Abstract
We report a method for studying global DNA methylation based on using bisulfite treatment of DNA and simultaneous PCR of multiple DNA repetitive elements, such as Alu elements and long interspersed nucleotide elements (LINE). The PCR product, which represents a pool of approximately 15 000 genomic loci, could be used for direct sequencing, selective restriction digestion or pyrosequencing, in order to quantitate DNA methylation. By restriction digestion or pyrosequencing, the assay was reproducible with a standard deviation of only 2% between assays. Using this method we found that almost two-thirds of the CpG methylation sites in Alu elements are mutated, but of the remaining methylation target sites, 87% were methylated. Due to the heavy methylation of repetitive elements, this assay was especially useful in detecting decreases in DNA methylation, and this assay was validated by examining cell lines treated with the methylation inhibitor 5-aza-2′deoxycytidine (DAC), where we found a 1–16% decrease in Alu element and 18–60% LINE methylation within 3 days of treatment. This method can be used as a surrogate marker of genome-wide methylation changes. In addition, it is less labor intensive and requires less DNA than previous methods of assessing global DNA methylation.
INTRODUCTION
The conversion of cytosine to 5-methylcytosine is an important epigenetic change in the vertebrate genome (1). DNA methyltransferase can transfer a methyl group from S-adenosyl-methionine to cytosine in CpG dinucleotides. This methylation of cytosine is associated with gene silencing, and genes with abundant 5-methylcytosine in their promoter region are usually transcriptionally silent (2). DNA methylation is vital during development, and aberrant DNA methylation, both hypermethylation and hypomethylation, has been associated with aging, cancer and other diseases (3–5). In addition, DNA methylation inhibitors can be used to treat cancer (6,7). Therefore, methods to study DNA methylation are important tools in biological research.
There are multiple methods to study DNA methylation. Most of these methods take advantage of a chemical reaction using sodium bisulfite, which can selectively deaminate cytosine but not 5-methylcytosine to uracil (8). This leads to a primary sequence change in the DNA that will allow distinguishing cytosine from 5-methylcytosine. Once this conversion has taken place the sequence differences between a methylated and unmethylated cytosine can be exploited by either direct sequencing, restriction digestion (COBRA) (9), nucleotide extension assays (MS-SnuPE) (10), primer-specific PCR (MSP) (11) or pyrosequencing (12). These methods are valuable in that they are not labor intensive and require smaller amounts of DNA. However, these methods are usually limited in that they can only study a single gene or locus at a time. Earlier methods of using methylation-sensitive restriction enzymes and Southern blotting to determine gene-specific DNA methylation have largely been replaced by these more convenient methods.
Gene-specific DNA methylation analysis does not provide a global picture of DNA methylation changes within a genome. However, there are several methods of detecting total 5-methylcytosine content in the genome. DNA can be digested into single nucleotides and total genomic 5-methylcytosine can be quantitated by either high-performance liquid chromatography (13,14), thin-layer chromatography (15), or liquid chromatography/mass spectroscopy (16). Global methylation patterns can also be quantitated using restriction digestion and nearest-neighbor analysis of DNA (17). Chloracetaldehyde can be used in a fluorescent assay to detect DNA methylation levels (18). SssI DNA methyltransferase, which methylates all CpG sites, can be used in conjunction with tritium-labeled S-adenosyl methionine to calculate the amount of unmethylated CpG sites and the level of DNA methylation can be inversely determined (19). These methods give a sense of global DNA methylation changes, but have the disadvantage of being labor intensive and/or requiring large amounts of good quality DNA as they are not PCR based.
In this paper we report a quick and easy method to assess global DNA methylation changes. This method is based on bisulfite treatment of DNA and PCR amplification of repetitive DNA elements. There are ∼1.4 million Alu repetitive elements in the human genome (20,21) and a half a million long interspersed nucleotide elements (LINE-1 elements) (22) that are normally heavily methylated, and it is estimated that more than one-third of DNA methylation occurs in repetitive elements (23–25). Thus, analyzing the methylation of repetitive elements can serve as a surrogate marker for global genomic DNA methylation. Genomic DNA is isolated and bisulfite treated using previously established methods. PCR is performed in non-stringent conditions using PCR primers designed from a consensus Alu or LINE repetitive element sequence that allows the amplification of a pool of several thousand repeats. The sequence difference in this pool of amplified repeats can be quantitated by a number of means to get a sense of global DNA methylation.
MATERIALS AND METHODS
DNA and cell lines
Hct116, RKO and SW48, colon cancer cell lines (American Type Culture Collection, Manassas, VA), were cultured using standard methods. Cells were treated with 5 µM 5-aza-2′deoxycytidine (DAC) for 3 days prior to being harvested.
DNA from cell lines and peripheral blood leukocytes was extracted using standard phenol–chloroform extraction methods.
Bisulfite treatment
Bisulfite modification of genomic DNA has been described previously (8). In brief, 1.5 µg of DNA was denatured in 50 µl of 0.2 M NaOH for 10 min at 37°C. Then, 30 µl of freshly prepared 10 mM hydroquinone (Sigma) and 520 µl of 3 M sodium bisulfite (Sigma) at pH 5.0 were added and mixed. The samples were overlaid with mineral oil to prevent evaporation and incubated at 50°C for 16 h. The bisulfite-treated DNA was isolated using Wizard DNA Clean-Up System (Promega). The DNA was eluted by 50 µl of warm water and 5.5 µl of 3 M NaOH were added for 5 min. The DNA was ethanol precipitated with glycogen as a carrier and resuspended in 20 µl of water. Bisulfite-treated DNA was stored at –20°C until ready for use.
PCR of repetitive elements
Methylation analysis of Alu repetitive elements was performed initially by the COBRA assay. A 25 µl PCR was carried out in 60 mM Tris–HCl pH 9.5, 15 mM ammonium sulfate, 5.5 mM MgCl2, 10% DMSO, 1 mM dNTP mix, 1 unit of Taq polymerase, 50 pmol of the forward primer (5′-GATCTTTTTATTAAAAATATAAAAATTAGT-3′), 50 pmol of the reverse primer (5′-GATCCCAAACTAAAATACAATAA-3′), and ∼50 ng of bisulfite-treated genomic DNA. The best COBRA results were obtained if the PCR primers contained a restriction site on the 5′ end that would be recognized by the COBRA restriction enzyme, which digested non-specific PCR products and helped prevent primer dimer formation. PCR cycling conditions were 96°C for 90 s, 43°C for 60 s and 72°C for 120 s for 27 cycles. The PCR product was then digested with 10 U of MboI. The digested PCR product was then separated by polyacrylamide gel electrophoresis or the PCR products were quantitated using a capillary electrophoresis system, an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
The Alu element PCR was modified for pyrosequencing based methylation analysis. A 50 µl PCR was carried out in 60 mM Tris–HCl pH 8.5, 15 mM ammonium sulfate, 2 mM MgCl2, 10% DMSO, 1 mM dNTP mix, 1 unit of Taq polymerase, 5 pmol of the forward primer (5′-GGGACACCGCTGATCGTATATTTTTATTAAAAATATAAAAATTAGT-3′), 50 pmol of the reverse primer (5′-CCAAACTAAAATACAATAA-3′), 50 pmol of biotinylated universal primer (5′-GGGACACCGCTGATCGTATA-3′) and ∼50 ng of bisulfite-treated genomic DNA. The forward primer has a 20 bp linker sequence on the 5′ end that is recognized by a biotin-labeled primer so the final PCR product can be purified using Sepharose beads. PCR cycling conditions were 96°C for 90 s, 43°C for 60 s and 72°C for 120 s for 40 cycles. The biotinylated PCR product was purified and made single-stranded to act as a template in a pyrosequencing reaction as recommended by the manufacturer using the Pyrosequencing Vacuun Prep Tool (Pyrosequencing, Inc., Westborough, MA). In brief, the PCR product was bound to Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden) and the Sepharose beads containing the immobilized PCR product were purified, washed, denatured using a 0.2 M NaOH solution, and washed again. Then, 0.3 µΜ pyrosequencing primer (5′-AATAACTAAAATTACAAAC-3′) was annealed to the purified single-stranded PCR product and pyrosequencing was performed using the PSQ HS 96 Pyrosequencing System (Pyrosequencing, Inc.). Methylation quantification was performed using the provided software.
Methylation of the LINE-1 promoter was also investigated by a similar COBRA assay. A 50 µl PCR was carried out in 60 mM Tris–HCl pH 8.8, 15 mM ammonium sulfate, 0.5 mM MgCl2, 1 mM dNTP mix, and 1 unit of Taq polymerase. Fifty picomoles of each PCR primer was used: 5′-TTGAGTTGTGGTGGGTTTTATTTAG-3′ and 5′-TCATCTCACTAAAAAATACCAAACA-3′. PCR cycling conditions were 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s for 35 cycles. The final PCR product was digested with the HinfI restriction enzyme. The digested PCR products were separated by electrophoresis on polyacrylamide gels. The cut bands representing methylated DNA were quantitated using densitometry.
Cloning and sequencing
PCR products were cloned using the TOPO-TA cloning kit (Stratagene) as per the manufacturer’s protocol. Mini-preps were prepared using QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). The M.D. Anderson Cancer Center Sequencing Core Facility performed all DNA sequencing.
RESULTS
PCR of bisulfite-treated Alu repetitive elements amplifies multiple unique Alu repetitive elements
We selected PCR primers that amplify an ∼150 bp fragment of bisulfite-treated DNA from Alu repetitive elements. The primers were designed to avoid potential CpG methylation sites in order to minimize amplification biases for methylated or unmethylated DNA; however, the primers used in this PCR will likely have biases for some Alu elements over others due to Alu element polymorphisms that will lead to primer annealing biases. In order to assure that a pool of different Alu elements was being amplified from the genome, the PCR product from four different human blood DNA samples was cloned using a TOPO TA-cloning kit (Invitrogen, Carlsbad, CA) and 60 individual clones representing individual PCR products were sequenced. The sequenced PCR products were all Alu elements, but all had unique sequences and no two clones showed an identical sequence. Representative data from one DNA sample are shown in Figure 1.
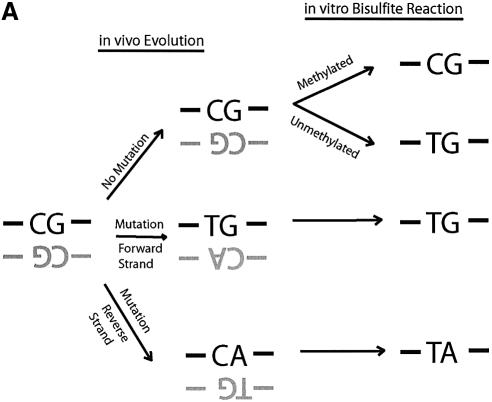
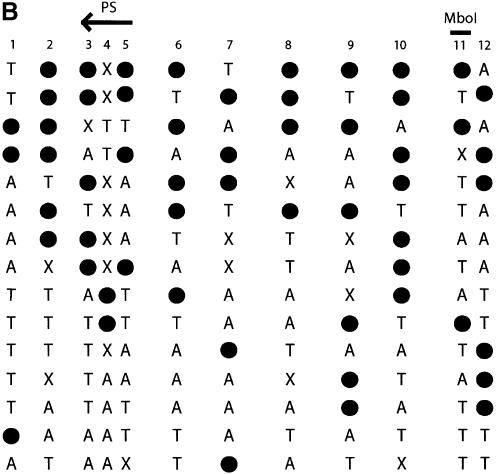
Figure 1.
Direct DNA sequencing of bisulfite repetitive element PCR of Alu elements. (A) Schematic of possible fates of Alu element CpG methylation sites. Due to the mutation of CpG sites via spontaneous deamination of 5-methylcytosine to T during evolution, a CpG site can be changed into a TpG or CpA dinucleotide. Neither TpG nor CpA are targets for methylation. Following bisulfite treatment, a methylated CpG will remain CpG. However, an unmethylated CpG will give rise to TpG, which is indistinguishable from a deamination mutation of the forward strand. Thus, three possibilities arise for our sequencing data: CpG that represents a methylated CpG site, TpG that represents either an unmethylated CpG site or a mutation of the forward strand, and TpA that represents a mutation of the reverse strand followed by conversion of the unmethylated C to T by bisulfite. (B) Sequencing data of bisulfite repetitive element PCR of Alu elements. Genomic DNA was isolated from peripheral human blood and bisulfite treated. Alu element PCR was performed and the PCR product was cloned, and 15 clones were sequenced. There were 12 potential CpG methylation sites per clone for a total of 180 potential methylation sites sequenced. Black circles represent methylated CpG sites (66/180 = 36.7%). ‘T’ represents TpG sites that were either unmethylated or mutated (53/180 = 24.4%). ‘A’ represents TpA sites that were mutated (41/180 = 22.8%). ‘X’ represents other mutations (20/180 = 11.1%). The CpG sites used for pyrosequencing (‘PS’) and COBRA (‘MboI’) are indicated.
The majority of potential CpG methylation sites are mutated
Based on the consensus sequence of Alu elements we would expect our 150 bp fragment to have 12 CpG sites. Therefore, examining sequence data from 15 Alu element clones, one would expect that, potentially, 180 CpG sites could be methylated. However, only 66 of 180 (36.7%) of the potential CpG sequences were maintained after bisulfite treatment and were therefore methylated. The remaining 114 CpG sites were either unmethylated or mutated either to TpG (53/180 = 24.4%), TpA (41/180 =22.8%) or other mutations (20/180 = 11.1%). The high frequency of TpG and TpA mutations can be explained by the spontaneous deamination of 5-methylcytosine to thymine, which is known to be mutagenic (26) (Fig. 1A). TpG is created either by mutation of 5-methylcytosine to thymine during evolution or conversion of an unmethylated cytosine to uracil (and then thymine during PCR) during bisulfite treatment. Unfortunately, our assay cannot distinguish the origin of TpG from either mutation of 5-methylcytosine or conversion of an unmethylated cytosine. TpA is created from the deamination of 5-methylcytosine occurring in the reverse strand of DNA that changes CpG to CpA, which destroys the methylation recognition sequence and during subsequent bisulfite treatment becomes TpA. If the assumption is made that deamination of 5-methylcytosine mutations occur with equal frequency on the forward and reverse strands of DNA, then it would be expected that TpG versus TpA frequency (53/180 versus 41/180) should be equal from mutation, and therefore the difference of 12 sites represents the approximate number of unmethylated CpG sites which were converted to TpG by bisulfite. The number of methylated CpG sites was 66, whereas the estimated number of unmethylated CpG sites was 12, and therefore, 84.6% of potential CpG sites were estimated to be methylated from our sequencing data. Thus, although almost two-thirds of the CpG sites are mutated and no longer have the potential to be methylated, the majority of remaining CpG sites are methylated. These calculations are based on a limited amount of sequencing data (180 potential CpG sites) and have the potential for being inaccurate. Therefore, other methods were employed to quantitate methylation and mutation.
Methylation could also be approximated by MboI digestion. In this analysis, restricted fragments are methylated while unrestricted fragments are either mutated or unmethylated. The use of restriction digestion simplified and expedited quantification with our assay, but limits the assay to a single restriction site that may not be reflective of the remainder of the repeat element. Analysis of normal human blood showed 27.2% methylation. The same DNA was treated with an excess of SssI methylase, which will methylate all CpG sites. Quantification of methylation of the SssI-treated DNA by MboI digestion showed only 29.2% methylation (data not shown). This means that 93% of potential CpG sites were methylated and the majority of CpG sites at the MboI recognition site had been mutated. Thus, these numbers were broadly similar to what was estimated by direct sequencing.
Bisulfite repetitive element PCR samples several thousand repetitive elements
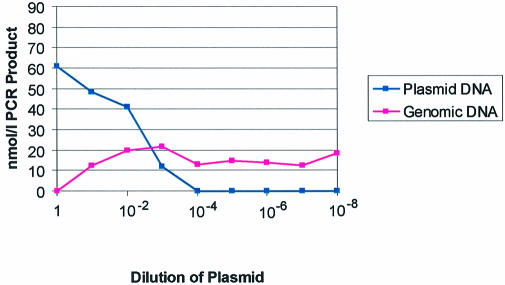
We wanted to estimate the number of Alu elements that are represented in our PCR product pool. One cloned and sequenced Alu element contained an internal tandem duplication and therefore created a PCR product which was larger than most of the amplified Alu elements. This allowed us to distinguish it from the PCR product of Alu elements being amplified from genomic DNA. Therefore, a competitive PCR experiment was performed where a fixed amount of bisulfite-treated genomic DNA was mixed with serial dilutions of our plasmid containing the larger Alu element (Fig. 2). From these experiments, we calculated that 25 ng of bisulfite genomic DNA gave equivalent PCR product as 0.5 ng of plasmid. With the assumptions that the human genome contains 3 × 109 bases and 9 × 1011 bases are in 1 ng of DNA, we calculated that there were ∼7500 genome equivalents in 25 ng of DNA. The plasmid vector was 4.2 kb in size and contained one Alu element per plasmid. We then calculated that 0.5 ng of plasmid contained 1 × 108 Alu elements. Therefore, a PCR starting from 7500 genomes is equivalent to a PCR starting from 1 × 108 cloned Alu elements, implying that ∼15 000 different Alu elements were being amplified from each genome. This diversity is borne out by the fact that the cloned Alu elements in Figure 1 were all unique.
Figure 2.
Calculation of the number of Alu elements being assessed by bisulfite Alu PCR. Competitive PCR was performed using a fixed amount of bisulfite-treated genomic DNA and a plasmid containing a cloned Alu element fragment with an internal duplication that gives a larger PCR product. A fixed amount of bisulfite-treated genomic DNA was mixed with serial dilutions of the plasmid which gave two PCR products, one from the genomic DNA and a larger one from the plasmid. The PCR products were quantitated using a capillary electrophoresis system, the Agilent 2100 Bioanalyzer (Agilent Technologies) and plotted above. From this experiment, it is approximated that 0.5 ng of plasmid gives equivalent PCR product to 50 ng of bisulfite-treated genomic DNA.
Bisulfite repetititve element PCR can detect DNA methylation decreases in cell lines treated with 5-aza-2′deoxycytidine
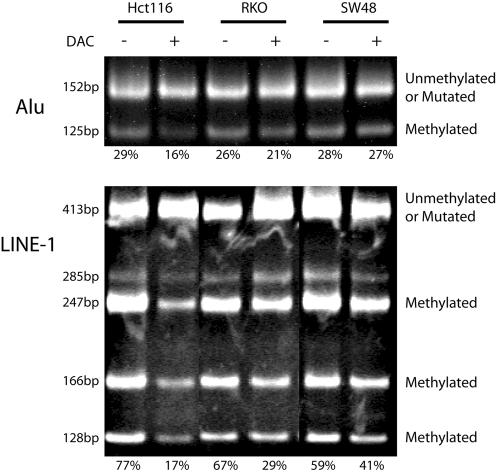
In order to test whether Alu element of LINE-1 methylation could approximate global methylation, we used COBRA restriction digestion of the repetitive element PCR product, to quantitate DNA methylation in cell lines treated with DAC (Fig. 3). The colon cancer cell line, Hct116, had 29% Alu element and 77% LINE-1 methylation prior to treatment, and 16% Alu element and 17% LINE-1 methylation after treatment with DAC. RKO cells treated with DAC showed a decrease from 26 to 21% in Alu element methylation, and a decrease from 67 to 29% in LINE-1 methylation. In contrast, SW48 cells showed little change in methylation with DAC treatment, with Alu methylation decreasing from 28 to 27% and LINE-1 methylation decreasing from 59 to 41%. Similar results were obtained for each cell line treated with DAC for both the Alu element and LINE-1 assays. Thus, both assays reliably detected inhibition of methylation induced by DAC. The Alu element assay showed that approximately two-thirds of the PCR product was not digested by MboI; however, this is not due to Alu elements being largely unmethylated. As found by our sequencing data, the majority of Alu elements, the CpG dinucleotide and methylation target had been mutated, and were no longer a target for DNA methylation. Overall, these data show that repetitive element PCR can be used as a marker of global DNA methylation.
Figure 3.
Quantitation of DNA methylation in cell lines treated with DAC using the bisulfite repetitive element PCR technique. Hct116, RKO and SW48 cell lines were treated with DAC. Genomic DNA was isolated from DAC-treated (+) and -untreated (–) controls. The genomic DNA was treated with sodium bisulfite and a non-specific PCR was performed, which amplified a pool of Alu or LINE-1 repetitive elements. The PCR product was then digested with MboI (Alu) or HinfI (LINE-1), which only cuts repetitive elements that were originally methylated. The Alu PCR assays a single methylation site and therefore the MboI digestion will cut the PCR product from 152 to 125 and 27 bp (data not shown). The LINE-1 PCR assays the methylation of two sites and therefore HinfI can generate five possible digestion products of 285, 247, 166, 128 and 38 bp (data not shown). The digested PCR product was separated by polyacrylamide gel electrophoresis and stained with ethidium bromide. The lower cut bands represent methylated repetitive elements. The upper band represents unmethylated repetitive elements or repetitive elements in which the restriction site has been mutated. The PCR bands were quantitated and the amount of methylation is shown below each gel lane. Similar results were obtained for both the Alu element and LINE-1 assays.
In order to test the reproducibility of our assay, we used COBRA to measure Alu element methylation on the same bisulfite-treated DNA sample eight separate times and the standard deviation was only 2%. In practice the quantification will be performed on DNA samples from different bisulfite treatments, and theoretically variability may arise with restriction digestion on PCR product from DNA unconverted by sodium bisulfite. In order to test for this possibility, we used both our Alu and LINE-1 PCR primers on genomic DNA not treated with bisulfite, and we were unable to obtain any PCR product. Therefore, our primers were specific for the bisulfite-treated DNA sequence.
Bisulfite repetitive element PCR can be quantitated using pyrosequencing
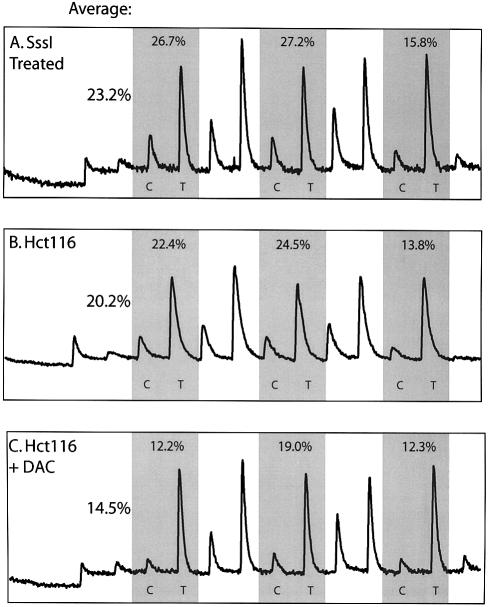
Bisulfite Alu PCR products could also be analyzed by pyrosequencing allowing for a rapid analysis of multiple CpG sites (12). Pyrosequencing is a direct sequencing by synthesis method originally developed to overcome artifacts of secondary structure and avoid gel electrophoresis (27). This method has the advantages of analyzing several methylation sites, is not restricted to restriction enzyme sites, avoids sequencing multiple clones, and allows accurate quantitation of multiple CpG methylation sites in the same reaction. Bisulfite Alu PCR products were pyrosequenced in an area that had three tandem CpG sites (Figs 1B and 4). This method was used to quantitate the decrease in methylation of the same Hct116 cells treated with DAC analyzed by our COBRA assay. In order to calculate the potential number of CpG sites that could be methylated, genomic DNA was double treated with an excess of SssI methylase, which will methylate all CpG sites, and bisulfite treated. By pyrosequencing, only 23.2% of potential CpG sites could be methylated, showing that most of the potential CpG sites had been mutated and could no longer be methylated (Fig. 4). Of these potential CpG sites, 20.2% were methylated in Hct116 cells, or ∼87% of the potential methylation sites. The difference between Alu methylation in genomic DNA and SssI methyltransferase-treated genomic DNA was very small. This is consistent with our previous results in which Alu elements were found to be 84.6% methylated in peripheral blood DNA (Fig. 1B). Treatment with DAC decreased methylation to 14.5%, or 62% of potential methylation sites. There was a small difference for methylation analysis of Hct116 cells using COBRA versus pyrosequencing. These differences are likely attributable to the fact that different CpG sites were analyzed (Fig. 1B) and by differences in the techniques.
Figure 4.
Quantitation of DNA methylation using bisulfite repetitive element PCR and pyrosequencing. (A) SssI methylase-treated DNA, (B) untreated Hct116 cell line DNA or (C) DAC-treated Hct116 cell line DNA was bisulfite treated and PCR of Alu repetitve elements was performed. The PCR product was purified and methylation was quantitated using the PSQ HS 96 Pyrosequencing System (Pyrosequencing, Inc.). The pyrogram quantitates C for methylated and T for unmethylated or mutated DNA. The shaded regions represent three tandem CpG sites quantitated in Alu elements, and the percent methylation at each site is shown above the peaks. The average methylation of the three sites is calculated on the left for each pyrogram. The maximum absolute methylation of 23.2% is calculated by SssI methylase-treated DNA (A), Hct116 cells have 20.2% methylation, and this methylation decreases to 14.5% after DAC treatment of Hct116 cells.
DISCUSSION
We report a simple method for assessing DNA methylation of several thousand loci in the genome simultaneously using bisulfite PCR of DNA repetitive element. This method can serve as a surrogate of global methylation, and has the advantage of being easier to perform than previous methods to quantitate total genomic 5-methylcytosine. This method has been used to look at two human repetitive elements, Alu and LINE-1 elements, and can be expanded to look at other human repetitive elements or repetitive elements from other species such as B1 elements in mice. We have quantitated repetitive element methylation using direct sequencing, COBRA and pyrosequencing. However, this method could be easily modified to Ms-SnuPE, MSP or MethyLight.
A drawback of this assay is that the mechanisms that control methylation of repetitive elements may differ from the rest of the genome and therefore this assay could not be a perfect representation of total genomic methylation (28). Neverthe less, the accessibility and ease of use of the approach will facilitate global methylation studies in normal and cancer tissues as well as in patients treated with DNA methylation inhibitors.
Acknowledgments
ACKNOWLEDGEMENTS
A.S.Y. is the recipient of an American Society of Clinical Oncology Young Investigator Award. M.R.H.E. is the recipient of FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil) grant 99/09368-1. This work was supported by grants P50CA100632 (SPORE) and R33CA89837 from the NIH. Core Grant CA16672 from the NIH supports DNA sequencing at the M.D. Anderson Cancer Center DNA analysis facility.
REFERENCES
- 1.Bird A. (1992) The essentials of DNA methylation. Cell, 70, 5–8. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.A. and Takai,D. (2001) The role of DNA methylation in mammalian epigenetics. Science, 293, 1068–1070. [DOI] [PubMed] [Google Scholar]
- 3.Jones P.A. and Baylin,S.B. (2002) The fundamental role of epigenetic events in cancer. Nature Rev. Genet., 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 4.Issa J.P. (2000) CpG-island methylation in aging and cancer. Curr. Top. Microbiol. Immunol., 249, 101–118. [DOI] [PubMed] [Google Scholar]
- 5.Richardson B. (2003) Impact of aging on DNA methylation. Ageing Res. Rev., 2, 245–261. [DOI] [PubMed] [Google Scholar]
- 6.Glover A.B., Leyland-Jones,B.R., Chun,H.G., Davies,B. and Hoth,D.F. (1987) Azacitidine: 10 years later. Cancer Treat. Rep., 71, 737–746. [PubMed] [Google Scholar]
- 7.Santini V., Kantarjian,H.M. and Issa,J.P. (2001) Changes in DNA methylation in neoplasia: pathophysiology and therapeutic implications. Ann. Intern. Med., 134, 573–586. [DOI] [PubMed] [Google Scholar]
- 8.Clark S.J., Harrison,J., Paul,C.L. and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Z. and Laird,P.W. (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res., 25, 2532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalgo M.L. and Jones,P.A. (1997) Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res., 25, 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman J.G., Graff,J.R., Myèohèanen,S., Nelkin,B.D. and Baylin,S.B. (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA, 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlmann K., Brinckmann,A., Toliat,M.R., Ritter,H. and Nurnberg,P. (2002) Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis, 23, 4072–4079. [DOI] [PubMed] [Google Scholar]
- 13.Wagner I. and Capesius,I. (1981) Determination of 5-methylcytosine from plant DNA by high-performance liquid chromatography. Biochim. Biophys Acta, 654, 52–56. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg A.P. and Vogelstein,B. (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature, 301, 89–92. [DOI] [PubMed] [Google Scholar]
- 15.Bestor T.H., Hellewell,S.B. and Ingram,V.M. (1984) Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol. Cell. Biol., 4, 1800–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friso S., Choi,S.W., Girelli,D., Mason,J.B., Dolnikowski,G.G., Bagley,P.J., Olivieri,O., Jacques,P.F., Rosenberg,I.H., Corrocher,R. et al. (2002) A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl Acad. Sci. USA, 99, 5606–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antequera F., Tamame,M., Villanueva,J.R. and Santos,T. (1984) DNA methylation in the fungi. J. Biol. Chem., 259, 8033–8036. [PubMed] [Google Scholar]
- 18.Oakeley E.J. (1999) DNA methylation analysis: a review of current methodologies. Pharmacol. Ther., 84, 389–400. [DOI] [PubMed] [Google Scholar]
- 19.Belinsky S.A., Nikula,K.J., Baylin,S.B. and Issa,J.P. (1996) Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc. Natl Acad. Sci. USA, 93, 4045–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwu H.R., Roberts,J.W., Davidson,E.H. and Britten,R.J. (1986) Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc. Natl Acad. Sci. USA, 83, 3875–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Z., Wang,H., Nekrutenko,A. and Li,W.H. (2000) Densities, length proportions and other distributional features of repetitive sequences in the human genome estimated from 430 megabases of genomic sequence. Gene, 259, 81–88. [DOI] [PubMed] [Google Scholar]
- 22.Kazazian H.H. Jr and Goodier,J.L. (2002) LINE drive. retrotransposition and genome instability. Cell, 110, 277–280. [DOI] [PubMed] [Google Scholar]
- 23.Kochanek S., Renz,D. and Doerfler,W. (1993) DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J., 12, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid C.W. (1998) Does SINE evolution preclude Alu function? Nucleic Acids Res., 26, 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bestor T.H. (1998) The host defence function of genomic methylation patterns. Novartis Found. Symp., 214, 187–195; discussion 195–199, 228–232. [DOI] [PubMed] [Google Scholar]
- 26.Rideout W.M. 3rd, Coetzee,G.A., Olumi,A.F. and Jones,P.A. (1990) 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science, 249, 1288–1290. [DOI] [PubMed] [Google Scholar]
- 27.Ronaghi M., Nygren,M., Lundeberg,J. and Nyren,P. (1999) Analyses of secondary structures in DNA by pyrosequencing. Anal. Biochem., 267, 65–71. [DOI] [PubMed] [Google Scholar]
- 28.Robertson K.D., Ait-Si-Ali,S., Yokochi,T., Wade,P.A., Jones,P.L. and Wolffe,A.P. (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet., 25, 338–342. [DOI] [PubMed] [Google Scholar]