 | ||||||||
| FaRIB413 | gene33863 | RNA interspacer (16S–23S) region | sense chain | ACCGTTGATTCGCACAATTGGTCATCG | 83,4 | 149 | 65 | 91 | 1,784±0,032 | [29] |
| anti-sense chain | TACTGCGGGTCGGCAATCGGACG | 81,9 | ||||||||
| FaWRKY1 | gene07210 | WRKY75 like transcription factor | sense chain | ACAGCAGTAAGATTAGGGATGAAGAAGGGAG | 76,2 | 196 | 65 | 85,5 | 1,824±0,025 | [30] |
| anti-sense chain | GCTTCTTCACATTGCAACCCTGATGCGTG | 83,8 |
(a) Accession number of genes found in Fragaria vesca genome (http://www.strawberrygenome.org/and http://www.rosaceae.org/) that may be amplified by the designed primer pairs. (b) Theoretical melting temperatures calculated by Oligo Primer Analisys software version 6.65 for each primer. (c) Recommended optimal annealing temperature was calculated by gradient PCR and subsequent PCR efficiency optimization. (d) PCR-product melting temperature as determined by melting curves (e) PCR efficiencies were calculated by LigRegPCR. (f) Known references for genes previously analyzed or described.
Primers designed of candidate reference genes
The RTqPCR primer pairs for each putative reference gene, as well as for FaWRKY1, were designed based on common criteria, and were tested to generate clear and unique PCR products in RTqPCR reactions (Table 1 and Figure S1).
All primers were designed from the CDS of the selected genes, avoiding regions of conserved sequence similarity to other genes. For genes belonging to gene families or with identified paralogs present in the genome of the diploid woodland strawberry (F. vesca) [35], the least conserved region was used to ensure amplification of a single gene by PCR. In four cases (FaEF1α, FaTUBα, FaTUBβ and FaACTIN), it was not possible to differentiate between either multicopy or nearly identical genes although unique amplicons were obtained. In six cases including the control gene (FaGAPDH1, FaTUBβ, FaBZIP1, FaTIM1, FaFHA1, FaWRKY1) primers were designed to span an exon-exon junction.
To ensure maximum specificity and efficiency during PCR amplification, primers were designed to have melting temperatures over 70 °C, and were required to generate short amplicons, usually between 100 and 200 bp (Table 1). The most appropriate annealing temperature for every primer pair was calculated by RTq-gradientPCR, and only primer pairs with optimal efficiency at annealing temperatures of above 65°C were considered for subsequent RTqPCR analyses. The primer pair for gene FaRIB413 was previously designed in our group [29], and tested to meet all of the above criteria. The specificity of the primers was tested by PCR using first-strand cDNAs synthesized from total RNA isolated from the biological samples.
The PCR efficiency of each primer pair was calculated using LinRegPCR, a method that utilizes absolute fluorescence data captured during the exponential phase of amplification of each real-time PCR reaction [36]. Table 1 shows the calculated PCR efficiencies for the primer pairs studied. Each efficiency value represents an average ± SD calculated from 192 amplification plots (i.e. two technical replicates of two biological replicates of a total of 48 different experimental conditions). For all primer pairs, values ranged from 1.712 to 1.925, with low standard deviation. These values indicated comparable amplification efficiencies among the 96 diverse cDNA samples tested (Table 1), and suggested that the designed primer pairs efficiently amplified their target genes. Therefore, the mean primer pair efficiency value was considered for all subsequent studies, including estimations of the relative expression level of the reference genes under evaluation.
Expression Stability of the Candidate Reference Genes under Different Experimental Conditions
Candidate reference genes were evaluated by RTqPCR analyses in response to the experimental conditions summarized in Table 2. Samples from different strawberry varieties were also examined. Two independent biological replicates were performed for each experimental condition. Between 10 and 18 independent samples per experiment were analyzed. In addition, two technical replicates corresponding to two biological replicates were used in this study. The generated results were subjected to the following analytical methods: analysis of ‘‘Stability index’’ [10], geNORM [4] implemented in qBASEplus software [37], NormFinder [9], BestKeeper [38], and the comparative Δ-Ct [39].
Table 2. Summary of strawberry varieties, tissues and experimental conditions used in this study.
| Biological process | Cultivar | Culture type/Tissue | Biological stages/Time points after treatments | Experimental conditions | |
| Ripening and senescence | Camarosa | Fruit | G, W, R, OR and SE | Fruit ripening in field | RCFa |
| Defense against fungal infection | Camarosa | Fruit | Red stage fruits: Mock/Infectedgrades 1, 2, 3 and 4 | Red fruit naturally infected withC. acutatum in field | FCFa, e |
| Defense against fungal infection | Camarosa | Crown | Mock: 1, 3, 5 and 7 dpi/Infected:1, 3, 5 and 7 dpi | Growth chamber C. acutatuminfection under controlledconditions | FCCb, e |
| Defense against fungal infection | Camarosa | Petiole | Mock: 1, 3, 5 and 7 dpi/Infected:1, 3, 5 and 7 dpi | Growth chamber C. acutatuminfection under controlledconditions | FCPb, d, e |
| Defense against fungal infection | Andana | Petiole | Mock: 3, 5 and 7 dpi/Infected:1, 3, 5 and 7 dpi | Growth chamber C. acutatuminfection under controlledconditions | FAPd |
| Hormone response | Camarosa | Young in-vitro plant | Mock: 12, 24, 48hpt/SA (5 mM):12, 24, 48hpt/MeJA (2 mM):12, 24, 48hpt | Mock, SA and MeJA treatment | HCYc, e |
| Hormone response | Chandler | Cellular suspensions | Mock: 4 and 6hpt/SA (0,75 mM):4 and 6hpt/MeJA (0,1 mM):4 and 6hpt | Mock, SA and MeJA treatment | HCCc |
RCF, Ripening-Camarosa-Fruit; FCF, Fungal-Camarosa-Fruit; FCC, Fungal-Camarosa-Crown; FCP, Fungal-Camarosa-Petiole; FAP, Fungal-Andana-Petiole; HCY, Hormone-Camarosa-Young-in-vitro; HCC, Hormone-Chandler-Cellular-suspensions. G1: small green, W: white, R: red, OR: over-ripened, SE: senescent.
(a) Comparison of gene expression between overripening-derived senescence and infection-derived necrosis, (b) Comparison of gene expression between cultivars under biotic stress, (c) Comparison of gene expression between cultivars under hormonal treatment, (d) Comparison of gene expression between different plant tissues, (e) Comparison of gene expression between infected and hormone treated plants.
Statistical analysis of gene expression by “stability index” calculation
Figure 1 shows the expression level of candidate reference genes in the seven experimental conditions named in Table 2. Mean Cq values for each transcript in every experimental condition, together with coefficient of variation (CV), slope, and stability index (SI), according to Brunner, (2004) [10] are given in Table 3.
Figure 1. Expression levels of candidate reference genes in different experimental sets.
Box plot graphs of Cq values for each reference gene tested in all strawberry samples and subsets. Cq values are inversely proportional to the amount of template and are shown as the first and third quartile. Vertical lines indicate the range of values, and median values are indicated by the black lines. Circles indicate outliers. RCF, Ripening-Camarosa-Fruit; FCF, Fungal-Camarosa-Fruit; FCC, Fungal-Camarosa-Crown; FCP, Fungal-Camarosa-Petiole; FAP, Fungal-Andana-Petiole; HCY, Hormone-Camarosa-Young-in-vitro; HCC, Hormone-Chandler-Cellular-suspensions; All, samples from all seven experiments analyzed together.
Table 3. Summary of statistics evaluating stability of gene expression.
| Mean b | SD | CV (%) | Slope c | Intercept | Stability index d | Mean b | SD | CV (%) | Slope c | Intercept | Stability index d | ||||
| Ripening-Camarosa-Fruit (n = 20) a | Fungal infection-Andana-Petiole (n = 28) | ||||||||||||||
| * | FaRIB413 | 8,341 | 0,239 | 2,860 | 0,004 | 8,329 | 0,011 | * | FaGAPDH1 | 26,129 | 0,632 | 2,418 | 0,004 | 26,143 | 0,009 |
| * | FaCHC1 | 23,085 | 0,201 | 0,869 | 0,021 | 23,024 | 0,018 | * | FaGAPDH2 | 18,484 | 0,374 | 2,024 | 0,025 | 18,585 | 0,051 |
| * | FaTUBβ | 22,334 | 0,359 | 1,609 | 0,015 | 22,289 | 0,024 | * | FaACTIN | 23,640 | 0,428 | 1,812 | 0,030 | 23,760 | 0,054 |
| FaACTIN | 23,894 | 0,309 | 1,294 | 0,144 | 24,326 | 0,186 | * | FaEF1α | 18,780 | 0,417 | 2,219 | 0,026 | 18,886 | 0,059 | |
| FaTIM1 | 22,602 | 0,359 | 1,587 | 0,151 | 23,054 | 0,239 | * | FaMT1 | 23,992 | 0,506 | 2,108 | 0,028 | 23,879 | 0,060 | |
| FaMT1 | 25,622 | 0,449 | 1,753 | 0,143 | 25,193 | 0,251 | * | FaFHA1 | 24,251 | 0,570 | 2,349 | 0,036 | 24,107 | 0,085 | |
| FaEF1α | 17,406 | 0,413 | 2,371 | 0,161 | 17,889 | 0,382 | FaTUBβ | 21,575 | 0,374 | 1,734 | 0,090 | 21,216 | 0,155 | ||
| FaFHA1 | 23,258 | 0,643 | 2,765 | 0,204 | 23,870 | 0,564 | FaCHC1 | 26,339 | 0,404 | 1,536 | 0,120 | 26,816 | 0,184 | ||
| FaTUBα | 22,899 | 1,174 | 5,128 | 0,556 | 24,567 | 2,851 | FaBZIP1 | 26,410 | 0,446 | 1,688 | 0,110 | 25,971 | 0,185 | ||
| FaBZIP1 | 30,089 | 1,485 | 4,936 | 0,607 | 28,270 | 2,994 | FaTIM1 | 26,864 | 0,481 | 1,790 | 0,116 | 26,401 | 0,207 | ||
| FaUBQ1 | 26,677 | 1,249 | 4,680 | 0,812 | 29,113 | 3,800 | FaUBQ1 | 27,650 | 0,642 | 2,322 | 0,115 | 27,085 | 0,266 | ||
| FaGAPDH2 | 17,073 | 1,071 | 6,274 | 0,622 | 18,939 | 3,903 | FaRIB413 | 8,790 | 0,444 | 5,053 | 0,061 | 9,036 | 0,310 | ||
| FaGAPDH1 | 24,080 | 1,715 | 7,120 | 1,115 | 27,425 | 7,939 | FaTUBα | 20,281 | 0,595 | 2,931 | 0,211 | 19,436 | 0,620 | ||
| Fungal infection-Camarosa-Fruit (n = 20) | Hormonal treatment-Camarosa-Young in-vitro plant (n = 36) | ||||||||||||||
| * | FaGAPDH1 | 23,530 | 0,316 | 1,345 | 0,005 | 23,545 | 0,007 | * | FaGAPDH1 | 25,817 | 0,479 | 1,856 | 0,024 | 25,938 | 0,045 |
| * | FaTUBα | 21,462 | 0,322 | 1,499 | 0,019 | 21,518 | 0,028 | * | FaUBQ1 | 28,954 | 0,518 | 1,789 | 0,026 | 29,085 | 0,047 |
| * | FaUBQ1 | 25,599 | 0,405 | 1,583 | 0,047 | 25,458 | 0,074 | * | FaGAPDH2 | 19,183 | 0,278 | 1,451 | 0,038 | 18,993 | 0,055 |
| FaGAPDH2 | 16,274 | 0,331 | 2,031 | 0,062 | 16,090 | 0,125 | * | FaRIB413 | 8,838 | 0,523 | 5,912 | 0,016 | 8,760 | 0,093 | |
| FaACTIN | 23,539 | 0,314 | 1,335 | 0,136 | 23,133 | 0,181 | FaCHC1 | 26,297 | 0,482 | 1,832 | 0,080 | 25,895 | 0,147 | ||
| FaEF1α | 16,556 | 0,250 | 1,510 | 0,130 | 16,166 | 0,196 | FaTUBα | 23,058 | 0,586 | 2,542 | 0,093 | 22,591 | 0,237 | ||
| FaTIM1 | 24,031 | 0,372 | 1,549 | 0,131 | 23,638 | 0,203 | FaFHA1 | 25,649 | 0,615 | 2,396 | 0,101 | 25,145 | 0,242 | ||
| FaCHC1 | 23,929 | 0,467 | 1,953 | 0,121 | 23,568 | 0,235 | FaEF1α | 18,593 | 0,442 | 2,375 | 0,119 | 17,996 | 0,284 | ||
| FaTUBβ | 21,668 | 0,387 | 1,784 | 0,133 | 21,271 | 0,236 | FaMT1 | 25,669 | 0,726 | 2,829 | 0,165 | 24,846 | 0,466 | ||
| FaBZIP1 | 27,780 | 0,478 | 1,719 | 0,164 | 27,288 | 0,282 | FaTIM1 | 27,336 | 0,800 | 2,928 | 0,176 | 26,457 | 0,514 | ||
| FaFHA1 | 23,606 | 0,545 | 2,308 | 0,213 | 22,969 | 0,490 | FaTUBβ | 23,573 | 0,775 | 3,286 | 0,218 | 22,484 | 0,716 | ||
| FaRIB413 | 8,635 | 0,323 | 3,736 | 0,158 | 8,161 | 0,590 | FaBZIP1 | 27,459 | 0,845 | 3,079 | 0,262 | 26,150 | 0,806 | ||
| FaMT1 | 25,910 | 0,745 | 2,876 | 0,425 | 24,635 | 1,222 | FaACTIN | 25,122 | 0,979 | 3,899 | 0,325 | 23,499 | 1,265 | ||
| Fungal infection-Camarosa-Crown (n = 32) | Hormonal treatment-Chandler-Cellular suspensions (n = 24) | ||||||||||||||
| * | FaUBQ1 | 27,734 | 0,486 | 1,752 | 0,037 | 27,567 | 0,065 | * | FaTIM1 | 25,850 | 0,974 | 3,766 | 0,003 | 25,862 | 0,013 |
| * | FaRIB413 | 7,873 | 0,241 | 3,057 | 0,027 | 7,752 | 0,083 | * | FaGAPDH2 | 17,889 | 0,300 | 1,679 | 0,013 | 17,843 | 0,022 |
| FaGAPDH1 | 25,569 | 0,453 | 1,771 | 0,064 | 25,282 | 0,113 | * | FaRIB413 | 8,426 | 0,299 | 3,551 | 0,021 | 8,498 | 0,073 | |
| FaCHC1 | 24,988 | 0,492 | 1,968 | 0,067 | 24,687 | 0,131 | * | FaUBQ1 | 27,222 | 0,566 | 2,079 | 0,039 | 27,322 | 0,081 | |
| FaEF1α | 17,786 | 0,386 | 2,173 | 0,062 | 17,509 | 0,134 | FaCHC1 | 24,163 | 0,427 | 1,767 | 0,129 | 23,712 | 0,228 | ||
| FaGAPDH2 | 19,286 | 0,352 | 1,825 | 0,090 | 18,880 | 0,165 | FaBZIP1 | 25,344 | 0,523 | 2,063 | 0,163 | 25,914 | 0,336 | ||
| FaMT1 | 22,968 | 0,571 | 2,486 | 0,068 | 22,664 | 0,168 | FaEF1α | 16,478 | 0,413 | 2,505 | 0,151 | 15,950 | 0,377 | ||
| FaFHA1 | 23,875 | 0,651 | 2,728 | 0,062 | 24,156 | 0,170 | FaTUBα | 20,364 | 0,539 | 2,649 | 0,236 | 19,538 | 0,625 | ||
| FaTIM1 | 25,885 | 0,813 | 3,139 | 0,116 | 25,363 | 0,364 | FaMT1 | 24,533 | 0,550 | 2,240 | 0,282 | 23,545 | 0,632 | ||
| FaTUBβ | 22,086 | 0,622 | 2,818 | 0,136 | 21,472 | 0,384 | FaFHA1 | 23,116 | 0,612 | 2,646 | 0,246 | 22,256 | 0,650 | ||
| FaTUBα | 20,298 | 0,585 | 2,883 | 0,136 | 19,687 | 0,392 | FaACTIN | 23,145 | 0,776 | 3,352 | 0,343 | 21,943 | 1,151 | ||
| FaACTIN | 24,440 | 0,563 | 2,303 | 0,211 | 23,493 | 0,485 | FaGAPDH1 | 20,908 | 0,836 | 3,999 | 0,401 | 22,313 | 1,605 | ||
| FaBZIP1 | 25,229 | 0,929 | 3,682 | 0,279 | 23,975 | 1,026 | FaTUBβ | 20,592 | 0,964 | 4,681 | 0,436 | 19,067 | 2,040 | ||
| Fungal infection-Camarosa-Petiole (n = 32) | All seven experiments (n = 192) | ||||||||||||||
| * | FaTUBα | 20,767 | 0,423 | 2,036 | 0,007 | 20,737 | 0,014 | * | FaACTIN | 24,011 | 0,883 | 3,676 | 0,004 | 23,905 | 0,015 |
| * | FaACTIN | 23,676 | 0,364 | 1,536 | 0,013 | 23,528 | 0,020 | FaRIB413 | 8,542 | 0,490 | 5,736 | 0,056 | 8,306 | 0,323 | |
| * | FaRIB413 | 8,816 | 0,237 | 2,685 | 0,027 | 8,695 | 0,072 | FaTUBβ | 22,073 | 1,067 | 4,835 | 0,069 | 22,252 | 0,333 | |
| * | FaBZIP1 | 24,874 | 0,620 | 2,493 | 0,034 | 25,025 | 0,084 | FaEF1α | 17,716 | 0,904 | 5,100 | 0,082 | 17,270 | 0,416 | |
| * | FaEF1α | 17,574 | 0,437 | 2,485 | 0,034 | 17,727 | 0,084 | FaMT1 | 24,338 | 1,399 | 5,747 | 0,097 | 24,857 | 0,560 | |
| FaGAPDH2 | 18,321 | 0,357 | 1,946 | 0,064 | 18,611 | 0,125 | FaFHA1 | 24,140 | 1,037 | 4,298 | 0,144 | 23,426 | 0,619 | ||
| FaMT1 | 22,583 | 0,517 | 2,288 | 0,065 | 22,293 | 0,148 | FaTUBα | 21,292 | 1,305 | 6,131 | 0,158 | 21,937 | 0,970 | ||
| FaTUBβ | 22,009 | 0,574 | 2,609 | 0,100 | 21,561 | 0,260 | FaGAPDH1 | 24,722 | 1,856 | 7,509 | 0,157 | 25,119 | 1,175 | ||
| FaCHC1 | 24,874 | 0,735 | 2,954 | 0,139 | 25,499 | 0,411 | FaUBQ1 | 27,492 | 1,134 | 4,124 | 0,295 | 26,149 | 1,217 | ||
| FaUBQ1 | 27,470 | 0,766 | 2,788 | 0,169 | 28,246 | 0,472 | FaGAPDH2 | 18,270 | 1,063 | 5,818 | 0,267 | 17,007 | 1,551 | ||
| FaTIM1 | 25,223 | 0,755 | 2,993 | 0,193 | 26,091 | 0,577 | FaCHC1 | 25,000 | 1,189 | 4,756 | 0,333 | 23,479 | 1,583 | ||
| FaFHA1 | 24,264 | 0,774 | 3,191 | 0,224 | 25,270 | 0,713 | FaBZIP1 | 26,547 | 1,789 | 6,741 | 0,489 | 28,697 | 3,297 | ||
| FaGAPDH1 | 25,419 | 1,120 | 4,405 | 0,263 | 26,600 | 1,156 | FaTIM1 | 25,650 | 1,591 | 6,201 | 0,619 | 22,923 | 3,839 | ||
Genes are ordered into each experiment analyzed, top to bottom, from those tending to show the highest stability to those showing the lowest, based on the stability index. a) "n" represents the number of individuals analyzed from each experiment, (four data points per sample, two biological and two technical replicates of each). b) Data based on analysis of Cq values. SD, standard deviation. CV, Coeficient of variation. c) Slope of regression of gene means. Intercepts are also given for the estimated regression lines. d) Stability index is the product of CV and slope (multiplication of columns 3 and 4). Transcripts with lower slope are preferred as controls. Asterisks mark the best candidate genes with stability index below 0.0×.
The analysis of variation, as reflected in the coefficient of variation (CV), showed highly predictability of all candidate reference genes in every of the seven experimental conditions. Considering them together, almost all CV values were below 6%. Exceptions were genes FaGAPDH1 and FaGAPDH2, which deviated substantially during ripening, and genes FaTUBα, FaGAPDH1, FaBZIP1 and FaTIM1, within the “all together” conditions (Table 3).
The mean expression level for each gene was regressed against the overall means for the different samples (Figure S2). The slope of the predicted regression lines provided an estimate of the degree to which the gene is sensitive to general expression-promoting conditions. Assuming that both consistent transcript levels among samples (low slope) and high predictability (low CV) are desired, the “stability index” (SI) (product of slope and CV) is used to describe transcript stability as in Brunner, (2004) [10]. Transcripts with the lowest stability index will usually provide the best reference genes or controls.
The results show that several predicted candidate genes show a favorable SI in each of the main areas studied (Table 3, marked by asterisks). During fruit ripening candidates FaRIB413, FaCHC1 and FaTUBβ showed low SI values (0.011, 0.018, and 0.024, respectively). Genes FaGAPDH1, FaTUBα and FaUBQ1 also appear to be excellent reference genes for fungal infection studies in red fruit (SI of 0.007, 0.028, and 0.074, respectively). In vegetative tissues challenged with the fungus, variations in number and diversity of convenient reference genes was also found. Thus, genes FaUBQ1 (SI, 0.065) and FaRIB413 (SI, 0.083), were found to be the best candidates for normalization on crown tissue of cultivar Camarosa but genes FaTUBα (SI, 0.014), FaACTIN (SI, 0.020), FaRIB413 (SI, 0.072), FaBZIP1 (SI, 0.084), FaEF1α (SI, 0.084) were also very good candidates on petiole tissue of this cultivar. However, on petiole tissue from cultivar Andana, the set of predicted good candidate reference genes is not the same. The best candidates were genes FaGAPDH1 (SI, 0.009), FaGAPDH2 (SI, 0.051), FaACTIN (SI, 0.054), FaEF1α (SI, 0.059), FaMT1 (SI, 0.060), and FaFHA1 (SI, 0.085). Only genes FaACTIN and FaEF1α were found to be the reasonable reference genes for normalization in petiole tissue of both strawberry cultivars. In addition, genes FaUBQ1, FaGAPDH2, and FaRIB413 were found to be the optimal reference genes for SA and JA studies either in in-vitro plants (SI, 0.047, 0.055, and 0.093, respectively) in cell suspension treatments (SI, 0.081, 0.022, and 0.073, respectively), as well as across different cultivars. Genes FaGAPDH1 (SI, 0.045) and FaTIM1 (SI, 0.013) were also found to be appropriate candidates for the in-vitro plants and cellular suspension experiments, respectively.
Also, we have considered an “all together” analysis where all seven experimental variables have been examined. In this analysis, gene FaACTIN showed the lowest stability index (SI, 0.015), and appears to be the best overall reference gene following this analytical method.
Expression stability and calculation of hypothetical normalization factor by geNormPLUS
The stability coeficient (M values) and the coefficient of variation (CV values) of each gene are inversely related to their expression stability. These values were calculated using qBase software [37] but taking into account the previously calculated specific PCR efficiency of each gene. The average stability coefficient (MA), defined as the average value of the M values (average pairwise variation of a gene with all other tested reference genes of all combinations of a gene and high-ranking reference genes) of the relative quantities of the thirteen genes under evaluation were analyzed with geNormPlus (qBase software, [4], [37]).
Figure 2 represents the average stability coefficients (MA) of the thirteen candidate reference genes tested from every analyzed condition. All thirteen genes showed acceptable expression stabilities (MA≤1), as described for heterogeneous samples [37], with the exception of genes FaBZIP1 and FaGAPDH1 when all seven experimental conditions were analyzed together.
Figure 2. Average expression stability value (MA) of each gene.
Specific MA values were calculated under seven single experimental conditions tested, as well as by combining all samples together. MA for genes tested are shown as derived by geNormPLUS analysis. The lowest MA value indicates the most stable expression.
Table 4 shows transcripts ranked by their MA and CV values. The MA results revealed that optimal candidate reference genes differed among the analyzed experimental conditions. Thus, FaACTIN (0.182) seems to be the most stable gene in fruit ripening analyses, meanwhile FaTIM1 (0.143) is in fruit natural infection, FaGAPDH2 (0.234) and FaRIB413 (0.300) in Camarosa crown and petiole infected tissues, respectively, FaMT1 (0.247) in Andana infected petiole, FaEF1α (0.242) in hormonal treatments of in-vitro plants, FaEF1α (0.242) and FaTUBα (0.242) in elicited cellular suspensions of cultivar Chandler, and finally, FaGAPDH2 (0.594) in the “all together” conditions. A similar result was obtained when CV values were considered.
Table 4. Reference genes ranked in order by their average expression stability (MA) and coefficient of variation (CV) respectively.
| Ranking by MA values from geNormPLUS | |||||||||||||
| RCF | FaGAPDH1 | FaBZIP1 | FaUBQ1 | FaTUBα | FaGAPDH2 | FaMT1 | FaTUBβ | FaCHC1 | FaRIB413 | FaTIM1 | FaEF1α | FaFHA1 | FaACTIN |
| (0.845) | (0.712) | (0.597) | (0.533) | (0.449) | (0.34) | (0.287) | (0.269) | (0.259) | (0.248) | (0.228) | (0.195) | (0.182) | |
| FCF | FaMT1 | FaCHC1 | FaUBQ1 | FaGAPDH1 | FaTUBα | FaFHA1 | FaBZIP1 | FaTUBβ | FaRIB413 | FaGAPDH2 | FaEF1α | FaACTIN | FaTIM1 |
| (0.361) | (0.325) | (0.31) | (0.289) | (0.267) | (0.248) | (0.232) | (0.211) | (0.196) | (0.175) | (0.163) | (0.156) | (0.143) | |
| FCC | FaBZIP1 | FaMT1 | FaFHA1 | FaTIM1 | FaACTIN | FaTUBβ | FaTUBα | FaGAPDH1 | FaUBQ1 | FaCHC1 | FaEF1α | FaRIB413 | FaGAPDH2 |
| (0.493) | (0.458) | (0.422) | (0.382) | (0.359) | (0.348) | (0.336) | (0.309) | (0.293) | (0.266) | (0.245) | (0.239) | (0.234) | |
| FCP | FaGAPDH1 | FaTIM1 | FaFHA1 | FaBZIP1 | FaCHC1 | FaUBQ1 | FaMT1 | FaTUBβ | FaGAPDH2 | FaEF1α | FaTUBα | FaACTIN | FaRIB413 |
| (0.645) | (0.563) | (0.541) | (0.515) | (0.488) | (0.454) | (0.41) | (0.379) | (0.357) | (0.345) | (0.324) | (0.302) | (0.3) | |
| FAP | FaGAPDH1 | FaUBQ1 | FaTUBα | FaBZIP1 | FaTIM1 | FaTUBβ | FaGAPDH2 | FaRIB413 | FaCHC1 | FaEF1α | FaFHA1 | FaACTIN | FaMT1 |
| (0.474) | (0.445) | (0.421) | (0.405) | (0.399) | (0.385) | (0.369) | (0.348) | (0.329) | (0.31) | (0.291) | (0.263) | (0.247) | |
| HCY | FaACTIN | FaGAPDH1 | FaUBQ1 | FaMT1 | FaBZIP1 | FaTIM1 | FaTUBβ | FaRIB413 | FaTUBα | FaGAPDH2 | FaCHC1 | FaFHA1 | FaEF1α |
| (0.531) | (0.49) | (0.469) | (0.444) | (0.426) | (0.393) | (0.362) | (0.337) | (0.319) | (0.293) | (0.266) | (0.248) | (0.242) | |
| HCC | FaGAPDH1 | FaTIM1 | FaTUBβ | FaBZIP1 | FaACTIN | FaUBQ1 | FaRIB413 | FaGAPDH2 | FaCHC1 | FaFHA1 | FaMT1 | FaTUBα | FaEF1α |
| (0.651) | (0.581) | (0.516) | (0.476) | (0.445) | (0.413) | (0.38) | (0.359) | (0.323) | (0.304) | (0.262) | (0.247) | (0.247) | |
| All samples | FaGAPDH1 | FaBZIP1 | FaMT1 | FaTIM1 | FaTUBα | FaRIB413 | FaTUBβ | FaACTIN | FaFHA1 | FaCHC1 | FaEF1α | FaUBQ1 | FaGAPDH2 |
| (1.174) | (1.079) | (0.987) | (0.901) | (0.835) | (0.764) | (0.738) | (0.717) | (0.681) | (0.669) | (0.631) | (0.603) | (0.594) | |
| Ranking by CV values from geNormPLUS | |||||||||||||
| RCF | FaGAPDH1 | FaBZIP1 | FaUBQ1 | FaMT1 | FaTUBα | FaGAPDH2 | FaTUBβ | FaCHC1 | FaRIB413 | FaTIM1 | FaFHA1 | FaACTIN | FaEF1α |
| (0.844) | (0.703) | (0.493) | (0.487) | (0.441) | (0.337) | (0.299) | (0.292) | (0.234) | (0.17) | (0.17) | (0.155) | (0.093) | |
| FCF | FaMT1 | FaGAPDH1 | FaTUBα | FaCHC1 | FaUBQ1 | FaFHA1 | FaTIM1 | FaACTIN | FaBZIP1 | FaTUBβ | FaRIB413 | FaGAPDH2 | FaEF1α |
| (0.352) | (0.236) | (0.219) | (0.206) | (0.188) | (0.171) | (0.144) | (0.139) | (0.118) | (0.116) | (0.116) | (0.1) | (0.073) | |
| FCC | FaBZIP1 | FaMT1 | FaFHA1 | FaTIM1 | FaUBQ1 | FaCHC1 | FaACTIN | FaGAPDH1 | FaTUBβ | FaGAPDH2 | FaTUBα | FaEF1α | FaRIB413 |
| (0.374) | (0.369) | (0.311) | (0.278) | (0.244) | (0.204) | (0.204) | (0.201) | (0.164) | (0.147) | (0.145) | (0.136) | (0.111) | |
| FCP | FaGAPDH1 | FaMT1 | FaFHA1 | FaCHC1 | FaTIM1 | FaTUBβ | FaBZIP1 | FaACTIN | FaUBQ1 | FaTUBα | FaEF1α | FaRIB413 | FaGAPDH2 |
| (0.758) | (0.372) | (0.325) | (0.308) | (0.304) | (0.297) | (0.257) | (0.246) | (0.237) | (0.205) | (0.191) | (0.166) | (0.136) | |
| FAP | FaGAPDH1 | FaUBQ1 | FaTUBα | FaRIB413 | FaACTIN | FaCHC1 | FaGAPDH2 | FaMT1 | FaBZIP1 | FaTIM1 | FaTUBβ | FaFHA1 | FaEF1α |
| (0.393) | (0.319) | (0.266) | (0.24) | (0.239) | (0.215) | (0.206) | (0.205) | (0.201) | (0.183) | (0.175) | (0.154) | (0.12) | |
| HCY | FaACTIN | FaGAPDH1 | FaTIM1 | FaUBQ1 | FaMT1 | FaBZIP1 | FaRIB413 | FaTUBβ | FaTUBα | FaGAPDH2 | FaFHA1 | FaCHC1 | FaEF1α |
| (0.375) | (0.373) | (0.34) | (0.295) | (0.282) | (0.277) | (0.224) | (0.203) | (0.181) | (0.18) | (0.173) | (0.15) | (0.122) | |
| HCC | FaGAPDH1 | FaTIM1 | FaTUBβ | FaACTIN | FaBZIP1 | FaMT1 | FaGAPDH2 | FaUBQ1 | FaFHA1 | FaRIB413 | FaTUBα | FaEF1α | FaCHC1 |
| (0.685) | (0.47) | (0.406) | (0.314) | (0.282) | (0.254) | (0.229) | (0.223) | (0.209) | (0.2) | (0.176) | (0.159) | (0.155) | |
| All samples | FaGAPDH1 | FaTIM1 | FaMT1 | FaBZIP1 | FaCHC1 | FaTUBα | FaGAPDH2 | FaUBQ1 | FaRIB413 | FaTUBβ | FaACTIN | FaFHA1 | FaEF1α |
| (1.703) | (1.072) | (0.721) | (0.595) | (0.581) | (0.533) | (0.521) | (0.474) | (0.419) | (0.376) | (0.332) | (0.279) | (0.259) | |
Increasing stability from left to right. See Table 2 for experimental description.
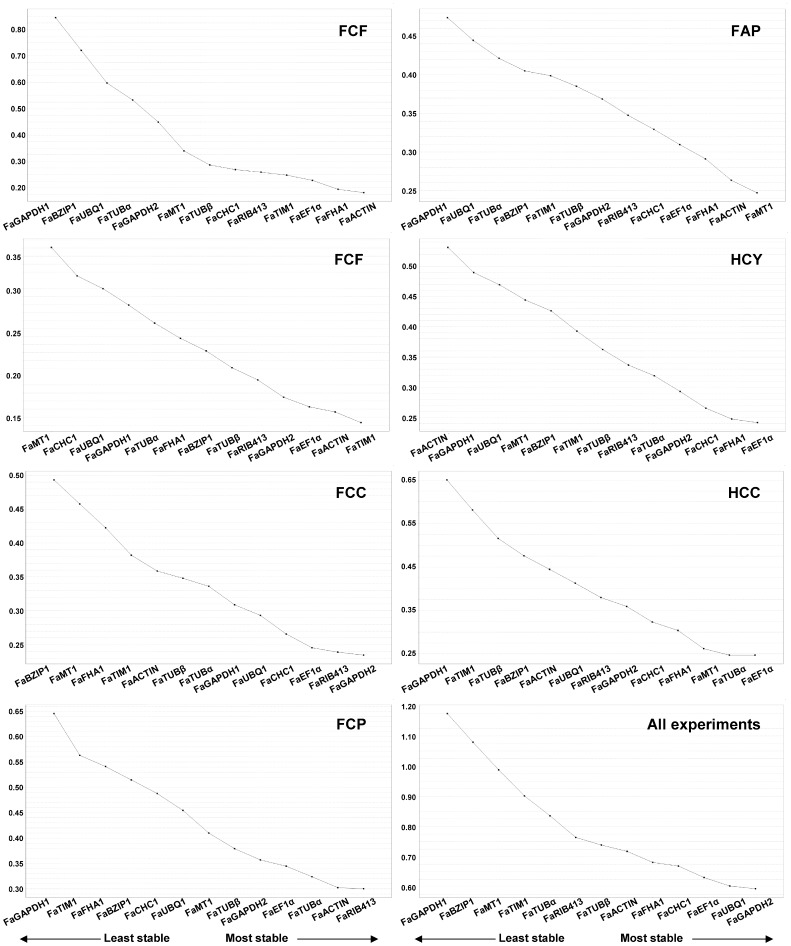
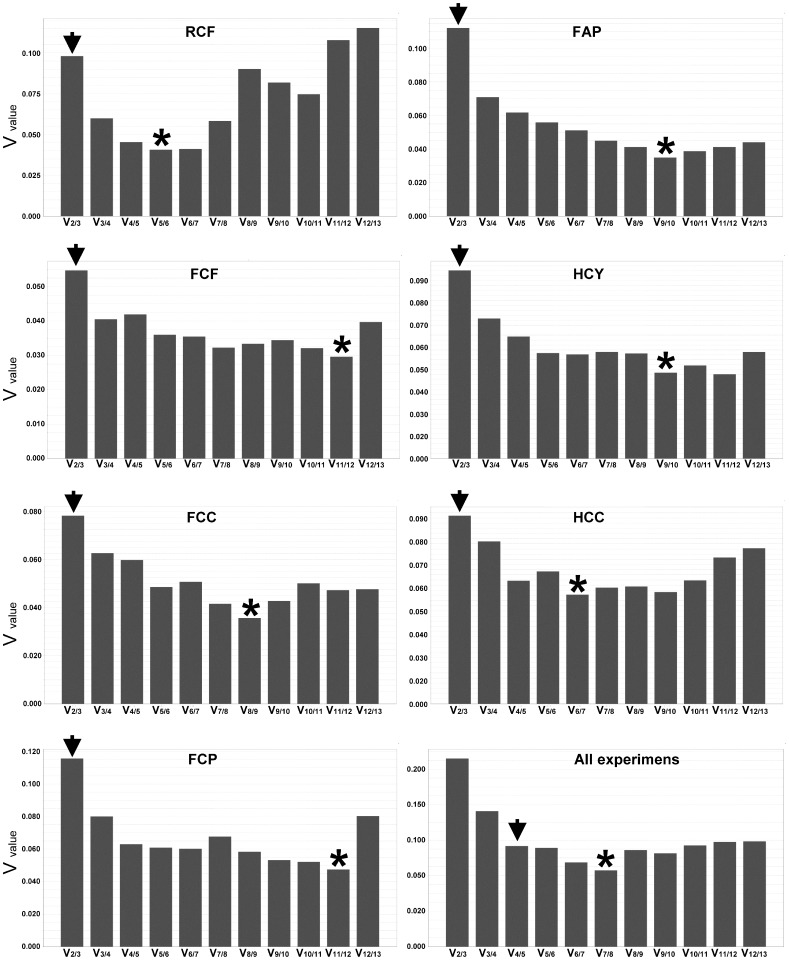
The optimal and the minimal number of reference genes needed to calculate a hypothetical optimal normalization factor suitable in each analyzed condition was determined, as described by Vandesompele [4]. Figure 3, shows that the optimal number (V) of these needed reference genes differed in each experimental conditions but a combination of them is assumed to be an ideal reference gene. Thus, in fruit ripening analyses, V5/6 was the lowest pairwise variation value (0.041). Therefore, the hypothetical normalization factor in these experimental conditions would be the geometric mean of the five or six more stable genes (see Figure 2 and Table 4, for the ranking of more stable genes for this and other experimental condition). Other lowest pairwise variation values were, V11/12 (0.03) for the infected fruit experiment, V8/9 (0.036) and V11/12 (0.047) for Camarosa crown and petiole infected tissues, respectively, V9/10 (0.035) for Andana infected petioles, V9/10 (0.043) for hormonal treatment of in-vitro plants experiment, V6/7 (0.053) for elicited cellular suspensions, and finally, V7/8 (0.086) when all experiments were considered together.
Figure 3. Determination of the number of genes required to calculate a hypothetical normalization factor.
Pairwise variation (Vn/n+1) analysis was carried out to determine the number of reference genes required for accurate normalization. An asterisk indicates the lowest V value in each experiment. An arrowhead indicates the minimum number of genes required to pass the suggested cut-off value (0.15) [4]. See Table 2 for experiment description.
In practice, however, the number of genes required should be low enough to make experimental procedures affordable, yet high enough to merit confidence in the conclusions. This means that if the pairwise variation value for n genes is below the recommended cut-off of 0.15, additional genes will not likely contribute to improved normalization [4]. Thus, the minimal number of reference candidates in each single experiment was determined to be two, for all the experimental conditions tested (marked with an arrowhead in Figure 3), but four in the all-together conditions. In each experimental condition, these genes were FaACTIN and FaFHA1 (V2/3 value of 0.098) for fruit ripening, FaTIM1 and FaACTIN (V2/3 value of 0.055) for fruit infection, FaGAPDH2 and FaRIB413 (V2/3 value of 0.078) for Camarosa crown infection, FaRIB413 and FaACTIN (V2/3 value of 0.116) for Camarosa petiole infection, FaMT1 and FaACTIN (V2/3 value of 0.112) for Andana petiole infection, FaEF1α and FaFHA1 (V2/3 value of 0.095) for in-vitro plants treated with hormones, and FaEF1α and FaTUBα (V2/3 value of 0.091) for elicited cellular suspensions. For the all-together conditions the minimal reference genes were FaGAPDH2, FaUBQ1, FaEF1α, and FaCHC1 (V4/5 value of 0.113).
Evaluation of expression stability by ΔCt method, Normfinder and BestKeeper approaches
In order to accurately assess the usefulness of the thirteen candidate reference genes, other analytical methods were applied to the same data set. These include the comparative ΔCt method [39], which ranks the reference genes by their mean standard deviation in the pairwise comparisons. The NormFinder [9] method was also used. NormFinder ranks the set of candidate normalization genes according to their expression stability in a given sample set and a given experimental design. The Bestkeeper algorithm [38] performs pairwise comparison using the geometric mean of the Cp (Cq), values, and this one was also implemented.
Table 5 shows the results obtained from all three methods. Both ΔCt and NormFinder analyses indicated a similar set of ideal reference genes for each experimental condition. Essentially, the best were FaTIM1 for ripening, FaEF1α for infected fruits, FaEF1α and FaGAPDH2 for Camarosa crown and petiole infected tissues, respectively, FaACTIN for Andana infected petioles, FaRIB413 for in-vitro hormone-treated plants, FaRIB413 for cellular suspension treatments, and finally, FaEF1α when all the experiments were analyzed together. Similar results were also obtained when BestKeeper algorithm was used.
Table 5. Ranking of candidate reference genes based on expression stability as assessed by ΔCt, Normfinder and BestKeeper methods.
| Ranking by STDEV values from ΔCt | |||||||||||||
| RCF | FaBZIP1 | FaGAPDH1 | FaUBQ1 | FaTUBα | FaCHC1 | FaMT1 | FaFHA1 | FaGAPDH2 | FaTUBβ | FaEF1α | FaRIB413 | FaACTIN | FaTIM1 |
| (1.64) | (1.60) | (1.29) | (1.15) | (1.03) | (0.99) | (0.99) | (0.96) | (0.93) | (0.85) | (0.84) | (0.82) | (0.82) | |
| FCF | FaBZIP1 | FaMT1 | FaFHA1 | FaTIM1 | FaCHC1 | FaUBQ1 | FaGAPDH2 | FaTUBβ | FaTUBα | FaRIB413 | FaGAPDH1 | FaACTIN | FaEF1α |
| (0.72) | (0.69) | (0.65) | (0.62) | (0.62) | (0.61) | (0.59) | (0.57) | (0.57) | (0.54) | (0.54) | (0.50) | (0.47) | |
| FCC | FaBZIP1 | FaFHA1 | FaGAPDH1 | FaTIM1 | FaTUBβ | FaCHC1 | FaACTIN | FaRIB413 | FaMT1 | FaGAPDH2 | FaTUBα | FaUBQ1 | FaEF1α |
| (1.12) | (1.08) | (1.07) | (1.04) | (0.99) | (0.96) | (0.96) | (0.95) | (0.94) | (0.88) | (0.88) | (0.88) | (0.82) | |
| FCP | FaGAPDH1 | FaFHA1 | FaTIM1 | FaBZIP1 | FaTUBβ | FaCHC1 | FaMT1 | FaACTIN | FaUBQ1 | FaEF1α | FaTUBα | FaRIB413 | FaGAPDH2 |
| (1.28) | (1.11) | (1.07) | (1.04) | (1.02) | (1.00) | (0.96) | (0.90) | (0.89) | (0.89) | (0.87) | (0.83) | (0.76) | |
| FAP | FaGAPDH1 | FaFHA1 | FaUBQ1 | FaBZIP1 | FaTIM1 | FaRIB413 | FaGAPDH2 | FaCHC1 | FaMT1 | FaTUBα | FaEF1α | FaTUBβ | FaACTIN |
| (0.94) | (0.82) | (0.80) | (0.75) | (0.74) | (0.74) | (0.72) | (0.70) | (0.68) | (0.68) | (0.60) | (0.59) | (0.59) | |
| HCY | FaACTIN | FaTIM1 | FaBZIP1 | FaGAPDH1 | FaTUBβ | FaMT1 | FaUBQ1 | FaFHA1 | FaCHC1 | FaTUBα | FaEF1α | FaGAPDH2 | FaRIB413 |
| (0.89) | (0.85) | (0.83) | (0.83) | (0.82) | (0.82) | (0.80) | (0.80) | (0.73) | (0.71) | (0.71) | (0.70) | (0.68) | |
| HCC | FaGAPDH1 | FaTIM1 | FaTUBβ | FaACTIN | FaUBQ1 | FaMT1 | FaGAPDH2 | FaBZIP1 | FaFHA1 | FaTUBα | FaCHC1 | FaEF1α | FaRIB413 |
| (1.14) | (1.13) | (1.06) | (0.92) | (0.89) | (0.84) | (0.81) | (0.79) | (0.78) | (0.75) | (0.72) | (0.70) | (0.70) | |
| All samples | FaBZIP1 | FaGAPDH1 | FaMT1 | FaTIM1 | FaTUBα | FaCHC1 | FaGAPDH2 | FaUBQ1 | FaTUBβ | FaRIB413 | FaFHA1 | FaACTIN | FaEF1α |
| (2.02) | (1.90) | (1.79) | (1.70) | (1.50) | (1.39) | (1.37) | (1.34) | (1.34) | (1.32) | (1.28) | (1.24) | (1.21) | |
| Ranking by stability values from NormFinder | |||||||||||||
| RCF | FaBZIP1 | FaGAPDH1 | FaUBQ1 | FaTUBα | FaCHC1 | FaMT1 | FaFHA1 | FaTUBβ | FaGAPDH2 | FaRIB413 | FaEF1α | FaACTIN | FaTIM1 |
| (1.533) | (1.498) | (1.103) | (0.845) | (0.738) | (0.638) | (0.638) | (0.571) | (0.535) | (0.396) | (0.379) | (0.267) | (0.243) | |
| FCF | FaBZIP1 | FaMT1 | FaFHA1 | FaTIM1 | FaCHC1 | FaUBQ1 | FaGAPDH2 | FaTUBα | FaTUBβ | FaGAPDH1 | FaRIB413 | FaACTIN | FaEF1α |
| (0.610) | (0.565) | (0.523) | (0.466) | (0.466) | (0.444) | (0.430) | (0.397) | (0.387) | (0.343) | (0.341) | (0.277) | (0.177) | |
| FCC | FaBZIP1 | FaGAPDH1 | FaFHA1 | FaTIM1 | FaTUBβ | FaACTIN | FaCHC1 | FaRIB413 | FaMT1 | FaUBQ1 | FaGAPDH2 | FaTUBα | FaEF1α |
| (0.907) | (0.856) | (0.840) | (0.784) | (0.745) | (0.673) | (0.670) | (0.662) | (0.630) | (0.573) | (0.571) | (0.554) | (0.429) | |
| FCP | FaGAPDH1 | FaFHA1 | FaTIM1 | FaTUBβ | FaBZIP1 | FaCHC1 | FaMT1 | FaACTIN | FaEF1α | FaUBQ1 | FaTUBα | FaRIB413 | FaGAPDH2 |
| (1.119) | (0.890) | (0.821) | (0.807) | (0.800) | (0.723) | (0.673) | (0.605) | (0.559) | (0.552) | (0.543) | (0.429) | (0.272) | |
| FAP | FaGAPDH1 | FaFHA1 | FaUBQ1 | FaBZIP1 | FaTIM1 | FaRIB413 | FaGAPDH2 | FaCHC1 | FaTUBα | FaMT1 | FaEF1α | FaTUBβ | FaACTIN |
| (0.809) | (0.661) | (0.637) | (0.564) | (0.550) | (0.548) | (0.507) | (0.478) | (0.463) | (0.439) | (0.300) | (0.283) | (0.277) | |
| HCY | FaACTIN | FaTIM1 | FaGAPDH1 | FaBZIP1 | FaMT1 | FaUBQ1 | FaTUBβ | FaFHA1 | FaCHC1 | FaTUBα | FaEF1α | FaGAPDH2 | FaRIB413 |
| (0.726) | (0.643) | (0.639) | (0.633) | (0.616) | (0.614) | (0.600) | (0.581) | (0.461) | (0.460) | (0.432) | (0.425) | (0.413) | |
| HCC | FaGAPDH1 | FaTIM1 | FaTUBβ | FaACTIN | FaUBQ1 | FaMT1 | FaGAPDH2 | FaBZIP1 | FaFHA1 | FaTUBα | FaCHC1 | FaEF1α | FaRIB413 |
| (1.013) | (0.972) | (0.932) | (0.746) | (0.647) | (0.604) | (0.525) | (0.490) | (0.473) | (0.401) | (0.356) | (0.324) | (0.297) | |
| All samples | FaBZIP1 | FaGAPDH1 | FaMT1 | FaTIM1 | FaTUBα | FaCHC1 | FaGAPDH2 | FaUBQ1 | FaTUBβ | FaRIB413 | FaFHA1 | FaACTIN | FaEF1α |
| (1.795) | (1.626) | (1.493) | (1.397) | (1.075) | (0.932) | (0.918) | (0.840) | (0.787) | (0.734) | (0.686) | (0.578) | (0.538) | |
| Ranking by SD of Cp from BestKeeper | |||||||||||||
| RCF | FaGAPDH1 | FaBZIP1 | FaTIM1 | FaMT1 | FaTUBα | FaCHC1 | FaTUBβ | FaUBQ1 | FaFHA1 | FaGAPDH2 | FaEF1α | FaACTIN | FaRIB413 |
| (1.52) | (1.36) | (1.34) | (1.26) | (1.09) | (1.06) | (0.95) | (0.89) | (0.88) | (0.85) | (0.82) | (0.76) | (0.35) | |
| FCF | FaMT1 | FaBZIP1 | FaCHC1 | FaTIM1 | FaTUBβ | FaGAPDH2 | FaUBQ1 | FaRIB413 | FaFHA1 | FaGAPDH1 | FaACTIN | FaTUBα | FaEF1α |
| (0.60) | (0.56) | (0.48) | (0.48) | (0.42) | (0.32) | (0.32) | (0.32) | (0.20) | (0.18) | (0.18) | (0.18) | (0.00) | |
| FCC | FaBZIP1 | FaTUBβ | FaTIM1 | FaACTIN | FaFHA1 | FaGAPDH1 | FaGAPDH2 | FaCHC1 | FaMT1 | FaRIB413 | FaUBQ1 | FaEF1α | FaTUBα |
| (0.84) | (0.73) | (0.72) | (0.69) | (0.64) | (0.61) | (0.59) | (0.59) | (0.53) | (0.53) | (0.46) | (0.46) | (0.40) | |
| FCP | FaGAPDH1 | FaFHA1 | FaCHC1 | FaTIM1 | FaTUBβ | FaBZIP1 | FaUBQ1 | FaEF1α | FaACTIN | FaMT1 | FaTUBα | FaRIB413 | FaGAPDH2 |
| (0.88) | (0.73) | (0.69) | (0.66) | (0.63) | (0.63) | (0.47) | (0.47) | (0.41) | (0.38) | (0.33) | (0.30) | (0.22) | |
| FAP | FaGAPDH1 | FaFHA1 | FaMT1 | FaTIM1 | FaTUBα | FaRIB413 | FaUBQ1 | FaEF1α | FaACTIN | FaGAPDH2 | FaCHC1 | FaBZIP1 | FaTUBβ |
| (0.65) | (0.56) | (0.50) | (0.49) | (0.46) | (0.46) | (0.43) | (0.41) | (0.34) | (0.27) | (0.27) | (0.27) | (0.24) | |
| HCY | FaACTIN | FaBZIP1 | FaTUBβ | FaTIM1 | FaMT1 | FaTUBα | FaUBQ1 | FaRIB413 | FaEF1α | FaFHA1 | FaCHC1 | FaGAPDH1 | FaGAPDH2 |
| (0.78) | (0.73) | (0.72) | (0.60) | (0.57) | (0.56) | (0.49) | (0.48) | (0.46) | (0.44) | (0.43) | (0.40) | (0.35) | |
| HCC | FaTUBβ | FaTIM1 | FaACTIN | FaUBQ1 | FaGAPDH1 | FaMT1 | FaFHA1 | FaGAPDH2 | FaBZIP1 | FaTUBα | FaCHC1 | FaEF1α | FaRIB413 |
| (0.83) | (0.78) | (0.75) | (0.68) | (0.67) | (0.56) | (0.56) | (0.49) | (0.44) | (0.42) | (0.38) | (0.15) | (0.15) | |
| All samples | FaGAPDH1 | FaBZIP1 | FaTIM1 | FaMT1 | FaTUBα | FaCHC1 | FaTUBβ | FaUBQ1 | FaFHA1 | FaGAPDH2 | FaEF1α | FaACTIN | FaRIB413 |
| (1.52) | (1.36) | (1.34) | (1.26) | (1.09) | (1.06) | (0.95) | (0.89) | (0.88) | (0.85) | (0.82) | (0.76) | (0.35) | |
Increasing stability from left to right. STDEV and SD, represent standard deviation; Cp and Ct, represent Cq for different methods.
Combination of All Five Methods Used for Selective Classification of Reference Genes by RankAggreg
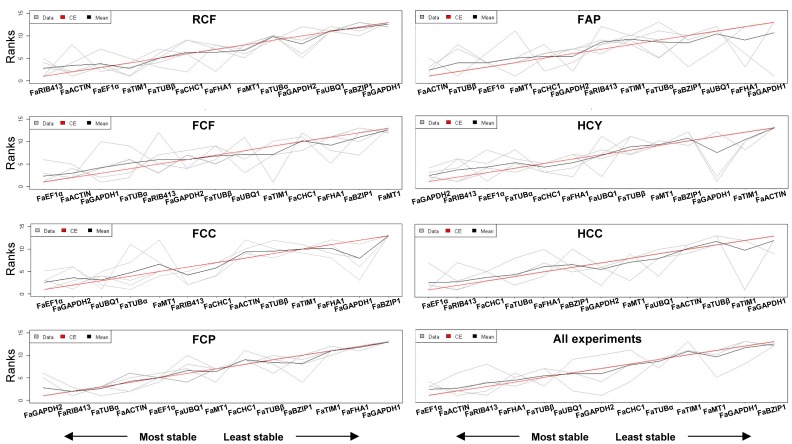
Combined stability measurements were generated by merging all five approaches (“stability index”, geNormPLUS, ΔCt method, Normfinder, and BestKeeper) to establish a consensus rank of reference genes by applying RankAggreg [40]. The input to this statistical package was a matrix of rank-ordered genes according to the different stability measurements previously computed by each of the five methods described above. RankAggreg calculated Spearman footrule distances and the software reformatted this distance matrix into an ordered list that matched each initial order as closely as possible. This consensus rank list was obtained by means of the Cross-Entropy Monte Carlo algorithm present in the software.
As shown in Figure 4, results of the merged data revealed that the most appropriate reference genes from all the preselected candidates tested for normalization are FaRIB413 and FaACTIN for analysis of strawberry fruit ripening, FaEF1α and FaACTIN for defense response studies in fruit, FaEF1α and FaGAPDH2, and FaGAPDH2 and FaRIB413, for defense response studies in crown and petiole, respectively, of cultivar Camarosa, FaACTIN and FaTUBβ, for defense response studies in petiole of cultivar Andana, FaGAPDH2 and FaRIB413 for SA and JA treatment of in-vitro plants, and FaEF1α and FaRIB413 for SA and JA treatment of cellular suspensions. Finally, FaEF1α and FaACTIN are the most stably expressed genes when all 48 experimental conditions are evaluated together.
Figure 4. Rank aggregation of gene lists using the Monte Carlo algorithm.
Visual representation of rank aggregation using Monte Carlo algorithm with the Spearman footrule distances. The solution of the rank aggregation is shown in a plot where genes are ordered based on their rank position according to their stability measurement (grey lines). Mean rank position of each gene is shown in black, as well the model computed by the Monte Carlo algorithm (red line). See Table 2 for experimental description.
The least stable, and therefore the least recommended reference genes are FaGAPDH1 and FaBZIP1 for analysis of strawberry fruit ripening, FaMT1 and FaBZIP1 for defense response studies in fruit, FaBZIP1 and FaGAPDH1, and FaGAPDH1 and FaFHA1 for defense response studies in crown and in petiole, respectively of cultivar Camarosa, FaGAPDH1 and FaFHA1 for defense response studies in petioles of cultivar Andana, FaACTIN and FaTIM1 for SA and JA treatment of in-vitro plants, and FaGAPDH1 and FaTIM1 for SA and JA treatment of cellular suspensions. Finally, FaBZIP1 and FaGAPDH1 was the least recommended when all the experiment are considered together.
Validation of the Selected Superior Reference Genes
In order to validate the selected superior reference genes, the relative expression level of the strawberry gene encoding the transcription factor FaWRKY1 (AtWRKY75 ortholog, [30]) was determined in all the experimental sets of evaluated conditions. The strawberry gene FaWRKY1 acts as positive regulator of defense response during compatible and incompatible interactions in Arabidopsis and, very likely, FaWRKY1 is an important element mediating defense responses to C. acutatum in strawberry. We also know that the FaWRKY1 gene is significantly upregulated in strawberry tissues under C. acutatum attack, and after SA and MeJA treatments ([30], Amil-Ruiz et al., unpublished data).
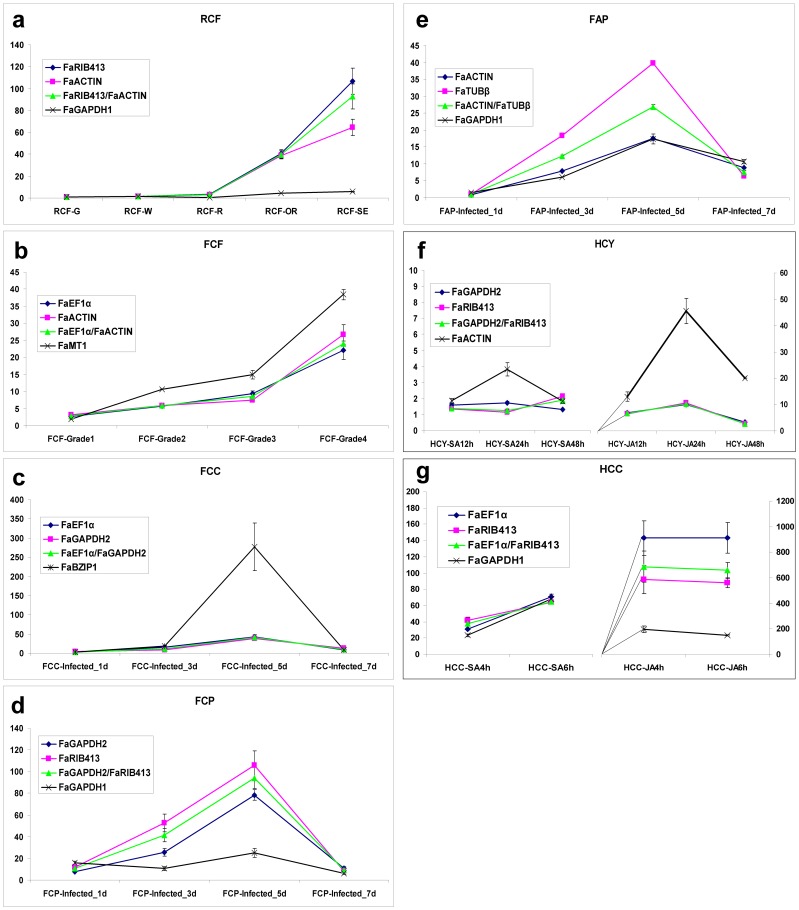
To analyze the bias effect on target expression analysis by selection of an inappropriate reference gene, FaWRKY1 was normalized to either a combination of the two best candidates ranked by RankAgreg algorithm as recommended by geNorm (Figures 3 and 4), or the least recommended one. FaWRKY1 primer sequences and other characteristics are listed in Table 1. As predicted, the reported expression profile of FaWRKY1 is strongly affected by the choice of the reference gene. Thus, in the strawberry fruit ripening conditions (RCF) as well as for infected petioles of cultivar Camarosa (FCP) and elicited cellular suspensions (HCC), the expression level values were similar to those previously reported ([30]) when the reference genes were the two most recommended ones (FaRIB413 and FaACTIN, FaGAPDH2 and FaRIB413, FaEF1α and FaRIB413, respectively), either individually or combined as geometric mean (Figures 5a, 5d and 5g). To the contrary, a strong bias in the FaWKRY1 expression pattern was obtained when the least recommended gene (FaGAPDH1 in all three cases) was used for normalization. From these data the use of FaGAPDH1 as reference gene somehow neutralizes the detectable induction of FaWRKY1 during fruit ripening and senescence, in the response to infection and after elicitation with SA and MeJA compounds.
Figure 5. Transcript level relative quantification of the FaWRKY1 transcription factor.
FaWRKY1 gene expression was analyzed in strawberry under the seven independent experimental conditions used in this study. Error bars show standard deviation calculated from two biological replicates. Normalization factors were calculated as the geometric mean of the expression levels of the two most stable reference genes as recommended in Figure 4 for each single experiment. Normalization to each gene individually is also shown. Additionally, the least stable reference gene was used for normalization of each experiment to demonstrate the effect of unstable reference genes in the quantification of the relative amount of target gene mRNA. Every sample was calibrated with their corresponding mock sample (see Table 2 for experimental details). Black lines linked to the X axis have been added to f and g to illustrate range of gene induction.
Interestingly, in other three experimental conditions (FCF, FCC and HCY) the use of the least stable reference gene (FaMT1, FaBZIP1 and FaACTIN respectively) seemed to have opposite influence in the detection of accurate expression values of the FaWRKY1 target gene. In this case the induction of this target gene was artificially high (Figures 5b, 5c and 5f). This is probably due to slightly but opposite variation of the corresponding reference mRNA levels during the analyzed process. These variations have significant impact in the final relative quantification of the expression of the target gene. Only ‘Andana’ petioles under fungal infection (FAP experiment) showed insignificant differences in the FaWRKY1 expression when either the best (FaACTIN) or the least effective (FaGAPDH1) reference candidates were considered.
Discussion
This work has mainly been focused to the evaluation of a set of potential strawberry reference genes for plant-defense response studies. Therefore, a variety of biological samples representing experimental conditions used to evaluate plant defense responses were used. The effect of natural pathogen infection as well as fruit senescence and decay are represented by experiments in this report through the analysis of fruit ripening and fruit natural infection by C. acutatum. Other tissues from Camarosa and Andana strawberry cultivars under fungal infection conditions were also included in this study, allowing comparisons between vegetative tissues within a cultivar, and between same tissues in different cultivars. Also, strawberry cultivars grown under contrasting contexts (in-vitro plants and cellular suspensions) were compared after treatment with either SA or JA, two phytohormones implicated in the activation of two well-known plant defense signaling pathways. A reference candidate with stability across such a range of conditions would be likely to perform well in narrower comparisons.
Several statistical procedures and software packages have been implemented to test which reference gene is best suited for transcript normalization in a given subset of biological samples. Each algorithm has its own strengths and limitations, so consensus among multiple tests provides great confidence that the results will be accurate and widely applicable.
Two methods, ΔCt and “stability index”, perform studies about the variation of ΔCt in pairwise genes or simple Ct, respectively. The comparative ΔCt method ranks the reference genes by their mean standard deviation in the pairwise comparisons, while the “stability index” approach introduces statistics and linear regression analysis to rank the candidates by the product of the coefficient of variation and slope of regression of gene means against overall means for the different samples. In the latter method (Table 3, Figure S2), although genes with the lowest “stability index” values represent the best option for normalization, many of the other strawberry candidate genes may also be considered acceptable as controls based on the SI value obtained in this study. In addition, the level of expression of the reference genes compared to that of the genes being analyzed is an important factor to be considered in certain cases [10]. Thus, the two most stably expressed strawberry genes in all seven experiments together exhibited the greatest range in steady-state transcript accumulation. FaRIB413 was detected at relatively high levels due to its role as a structural component of the ribosome (mean Cq = 8.542), whereas FaACTIN was expressed at a much lower level (mean Cq = 24.011). Therefore, they may be considered as appropriate reference genes to test target genes with high or low transcript levels, respectively. Indeed, the FaRIB413 RNA has been demonstrated to be an appropriate internal control for strawberry expression studies across several tissues and experimental conditions, using RNA-gel blots or RTqPCR analyses [41], [29], [30]. FaRIB413 has also been recommended for studies of strawberry genes expressed at relatively low levels, but it must be diluted up to 4000 times in order to equilibrate this transcript to general expression levels an achieve comparative Cq analyses [29]. Using the “stability index” method, it appears clear that FaACTIN may serve as a non-diluted reference instead of FaRIB413.
The geNORM program (Table 4) uses pair-wise comparisons and geometric averaging across a matrix of reference genes and biological samples to determine the best reference. The program calculates the expression stability value (MA) and allows accurate normalization of RTqPCR data [4], [37]. However, this approach leaves the method vulnerable to errors due to co-regulation, which tends to select those genes with the highest degree of similarity in their expression profiles [9]. On the other hand, it has the advantages that it is minimally affected by expression intensity of the reference genes [42] and it can determine the optimal number of genes (V) required to accurately normalize RTqPCR data based in pairwise variation [4]. Accordingly, two common well established sets of candidates with relatively high and low stability values were detected in all experimental conditions. FaEF1α always appears well positioned in the experimental conditions tested, and FaACTIN is stably expressed in ripening and mostly all infection conditions (except in crown tissue of cultivar Camarosa).
In contrast, the FaGAPDH1 and FaBZIP1 transcripts mostly showed high MA values (a lower stability) in all conditions. FaFHA1 is stably expressed in all conditions except in all infected tissues from cultivar Camarosa, and FaRIB413 is also stable but only in infected crown and petiole tissues from the same cultivar. On the other hand, the FaTIM1 transcript presented high MA values in all conditions except the two fruit experiments, where its accumulation was stable. The FaMT1 transcript presented low stability in all ‘Camarosa’ experimental conditions, but low MA values when cultivar Andana and Chandler are considered.
Unlike geNORM, NormFinder is not affected by correlated expression of the candidate genes (Table 5). However, the latter gains in robustness as the sample number is increased, while geNorm doesn’t need large sample size since it uses pair-wise comparisons. The Bestkeeper algorithm also performs pairwise comparisons using the geometric mean of the Cp (Cq) values, but different expression levels can generate heterogeneous variance between groups, and this can invalidate the use of Pearson correlation coefficient [43], [5]. The results from these two methodologies coincide with that of ΔCt method, and taken together these results indicate that gene FaEF1α seemed to be the most stably expressed reference gene meanwhile genes FaGAPDH1 and FaBZIP1 were the least stable ones.
Recommended Reference Genes in a Strawberry-defense Response Context
We have applied RankAggreg [40] to establish a consensus rank of reference genes by combination of all five above methods. This approach strengthens the value of the recommended candidates to normalize target gene expression in any of the conditions here described. Thus, results in Figure 4 show genes recommended in each particular experiment, suggesting they can be used as superior reference genes for transcript quantitation. Taken together, we propose genes FaRIB413, FaACTIN, FaEF1α and FaGAPDH2 as superior reference genes for accurate transcript normalization in strawberry (Fragaria × ananassa) under the described experimental conditions.
The genes proposed here have been reported in previous strawberry studies (see Table 1), although no experimental work was performed to validate their usefulness as RTqPCR reference genes in a variety of tissues, treatments or conditions. As previously stated, the FaRIB413 gene has been extensively used for northern and RTqPCR normalization in strawberry [41], [29], [44], [30], [45], [46]. However, FaRIB413 encodes a highly abundant ribosomal RNA (Cq around 8 in our study, Table 3), which does not contain a poly(A) tail, making it unsuitable for RTqPCR analysis aimed at differentiating the expression levels of rare genes, and also for the synthesis with cDNA using oligo(dT) primers. Although FaRIB413 presents good values of expression stability in almost all of the experiments analyzed by RankAggreg (Figure 4), it is strongly recommended that an alternative strawberry reference with Cq values as close as possible to the Cq values showed by the target gene be used.
An actin transcript was used by Lin-Wang et al. (2010) [28], for normalization of RTqPCR studies in different strawberry plant tissues. The authors selected this gene as a reference gene “because of its consistent transcript level throughout fruits and leaves”. From our results, FaACTIN presents high stability in all fruit experimental conditions, such as ripening and infection, in ‘Andana’ petiole tissues, and also considering all the experiments together. These data match well with the analysis reported by Lin-Wang et al. (2010) [28]. However, this FaACTIN gene was not appropriate when vegetative tissues of cultivar Camarosa (crown and petioles) were exposed to fungal infection, or by phytohormone elicitation either of strawberry plants or cellular suspensions.
Also, a strawberry elongation factor 1α gene (EF1a) was used by Guidarelli et al. (2011), to normalize raw expression data in an RTqPCR experiment with fruits of the very susceptible strawberry cultivar Alba inoculated with C. acutatum. [27]. Although authors did not assess the stability of expression of this gene by none of the available methods, they detected that this gene had “the most constant expression levels (absolute ΔCt <1 among treatments)”, and assumed this candidate gene for data normalization. From our results, FaEF1α is indeed recommended as the best candidate for normalization of experiments based on strawberry fruits under biotic interaction. Therefore, our analysis agrees with the controls used by Guidarelli et al. (2011).
In addition, FaGAPDH1 and FaGAPDH2 genes have been previously used as reference genes in plant-pathogen interaction studies [26], [47], [48], [49]. In the case of FaGAPDH2 gene reported by Khan et al. (2004), our results support the use of this gene as control in the experimental conditions reported by these authors, (i.e. strawberry vegetative tissues inoculated with Colletotrichum) (see Figure 4). The FaGAPDH1 reference gene has been reported for use in strawberry experimental treatments with phytohormones or after fungal inoculation, as reported Grellet-Bournonville et al. (2012), Mamaní et al. (2012) and Zamora et al. (2012). The data in the current report indicate that this reference may not have been the best choice as this transcript has shown the lowest values of stability in almost all the experimental conditions.
The comparative analysis between using the most and the least appropriate reference gene in a given experiment (Figure 5) illustrates the magnitude of the bias produced by normalization with an unstable gene, and also highlights how the incorrect use of reference genes without any previous validation can lead to misinterpretation of data. For this reason we strongly recommend to perform a validation of the putative reference genes prior any quantitative expression studies, as recommended elsewhere [50], [23], [51], [52], [19].
It is important to note that in certain species even the best reference candidates show some variation across the different tissues, developmental stages and environmental conditions [10]. Differences in the defense gene expression patterns have been reported across different strawberry tissues and cultivars challenged with C. acutatum [29]. These observations indicate that the first step in any gene expression experiment should be to test reference candidates in the specific genetic background and in the same experimental conditions. This validation is especially important when testing effects of strong biotic or abiotic stresses, such as pathogen challenge.
In conclusion, stably expressed genes were selected from two independent strawberry biological replicates of a total of forty eight samples, representing seven different experimental conditions. Our results represent a relevant contribution to the scientific plant community as the best candidates for reference genes in strawberry. The candidates have been ranked accordingly to their respective expression stability in a variety of samples representing major conditions typically used in a plant-defense context. The identification of other stable reference pools under different experimental conditions would build a useful community resource for gene expression analysis in this crop.
Materials and Methods
Plant Materials and Growth Conditions
Plant material, Fragaria × ananassa cultivars Chandler, Camarosa and Andana were used. Colletotrichum acutatum, a major strawberry pathogen was used for natural infection and controlled inocculation. All the plant culture and growth conditions, C. acutatum experimental conditions, and treatments with chemicals have been previously described [29], [30], and are summarized in Table 2. Briefly, strawberry cellular suspensions (cv. Chandler) were prepared from in vitro growing calli. Five days old cell suspensions were treated with MeJa (0.1 mM), SA (0.75 mM) or water (as control). Alicuots were taken at 2 hour intervals and cells were frozen in liquid nitrogen. Samples at 4 and 6 hours were used in this work because they match with a strong relative expression of the FaWRKY1 target gene, and many other strawberry genes currently under study in our lab. Axenic in-vitro plants from cv. Camarosa were aseptically sprayed with water, MeJa (2 mM) and SA (5 mM) solutions and collected at 12, 24 and 48 hours post-treatment. Strawberry fruits were collected from a growing field in several ripening stages and pooled by stage. Red stage strawberry fruits naturally-infected by Colletotrichum acutatum and exhibiting different increasing degrees of fungal necrotic lesions were collected and fruits having similar symptoms were pooled. No specific permissions were required for these activities. None human manipulation was applied to strawberry field prior to sample collection. Field studies did not involve endangered or protected species. Eight-week-old strawberry plantlets were placed in 20 cm diameter plastic pots containing sterilized peat and grown for a minimum of six additional weeks prior to mock or pathogen inoculation by spraying a spore suspension of 106 CFU ml−1. Crowns and petioles were collected 1, 3, 5 and 7 days after treatment. All samples were flash frozen in liquid nitrogen and stored at −80°C until needed.
RNA Preparation for RTqPCR
Total RNA from strawberry fruits and vegetative tissues, as well as cell suspension cultures, was isolated according to Manning [53], treated with DnaseI (Invitrogen) to remove the residual contaminating DNA, and further purified with the RNeasy MinElute Cleanup Kit (QIAGEN). Extracted RNA samples showed high degree of purity, without residual contamination by organic compounds, accordingly to Accerbi et al. (2010) [54]. RNA samples were tested to be free of genomic DNA contamination after DNase I treatment by performing a qPCR analysis using the primer pairs corresponding to the FaGAPDH2 and FaRIB413 genes. Amplicons corresponding to these two genes were undetectable in the RNA samples after 40 cycles as confirmed by qPCR or by agarose gel electrophoresis (data not shown). These results indicated that amplicons generated by RTqPCR analysis were produced only from synthesized cDNA. Purified RNA was quantified by the NanoDrop 1000 Spectrophotometer (Thermo scientific) and the integrity checked by agarose gel electrophoresis and the Agilent 2100 Bioanalyzer (Agilent Technologies, Deutschland). All the samples showed RIN values over 8 (data not shown) and therefore were deemed suitable for RTqPCR analysis.
To ensure equal concentrations of RNA in all samples prior to the RT reactions, samples were diluted to 200 ng/ul and reassessed three times in a serial dilution of 1∶0, 1∶5 and 1∶25, to ensure fidelity of the measure. First-strand cDNA synthesis was carried out by the iScript cDNA Synthesis kit (Bio-Rad) using as template 1 µg of purified total RNA per 20 µL of reaction volume. RT reactions were diluted 5-fold with nuclease-free water prior to be used in the qPCR.
Real-time qPCR
Specific primer pairs set for the genes tested were designed using Oligo Primer Analysis software version 6.65, tested by dissociation curve analysis, and verified for the absence of non-specific amplification. More details are provided in results. RTqPCR runs were performed in MyIQ and iCycler real-time PCR systems (Bio-Rad) using 96-well plates and 20 µL final reaction volume per well. Two µL template cDNA was added to the PCR reaction mixture containing 0.4 µM of each primer and 10 µL of 2× SsoAdvanced™ SYBR® Green supermix (Bio-Rad). The protocol was: an inicial step of enzyme activation/DNA denaturation of 95°C for 1 min, followed by 40 cycles of 95°C for 15 sec, 65°C for 15 sec and 72°C for 15 sec, and a final standard dissociation protocol to obtain the melting profiles. Data were adquired by means of the MyIQ v1.004 and iCycler v3.1 softwares (Bio-Rad).
Computational Data Analysis
Data analysis strategy is described in detail in the results section. Reaction efficiency calculus was done using LinRegPCR version 2012.3 [36], [55]. Resulting mean PCR efficiencies per amplicon were taken. Reference genes validation was performed using previously described software applications, included the MS Excel VBA applets NormFinder v0.953 [9] and BestKeeper v1 [38], and the geNorm [4] algorithm provided in qBasePlus v2.4 package [37]. Other statistical procedures were performed with the free software R v2.15.2 (http://www.R-project.org), with the packages RankAggreg 0.4–3, clValid 0.6–4 and gtools 2.7.0; and SPSS software ver 15.0 for Windows.
Supporting Information
Dissociation curves and agarose gel analysis of the amplicons tested in this study. (a) Melting curve analysis of 13 potential reference genes along with control gene for validation (FaWRKY1) was carried out to confirm the absence of multiple amplicon species after RTqPCR. Each line represents a melting curve of amplicons from two technical replicates of two biological replicates in the given experiments. (b) Agarose gel electrophoresis of RTqPCR products after 40 cycles of PCR.
(TIF)
Regression analysis for several genes showing predicted regression lines and actual means over all experiments. The most stable and consistent control genes would have the lowest slope and closest fit to the regression line. (a) FaACTIN (first in top) had the highest stability and FaRIB413, as well as FaEF1α and FaTUBβ, have also very good values of stability (from first in bottom to second in top). (b) Genes FaBZIP1 and FaTIM1 had the lowest stability index. See Table 2 for descriptions of tissue samples, represented here by abbreviations.
(TIF)
Acknowledgments
Authors want to thank Fernando Pliego-Alfaro, Fernando Romero-Muñoz and Berta de los Santos for providing part of plant materials used in this study.
Funding Statement
This work was supported by Junta de Andalucía (Proyecto de Excelencia P07-AGR-02482, and grants to Grupo-BIO278). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Czechowski T, Bari RP, Stitt M, Scheible W-R, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. The Plant Journal 38: 366–379. [DOI] [PubMed] [Google Scholar]
- 2. Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? Journal of Experimental Botany 55: 1445–1454. [DOI] [PubMed] [Google Scholar]
- 3. Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR – a perspective. Journal of Molecular Endocrinology 34: 597–601. [DOI] [PubMed] [Google Scholar]
- 4. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 6. Goidin D, Mamessier A, Staquet M-J, Schmitt D, Berthier-Vergnes O (2001) Ribosomal 18S RNA Prevails over Glyceraldehyde-3-Phosphate Dehydrogenase and β-Actin Genes as Internal Standard for Quantitative Comparison of mRNA Levels in Invasive and Noninvasive Human Melanoma Cell Subpopulations. Analytical Biochemistry 295: 17–21. [DOI] [PubMed] [Google Scholar]
- 7. Bustin S (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Journal of Molecular Endocrinology 29: 23–39. [DOI] [PubMed] [Google Scholar]
- 8. Kim B-R, Nam H-Y, Kim S-U, Kim S-I, Chang Y-J (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnology Letters 25: 1869–1872. [DOI] [PubMed] [Google Scholar]
- 9. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Research 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 10. Brunner A, Yakovlev I, Strauss S (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, et al. (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques 37: 112–119. [DOI] [PubMed] [Google Scholar]
- 12. Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, et al. (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications 313: 856–862. [DOI] [PubMed] [Google Scholar]
- 13. Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, et al. (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of Experimental Botany 60: 487–493. [DOI] [PubMed] [Google Scholar]
- 14. Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiology 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre J-F, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal 6: 609–618. [DOI] [PubMed] [Google Scholar]
- 16. Udvardi MK, Czechowski T, Scheible W-R (2008) Eleven Golden Rules of Quantitative RT-PCR. The Plant Cell Online 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH (2010) Identification and Testing of Superior Reference Genes for a Starting Pool of Transcript Normalization in Arabidopsis. Plant and Cell Physiology 51: 1694–1706. [DOI] [PubMed] [Google Scholar]
- 18. Die J, Román B, Nadal S, González-Verdejo C (2010) Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232: 145–153. [DOI] [PubMed] [Google Scholar]
- 19. Podevin N, Krauss A, Henry I, Swennen R, Remy S (2012) Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa . Molecular Breeding 30: 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L, Zhong H-y, Kuang J-f, Li J-g, Lu W-j, et al. (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234: 377–390. [DOI] [PubMed] [Google Scholar]
- 21. Cordoba EM, Die JV, González-Verdejo CI, Nadal S, Román B (2011) Selection of reference genes in Hedysarum coronarium under various stresses and stages of development. Analytical Biochemistry 409: 236–243. [DOI] [PubMed] [Google Scholar]
- 22. Obrero An, Die JV, Román Bn, Gómez P, Nadal S, et al. (2011) Selection of Reference Genes for Gene Expression Studies in Zucchini (Cucurbita pepo) Using qPCR. Journal of Agricultural and Food Chemistry 59: 5402–5411. [DOI] [PubMed] [Google Scholar]
- 23. Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, et al. (2012) Reference Genes for Accurate Transcript Normalization in Citrus Genotypes under Different Experimental Conditions. Plos One 7: e31263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mezzetti B (2009) GMO strawberry: Methods, risk and benefits. In: Folta KM, Gardiner SE, editors. Genetics and genomics of rosaceae: Springer Science+Business Media, LLC.
- 25. Amil-Ruiz F, Blanco-Portales R, Muñoz-Blanco J, Caballero JL (2011) The Strawberry Plant Defence Mechanism: A Molecular Review. Plant and Cell Physiology 52: 1873–1903. [DOI] [PubMed] [Google Scholar]
- 26. Khan AA, Shih DS (2004) Molecular cloning, characterization, and expression analysis of two class II chitinase genes from the strawberry plant. Plant Science 166: 753–762. [Google Scholar]
- 27. Guidarelli M, Carbone F, Mourgues F, Perrotta G, Rosati C, et al. (2011) Colletotrichum acutatum interactions with unripe and ripe strawberry fruits and differential responses at histological and transcriptional levels. Plant Pathology 60: 685–697. [Google Scholar]
- 28. Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, et al. (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biology 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Casado-Díaz A, Encinas-Villarejo S, Santos Bdl, Schilirò E, Yubero-Serrano E-M, et al. (2006) Analysis of strawberry genes differentially expressed in response to Colletotrichum infection. Physiologia Plantarum 128: 633–650. [Google Scholar]
- 30. Encinas-Villarejo S, Maldonado AM, Amil-Ruiz F, de los Santos B, Romero F, et al. (2009) Evidence for a positive regulatory role of strawberry (Fragaria x ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. Journal of Experimental Botany 60: 3043–3065. [DOI] [PubMed] [Google Scholar]
- 31. Wong ML, Medrano JF (2005) Real-time PCR for mRNA quantitation. BioTechniques 39: 75–85. [DOI] [PubMed] [Google Scholar]
- 32. Hruz T, Wyss M, Docquier M, Pfaffl M, Masanetz S, et al. (2011) RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 12: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amil-Ruiz F, Encinas-Villarejo S, de los Santos B, Muñoz-Mérida A, Mercado JA, et al. (2012) Distinctive Transcriptome Response of Two Strawberry (Fragaria x ananassa) Cultivars to Colletotrichum acutatum Infection. Acta Hort (ISHS) 929: 47–50. [Google Scholar]
- 34. Fan HPY, Di Liao C, Fu BY, Lam LCW, Tang NLS (2009) Interindividual and Interethnic Variation in Genomewide Gene Expression: Insights into the Biological Variation of Gene Expression and Clinical Implications. Clinical Chemistry 55: 774–785. [DOI] [PubMed] [Google Scholar]
- 35. Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nature Genetics 43: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 37. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnology Letters 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 39. Silver N, Best S, Jiang J, Thein S (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pihur V, Datta S, Datta S (2009) RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benítez-Burraco A, Blanco-Portales R, Redondo-Nevado J, Bellido ML, Moyano E, et al. (2003) Cloning and characterization of two ripening-related strawberry (Fragaria × ananassa cv. Chandler) pectate lyase genes. Journal of Experimental Botany 54: 633–645. [DOI] [PubMed] [Google Scholar]
- 42. Mehta R, Birerdinc A, Hossain N, Afendy A, Chandhoke V, et al. (2010) Validation of endogenous reference genes for qRT-PCR analysis of human visceral adipose samples. BMC Molecular Biology 11: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lefever S, Hellemans J, Pattyn F, Przybylski DR, Taylor C, et al. (2009) RDML: structured language and reporting guidelines for real-time quantitative PCR data. Nucleic Acids Research 37: 2065–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, et al. (2008) Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). The Plant Journal 54: 43–55. [DOI] [PubMed] [Google Scholar]
- 45. Csukasi F, Osorio S, Gutierrez JR, Kitamura J, Giavalisco P, et al. (2011) Gibberellin biosynthesis and signalling during development of the strawberry receptacle. New Phytologist 191: 376–390. [DOI] [PubMed] [Google Scholar]
- 46. Moyano-Cañete E, Bellido ML, García-Caparrós N, Medina-Puche L, Amil-Ruiz F, et al. (2013) FaGAST2, a Strawberry Ripening-Related Gene, Acts Together with FaGAST1 to Determine Cell Size of the Fruit Receptacle. Plant and Cell Physiology 54: 218–236. [DOI] [PubMed] [Google Scholar]
- 47. Grellet-Bournonville CF, Martinez-Zamora MG, Castagnaro AP, Díaz-Ricci JC (2012) Temporal accumulation of salicylic acid activates the defense response against Colletotrichum in strawberry. Plant Physiology and Biochemistry 54: 10–16. [DOI] [PubMed] [Google Scholar]
- 48. Mamaní A, Filippone MP, Grellet Bournonville CF, Welin B, Castagnaro AP, et al. (2012) Pathogen-Induced Accumulation of an Ellagitannin Elicits the Plant Defense Response. Molecular Plant-Microbe Interactions 25: 1430–1439. [DOI] [PubMed] [Google Scholar]
- 49. Zamora MGM, Bournonville CG, Castagnaro AP, Ricci JCD (2012) Identification and characterisation of a novel class I endo-β-1,3-glucanase regulated by salicylic acid, ethylene and fungal pathogens in strawberry. Functional Plant Biology 39: 412–420. [DOI] [PubMed] [Google Scholar]
- 50. Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, et al. (2012) Identification of Reference Genes for RT–qPCR Expression Analysis in Arabidopsis and Tomato Seeds. Plant and Cell Physiology 53: 28–37. [DOI] [PubMed] [Google Scholar]
- 51. Matta B, Bitner-Mathé B, Alves-Ferreira M (2011) Getting real with real-time qPCR: a case study of reference gene selection for morphological variation in Drosophila melanogaster wings. Development Genes and Evolution 221: 49–57. [DOI] [PubMed] [Google Scholar]
- 52. de Oliveira LA, Breton MC, Bastolla FM, Camargo SdS, Margis R, et al. (2012) Reference Genes for the Normalization of Gene Expression in Eucalyptus Species. Plant and Cell Physiology 53: 405–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manning K (1991) Isolation of nucleic acids from plants by differential solvent precipitation. Analytical Biochemistry 195: 45–50. [DOI] [PubMed] [Google Scholar]
- 54.Accerbi M, Schmidt S, Paoli E, Park S, Jeong D-H, et al.. (2010) Methods for Isolation of Total RNA to Recover miRNAs and Other Small RNAs from Diverse Species. In: Meyers BC, Green PJ, editors. Plant MicroRNAs: Humana Press. 31–50. [DOI] [PubMed]
- 55. Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, et al. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dissociation curves and agarose gel analysis of the amplicons tested in this study. (a) Melting curve analysis of 13 potential reference genes along with control gene for validation (FaWRKY1) was carried out to confirm the absence of multiple amplicon species after RTqPCR. Each line represents a melting curve of amplicons from two technical replicates of two biological replicates in the given experiments. (b) Agarose gel electrophoresis of RTqPCR products after 40 cycles of PCR.
(TIF)
Regression analysis for several genes showing predicted regression lines and actual means over all experiments. The most stable and consistent control genes would have the lowest slope and closest fit to the regression line. (a) FaACTIN (first in top) had the highest stability and FaRIB413, as well as FaEF1α and FaTUBβ, have also very good values of stability (from first in bottom to second in top). (b) Genes FaBZIP1 and FaTIM1 had the lowest stability index. See Table 2 for descriptions of tissue samples, represented here by abbreviations.
(TIF)