Abstract
This study reports a novel microfluidic platform for rapid and long-ranged concentration of rare-pathogen from human blood for subsequent on-chip surface-enhanced Raman spectroscopy (SERS) identification/discrimination of bacteria based on their detected fingerprints. Using a hybrid electrokinetic mechanism, bacteria can be concentrated at the stagnation area on the SERS-active roughened electrode, while blood cells were excluded away from this region at the center of concentric circular electrodes. This electrokinetic approach performs isolation and concentration of bacteria in about three minutes; the density factor is increased approximately a thousand fold in a local area of ~5000 μm2 from a low bacteria concentration of 5 × 103 CFU/ml. Besides, three genera of bacteria, S. aureus, E. coli, and P. aeruginosa that are found in most of the isolated infections in bacteremia were successfully identified in less than one minute on-chip without the use of any antibody/chemical immobilization and reaction processes.
Rapid separation, concentration, and detection of pathogens in clinical diagnosis of bacteremia and sepsis infections have become increasingly important. Conventional bacteria identification assays typically require several hours for DNA amplification and hybridization, hours for antibody-antigen reaction, or even days for bacteria culture1,2,3,4. Raman spectroscopy is attractive for analysis of chemical and biological components without complicated sample preparations5,6,7. Unfortunately, Raman signals obtained from biological samples are usually very weak, especially in dilute samples6. Metallic nanoparticles (NPs) and roughened metallic substrates have been used to enhance the Raman signal via surface plasmon resonance in Surface-Enhanced Raman Spectroscopy (SERS)7,8,9,10,11,12. SERS is a useful tool that could identify and analyze cell/bacteria/virus/protein/molecule without DNA hybridization and antibody-antigen reaction13,14,15,16,17,18,19,20 if the bacteria concentration is increased by orders of magnitude through incubation. The coffee-ring effect due to surface tension and the evaporation of the sample liquid can concentrate the target sample around the edge of the evaporated droplet21. The effect has been widely utilized in SERS measurements, not only does it increases the local density of the bacteria, but it also makes bacteria contact the SERS substrate19,22. However, a high bacteria concentration of 109 CFU/ml is still required for the coffee-ring technique to achieve a dense bacteria ring for SERS measurement16,23. Unfortunately, in microbe infections, the typical target concentration is as low as 103 CFU/ml in the real sample. Therefore, a time consuming process of incubating bacteria to a high concentration of 109 CFU/ml is required. It is even more difficult to detect pathogens from a mixed sample, such as the detection of bacteria in a blood sample, which is very important for diagnosing bacteremia and sepsis15,24. The pathogenic bacteria must be isolated from human blood and their densities increased before Raman measurements so that a pure bacteria signal can be obtained without inferences from others such as the RBC and WBC signals. A novel method has been developed by Liu et al. using a van-coated SERS substrate to trap the bacteria from blood for SERS measurement25. However, immobilization of chemical/antibody on the SERS-active substrate surface, and long incubation time (hour level) for van-bacteria binding, and repeated wash steps to reduce interferences from non-specific bound cells are still required.
Dielectrophoresis (DEP) has been widely used for the separation, concentration, assembly and discrimination of particles/cells/bacteria/DNA15,26,27,28. The rapid electrokinetic separation and concentration of bacteria from blood via DEP has been previously developed29. Cheng J. et al. published DEP separation and concentration of 2 × 109 bacteria/ml from diluted blood (2 × 108 cells/ml). 109 bacteria/ml requires culture time for over 12 hours. The need for high bacteria concentration arises because DEP works only in short range due to the electric field decay with the distance away from the electrodes. DEP chips working in biological buffer with high conductivity have also been developed30,31,32, and thus sample dilution is not needed. However, the short-range separation mechanism of the pure DEP technique makes it difficult to use low bacteria density samples.
In this study, a compact hybrid electrokinetic mechanism - utilizes dielectrophoresis (DEP), electrophoresis, electrohydrodynamics (EHD) simultaneously to achieve concentration of bacteria from human blood using long range (~6 mm) transport. Our design combines a short-range DEP and long-range AC electroosmosis (ACEO) flows to rapidly and selectively concentrate the bacteria in the diluted human blood sample. AC electric field induced electrokinetic forces (ACEK) were utilized to separate and concentrate pathogens from blood for on-chip SERS measurements and analysis. We demonstrate that a hybrid AC electrokinetic strategy not only can separate bacteria and blood cells but it also can concentrate the rare bacteria from manifold and dense blood cells. This strategy would allow an extremely high density of bacteria aggregate to be obtained for effective measurements and very clear and specific SERS-fingerprints for pathogenic identification.
Results
Concentration of bacteria from human blood
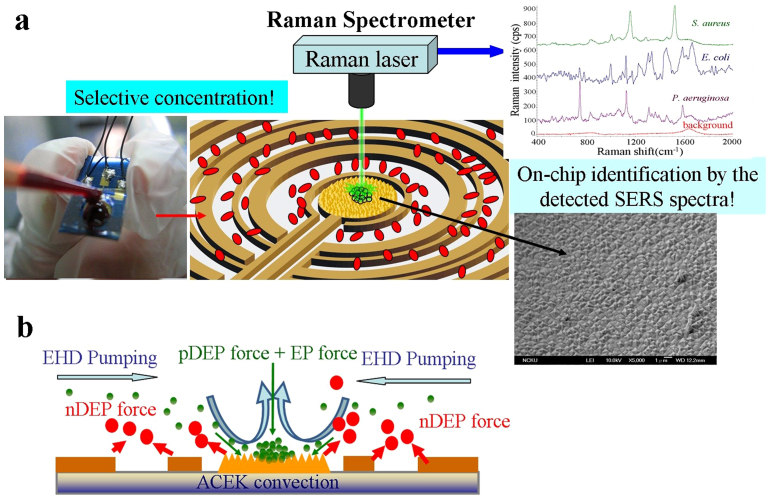
A concentric set of electrodes with a maximum diameter of 6 mm was designed for the wide-range selective concentration of bacteria from human blood, as shown in Fig. 1(a and b). AC voltages of 12 Vpp, 8 Vpp and a ground with a DC bias of 0.5 V were applied to the wider ring and the narrower ring electrodes, and the central circle-shaped electrode, respectively.
Figure 1.
(a) Experimental setup. The SEM image shows the roughened Au surface at the centre electrode. AC electric field induced dielectrophoresis and electrohydrodynamics were used to rapid concentrate bacteria form human blood. SERS Raman spectroscopy fingerprint of the concentrated bacteria identifies the bacteria. (b) Illustration of the hybrid mechanism of selective concentration over a wide range asymmetric electrode array.
Rapid separation and concentration of the rare pathogen from blood are the critical needs before identifying the specific genus. In approximately 90% of bacteremia patients, only single pathogenic species can be found at a time24; it is rare to see the mixed infection in a blood sample4. Thus effectively concentrating bacteria in a blood sample is a key technique for further identification. There is an optimal AC voltage frequency range for selectively concentrating bacteria in a blood sample electrohydrodynamic flow transports the bacteria toward a central stagnation area; where the pathogen was trapped onto the roughened Au surface by AC DEP forces (Fig. 1(a and b)).
Figure 2(a) shows that at low frequencies, the blood cells in fact experience opposite DEP force than the bacteria, furthermore, the blood cells (d = 6–8 μm) and bacteria (d = 0.5–2 μm) experience very different magnitudes in the induced DEP forces. Therefore, by tuning the applied frequency, EHD forces could be adjusted to be comparable to DEP forces; this effect allows us to separate the bacteria and the blood cells in addition to concentrating the bacteria.
Figure 2.
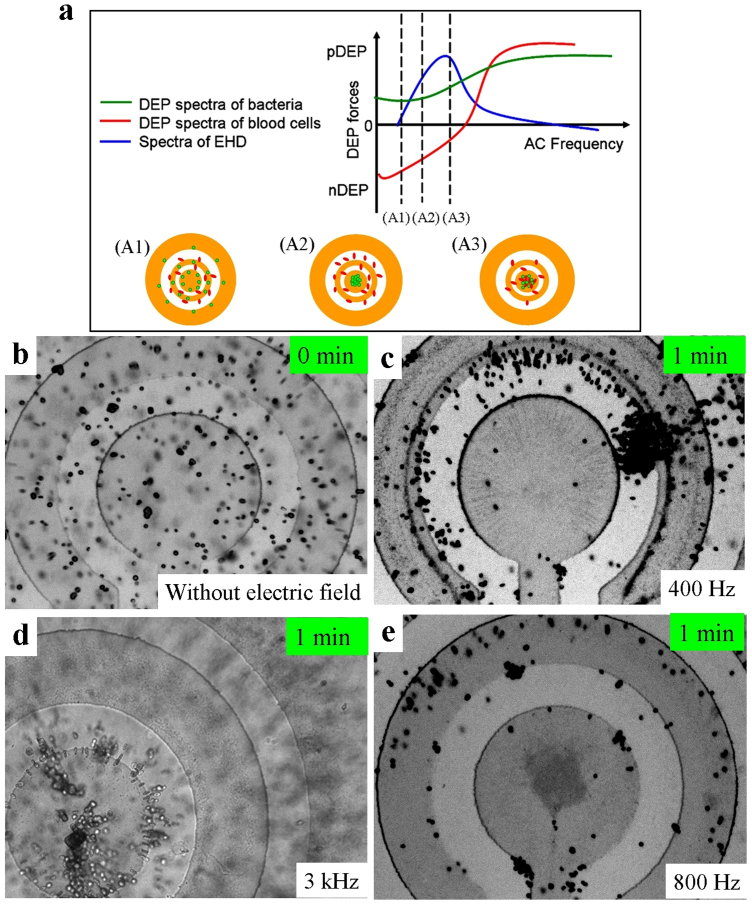
(a) Schematic illustration of the selective concentration mechanism. When 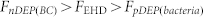 , electrohydrodynamic transport is higher than the positive DEP force on the bacteria and lower than the opposite DEP force on the blood cell that allows the selective concentration of bacteria at some specific frequencies of the applied voltage. (b) Bacteria and blood cells were random distribution before a voltage is applied. (c) Neither Bacteria nor blood cells were conveyed to the centre electrode when the applied frequency was 400 Hz and the ACEO drag is low. (d) At a relative higher frequency, bacteria and blood cells were both concentrated into the centre electrode. (e) In the frequency range between 600 Hz to 1.5 kHz, bacteria were conveyed and concentrated onto the centre electrode and blood cells were excluded from electrodes.
, electrohydrodynamic transport is higher than the positive DEP force on the bacteria and lower than the opposite DEP force on the blood cell that allows the selective concentration of bacteria at some specific frequencies of the applied voltage. (b) Bacteria and blood cells were random distribution before a voltage is applied. (c) Neither Bacteria nor blood cells were conveyed to the centre electrode when the applied frequency was 400 Hz and the ACEO drag is low. (d) At a relative higher frequency, bacteria and blood cells were both concentrated into the centre electrode. (e) In the frequency range between 600 Hz to 1.5 kHz, bacteria were conveyed and concentrated onto the centre electrode and blood cells were excluded from electrodes.
Figure 2(b) shows that the blood cells and bacteria are distributed randomly when no electric field was applied to the electrodes. After applying an AC voltage at a frequency of 400 Hz, Fig. 2(c) shows that neither bacteria nor blood cells were conveyed to the centre area. This is because both types of particles experience stronger DEP forces repelling blood cells away from the electrodes and attracting bacteria at the rings than the EHD force that moves them toward the central region, as the state A1 in Fig. 2(a), 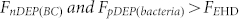 . When a 3 kHz voltage is applied, Fig. 2(d) shows that bacteria and blood cells were both concentrated onto the central electrode due to a strong biased ACEO flow (
. When a 3 kHz voltage is applied, Fig. 2(d) shows that bacteria and blood cells were both concentrated onto the central electrode due to a strong biased ACEO flow (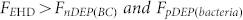 ), as the state A3 in Fig. 2(a). Under the optimal frequency range between 600 Hz and 1.5 kHz, as shown in Fig. 2(e) for 800 Hz and after only one minute, bacteria are effectively conveyed and concentrated onto the central electrode while blood cells were excluded from the electrodes, as the state A2 in Fig. 2(a). At 800 Hz the blood cells experience a negative DEP force that is stronger than the ACEO drag (
), as the state A3 in Fig. 2(a). Under the optimal frequency range between 600 Hz and 1.5 kHz, as shown in Fig. 2(e) for 800 Hz and after only one minute, bacteria are effectively conveyed and concentrated onto the central electrode while blood cells were excluded from the electrodes, as the state A2 in Fig. 2(a). At 800 Hz the blood cells experience a negative DEP force that is stronger than the ACEO drag (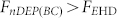 ) and pushes the blood cells away from the center region. On the other hand, the ACEO drag transports the bacteria toward the center region (
) and pushes the blood cells away from the center region. On the other hand, the ACEO drag transports the bacteria toward the center region (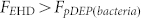 ) and the DEP force further pulls the bacteria into the stagnation area in the middle of the centre electrode, where the EHD force is the weakest, and the DEP and EP forces are the largest. Thus the bacteria are separated from the blood cell, concentrated, and effectively captured on the roughened Au surface. This hybrid AC electrokinetic technique enables separation and concentration of pathogens from a sample containing a mixture of blood cells and bacteria in three minutes.
) and the DEP force further pulls the bacteria into the stagnation area in the middle of the centre electrode, where the EHD force is the weakest, and the DEP and EP forces are the largest. Thus the bacteria are separated from the blood cell, concentrated, and effectively captured on the roughened Au surface. This hybrid AC electrokinetic technique enables separation and concentration of pathogens from a sample containing a mixture of blood cells and bacteria in three minutes.
Raman scattering detection after concentrating bacteria on the SERS-active surface
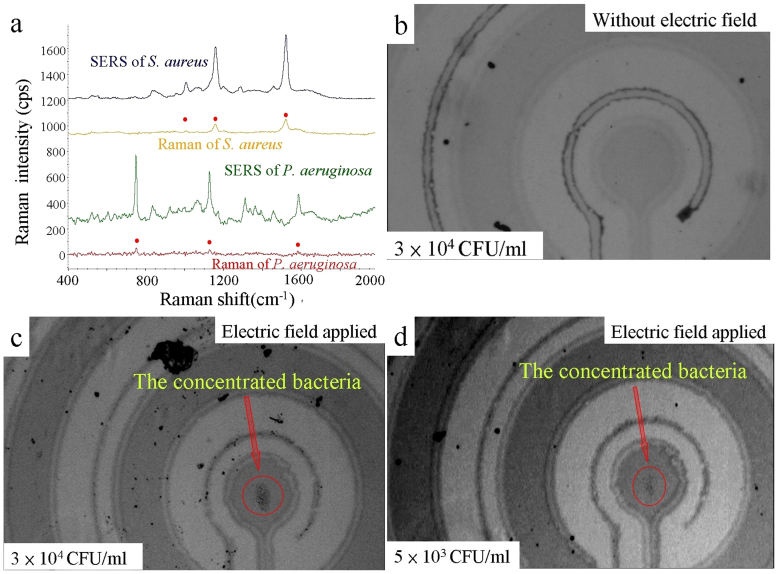
Figure 3(a) shows that the Raman spectra and the SERS spectra of S. aureus and P. aeruginosa. For comparison, bacteria suspended in buffer droplets with volume of 5 μl at the high concentration of 1 × 109 CFU/ml were used for coffee-ring formation on a smooth Au coated surface as a control. The Raman spectra of these formed coffee-ring samples are also shown in Fig. 3(a). To investigate the enhancement of ACEK-SERS, 5 μl buffer samples were dropped onto the chips and subjected to AC excitation at 800 Hz for 1 minute. After electrokinetically concentrating the dropped sample for 1 minute, the target bacteria had accumulated a sufficiently high volume onto the roughened SERS surface for on-chip Raman measurements. Clearly the ACEK-SERS signal is much stronger than the coffee ring samples (Fig. 3(a)).
Figure 3.
(a) The Raman spectra of S. aureus and P. aeruginosa obtained by a smooth Au substrate at high bacteria concentration by using the coffee-ring technique, which are much weaker than the ACEK-SERS spectra obtained by the roughened Au surface using ACEK concentration. (b) The bacteria are randomly distributed when no voltage is applied to the electrodes. (c) Result of a concentration of 3 × 104 CFU/ml bacteria suspended in a 50 μl buffer with an AC voltage at 800 Hz for 3 minutes. (d) Result of an ultra low bacteria concentration of 5 × 103 CFU/ml sample under the same condition.
For low bacteria concentrations (lower than 106 CFU/ml), 50 μl buffer samples were dropped onto the chips and subjected to AC excitation at 800 Hz for 3 minutes to accumulate a sufficiently high bacteria volume onto the detection area. Figure 3(b) shows the photography of the dropped bacteria sample at low bacteria concentration of 3 × 104 CFU/ml without electric field applied. Figure 3(c) and 3(d) demonstrate the applicability of the ACEK technique to samples of low bacteria concentrations of 3 × 104 and 5 × 103 CFU/ml, respectively. The concentrated bacteria aggregate on the centre electrode can be clearly observed. Approximately 40–50% capture efficiency onto the stagnation area was achieved.
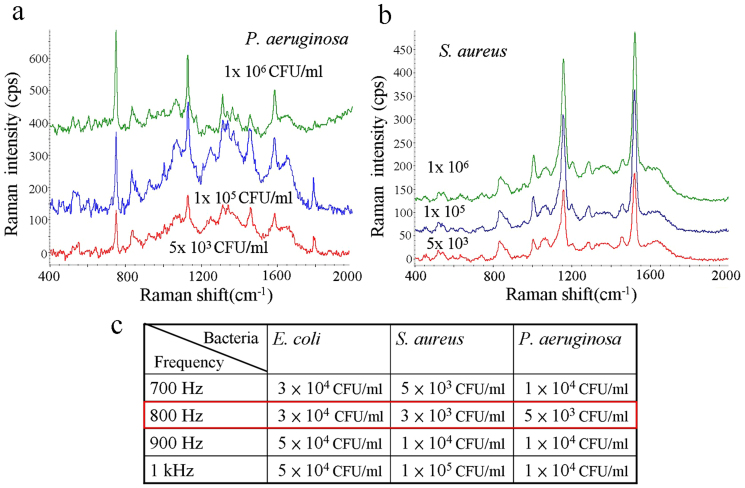
To investigate the effect of sample bacteria concentrations on SERS signals after electrokinetic concentration, Figure 4(a) shows the SERS spectra of P. aeruginosa for sample bacteria concentrations 1 × 106, 1 × 105, and 5 × 103 CFU/ml. The detection limit obtained by the coffee-ring method was approximately 1 × 108 CFU/ml for obtaining a discriminable SERS spectrum. In contrast, the SERS spectra of S. aureus of concentrations 1 × 106, 1 × 105, and 5 × 103 CFU/ml are shown in Figure 4(b). The detection limit is clearly lower than 5 × 103 CFU/ml, which is four orders of magnitude than the coffee ring technique. Bacteria concentrations ranging between 3 × 103 and 1 × 105 CFU/ml were used to investigate the detection limit at different AC frequencies for the different genera of bacterium. Figure 4 (c) shows that the optimal frequency is the same (about 800 Hz); under this condition, detection limits reached 3 × 103, 5 × 103, and 1 × 104 CFU/ml for S. aureus, P. aeruginosa and E. coli. These detection limits match the need for detecting bacteremia/sepsis samples after 2–3 hours of blood culturing and can be reduced with improved electrode array design.
Figure 4.
(a) The fingerprint of P. aeruginosa is detectable at a concentration as low as 5 × 103 CFU/ml. (b) S. aureus is detectable at very low concentration. (c) The summary of the detection limits at several AC frequencies for three genera of bacteria.
On-chip identification of bacteria from human blood
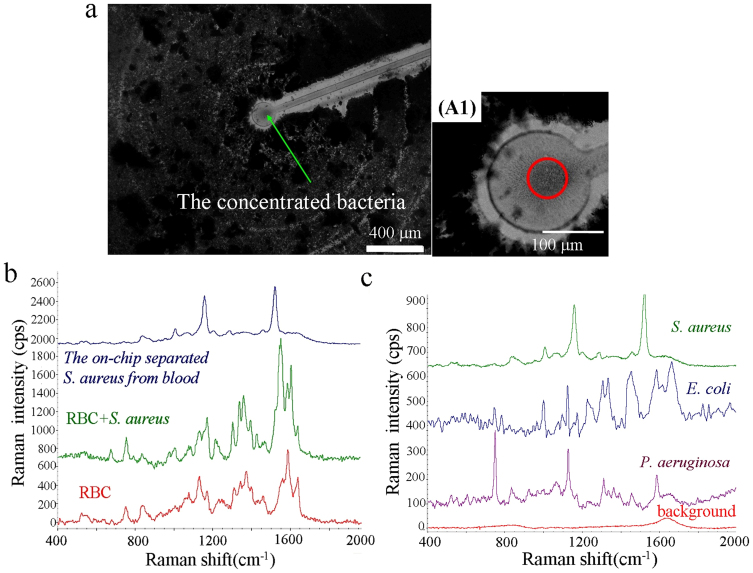
The coffee-ring formation contained a mixture of bacteria and RBCs (bacteria: 1 × 109 CFU/ml, blood cells: 3 × 107 cells/ml) after sample evaporation for 15 minutes. The results clearly indicate that blood cells could significantly influence SERS measurements. To demonstrate the selective concentration and detection capability of the ACEK-SERS approach, human whole blood spiked with S. aureus was tested. In order to avoid blood cell coagulation, human blood was diluted 20 times with the prepared bacteria solution, so the final blood cell concentration was approximately 2 × 108 cells/ml and the bacteria concentration was 1 × 105 CFU/ml. Figure 5(a) shows the center region of the chip after selectively concentrating S. aureus from the mixed sample containing blood cells and bacteria. S. aureus were concentrated at the stagnation area on the SERS-active roughened electrode by the combined EHD and DEP forces. In contrast, blood cells did not enter the central electrode because the induced repelling DEP force was higher than the EHD drag force. In Fig. 5(a), a dense dark layer of RBC can be seen everywhere except for a ~5000 μm2 area centered around the centre electrode. On the other hand, there is a high concentration of bacteria at the center of the centre electrode, as shown in A1 of Fig. 5(a). This visual observation is supported with Raman spectroscopy later.
Figure 5.
(a) On-chip selective concentration of 1 × 105 CFU/ml bacteria and clear separation from a very dense blood sample (RBC concentration ~ 2 × 108 cells/ml). The A1 shows a higher magnification photo of the centre area, there is a high concentration of bacteria without blood cells at the center of the centre electrode. (b) The SERS signatures of RBCs, RBCs-bacteria mixture, and the S. aureus electrokinetically separated from blood. (c) The measured clear SERS signatures of S. aureus, E. coli, P. aeruginosa, and background after ACEK concentration of bacteria from human blood.
The SERS signatures of RBCs, a RBCs-bacteria mixture (1:50), and the electrokinetically separated S. aureus from blood were measured and are shown in Figure 5(b). The results demonstrate that the spectrum of the RBC-bacteria mixture is similar to the RBC signature (most of the peaks are contributions from blood cells). This makes it difficult to detect the target bacteria without a separation procedure. On the other hand, a clear and pure bacteria SERS signature was obtained by directing a laser spot at the bacteria aggregate separated from the blood cells after applying a pre-determined electric excitation at a frequency of 1 kHz for 3 minutes, as shown in Figure 5(a).
Figure 5(c) shows very distinct fingerprints of S. aureus, E. coli, and P. aeruginosa that were detected after each was separated and concentrated from a bacteria-blood mixture. The background profile was measured from the human blood diluted with sucrose buffer without any bacteria after electrokinetically excluding the blood cells. The detected main characteristic peaks of S. aureus were at the wavenumbers of 1006, 1158 and 1522 cm−1, E. coli at 732, 782, 1006, 1123, 1334, 1584 and 1663, and P. aeruginosa at 750, 833, 923, 1125, 1310, 1455 and 1585 cm−1. The detected bacteria spectra are very consistent with the spectra libraries that are shown in previous reports33,34,35,36,37. In addition, Gram positive (S. aureus) and Gram negative (E coli and P. aeruginosa) bacteria discrimination are easy and rapid without requiring complex sample treatment. The results show that this technique is capable of capturing low concentration bacteria from a sample containing very dense blood cells to determine the blood infections (positive or negative), and can perform on-chip identification of pathogenic bacteria in bacteremia by comparing the detected SERS spectra with a spectra library.
Discussion
Here, blood cells (RBC and WBC) was induced a negative DEP force and bacteria was induced a positive DEP force by an AC electric field at the frequency range between 200 Hz and 20 kHz. The magnitude of DEP force is complex function of the particle size (FDEP ~ r3) and the applied frequency (FDEP ~ fCM(ω)). The EHD is a bulk fluid force, which acts on a particle via hydrodynamic drag. The DEP force is proportional to r3 while the EHD induced drag force is proportional to the particle radius r. Therefore, the scaling dependence of the relative influence of DEP and EHD for manipulating particle of different sizes 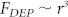 . Based on the above scaling dependence equation, the importance of EHD relative to DEP increases for smaller particles while DEP becomes the dominated dynamics for manipulating bigger particles. The DEP force is approximately constant when the applied frequency is below/above the crossover-frequency (cof), but the magnitude of DEP force has a large difference near the cof38. Besides, the magnitude of ACEO is also very sensitive to frequency of the applied voltage (eq. (1)). The high ACEO velocity appears near the charge relaxation characteristic frequency, and the velocity decays violently when the applied frequency deviates form the characteristic frequency.
. Based on the above scaling dependence equation, the importance of EHD relative to DEP increases for smaller particles while DEP becomes the dominated dynamics for manipulating bigger particles. The DEP force is approximately constant when the applied frequency is below/above the crossover-frequency (cof), but the magnitude of DEP force has a large difference near the cof38. Besides, the magnitude of ACEO is also very sensitive to frequency of the applied voltage (eq. (1)). The high ACEO velocity appears near the charge relaxation characteristic frequency, and the velocity decays violently when the applied frequency deviates form the characteristic frequency.
Therefore, for the bacteria-RBC mixture, the dominated separation mechanism could be achieved via changing the magnitude of the tunable frequency-dependency ACEO relative to size-dependency DEP force. The magnitude of EHD drag force compares to DEP force with two directions for the selectively concentrating bacteria to the centre of inner electrode, which can control smaller particles in one direction and the opposite direction for others.
Blood samples for in-filed testing usually have small sample volumes (<100 μl). With such small sample volumes, the conventional centrifugal methods have challenging limits in the separation purity and recovery rate, especially for samples with low bacteria concentrations. The proposed new technique significantly shortens the concentration and identification times from several hours to a few minutes, and can detect a low concentration bacteria of 3 × 103 CFU/ml among manifold and dense blood cells. It forms a bacterial aggregate with an extremely high density for on-chip SERS measurements. By comparing the detected SERS spectra with a spectra library, on-chip identification of bacteria is feasible. The combination of short-range DEP and wide-range electrohydrodynamic convection can selectively concentrate bacteria from several millimeters to a single small area. Therefore, the detection limit could be lower than pure DEP even with blood dilution28,31. Also, the biased ACEO provides a high shear rate that prevents the target cells being attracted to the electrode edges by the positive DEP. The detection limit can be further decreased by extending the asymmetric electrode array to be centimeter wide for increasing the sampling volume, and thus the detection limit could be one order lower.
For bacteria concentrations ranging from 1 × 107 to 3 × 104 CFU/ml (data not shown), the measured SERS signal did not vary significantly with bacteria concentration. Possibly the local density of bacteria aggregate from samples with bacteria concentrations larger than 3 × 104 CFU/ml all reached a saturation density on the roughened surface after 3 minutes of ACEK concentration. Varying the concentration time should be an effective way to quantify the bacterial concentration. The SERS spectra were not significantly different in magnitude at any detectable bacteria concentrations being tested after the electrokinetic concentration, showing the high detection capability and stability of this assay. Compared to the ELISA kits, this chip can isolate and identify bacteria in blood samples without the need for expensive antibody immobilizations, antibody-antigen binding, repeated wash steps, and secondary antibody reporting, thus significantly reducing the operation processes, detection time and the cost. The total detection and identification can be finished within 5 minutes. Sample dilution takes 1 min, electrokinetic selective concentration takes only 3 min, and Raman measurement is quite fast (~10 sec). The platform demonstrated excellent separation and concentration capabilities by AC electrokinetics and high bacteria discriminability by their measured SERS signatures with multiple characteristic peaks. For clinically bacteremia sample, the bacteria concentration in blood is in the range of 10–1000 CFU/mL39,40, particularly, often greater than 1,000 CFU/ml in pediatric patients.
The detection limit of this demonstration is hundreds bacteria in 50 μL sample or about 103 CFU/mL. Blood culturing for 2–3 hours is needed to increase the bacteria concentration over 5 × 104 CFU/ml detection of this demonstration. Thereafter, the pathogen in the diluted blood sample can be directly separated, concentrated, and identified within five minutes. In the future, larger range (centimeter size) electrode array could lower the detection limit of the pathogen concentration to match that of bacterimia. The combination of the hybrid ACEK and SERS technologies can rapidly determine the blood infections and identify the pathogens in human blood without the time consuming and complicated processes, such as immobilization of DNA/antibody, DNA-DNA/antibody-antigen reactions, and the repeated wash steps.
Methods
Theory, chip design and operation
The time averaged DEP force 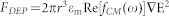 is dependent on the permittivity of the medium
is dependent on the permittivity of the medium 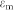 , the radius of the particle r3, the effective polarizability- Clausius-Mossotti (CM) factor fCM, and the magnitude of the electric field gradient
, the radius of the particle r3, the effective polarizability- Clausius-Mossotti (CM) factor fCM, and the magnitude of the electric field gradient 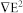 . If particles are more polarizable than the surrounding medium, they will be attracted to the region of relatively strong electrical field gradient (positive DEP, pDEP in Fig. 1 and 2a); if particles are less polarizable than the surrounding medium, they will be pushed to the region of relative weak electrical field gradient (negative DEP, nDEP in Fig. 1 and 2a). However, DEP works in a very short range due to the exponential decay of the electric field gradient with the distance from the electrode edge.
. If particles are more polarizable than the surrounding medium, they will be attracted to the region of relatively strong electrical field gradient (positive DEP, pDEP in Fig. 1 and 2a); if particles are less polarizable than the surrounding medium, they will be pushed to the region of relative weak electrical field gradient (negative DEP, nDEP in Fig. 1 and 2a). However, DEP works in a very short range due to the exponential decay of the electric field gradient with the distance from the electrode edge.
In contrast, AC electroosmosis (ACEO) is caused by the interaction between a non-uniform AC electric field and the electric double layer that is induced via accumulating charges near the surface of the polarized electrode. The ion migration drags the fluid to generate a surface-driven bulk flow over the electrodes. This effect is capable of generating a long-range convection flow without selectivity for the particles with proper designs. The flow velocity of ACEO can be described as follows41:
The magnitude of ACEO velocity is dependent on the applied AC voltage V0, the characteristic length of the electrode separation x, and the non-dimensional frequency Ω that is dependent on conductivity and permittivity of the medium and Debye length k. Biased ACEO combines a DC bias with AC signals to generate two electrode charging effects, capacitive charging and Faradaic charging on different electrodes. This asymmetric polarization is capable of inducing fluid convection more effectively and could be used in a wider range of buffer conductivities to induce sufficiently high convections42. In high ionic strength fluid, the major electrohydrodynamic influence changes to electrothermal AC electro-osmosis, and its working frequencies would shift to higher43,44.
EHD can induce a net flow to convey liquid samples in a wide range, but there is no specificity/selectivity for the particles in the fluid. By combining the short-range DEP that has a discriminating capability with DC-biased ACEO that has a wide-range transporting capability, pathogens in a blood sample can be selectively concentrated for subsequent SERS measurements and analysis. The positive DEP force also provides an attraction force to form a hard junction between the bacteria and the peaks of the roughened SERS-active substrate. The magnitude of DEP and EHD forces are very sensitive to the frequency of the applied AC voltage. Due to the difference in the polarizability and size of particles, the net force at a given locality can be in one direction for some particles and the opposite direction for others. For example a strong repulsive DEP force on the blood cell opposes and overwhelms the EHD convective force toward the center electrode, while an attractive DEP force on the bacteria that is lower than the EHD convective force to pull the bacteria onto the center electrode. Consequently, bacteria are continuously conveyed to the stagnation area on the central electrode while blood cells are repelled from the electrodes, as the state A2 of Fig. 2(a).
Bacteria culture and sample preparation
S. aureus, (BCRC 14957, Gram positive), P. aeruginosa (ATCC 27853, Gram negative), and E. coli (BCRC 15922, Gram negative) were cultured on tryptic soy agar (TSA) at 35°C. An isotonic solution, 300 mM sucrose solution with a low conductivity, was used as the suspending solution. Bacteria suspended in the isotonic solution with concentrations ranged from 3 × 103 to 1 × 106 CFU/ml was used to investigate the detection limit of this system. Human blood was spiked into the prepared bacteria solution in ratios of 1:20, giving a final blood cell concentration of 2 × 108 cells/ml. All of the Raman data were acquired with a 10X objective and a spot size of ~5 μm in diameter. A low laser power of 1 mW was used to avoid cell damage during Raman measurements. All spectra reported here had the exposure time set to 2 s and the signal was accumulated five times in a wavenumber range of 400–2000 cm−1. The total detection time is only 10 sec. Note, the background signal caused by the buffer increased after voltages were applied for a prolonged time (>15 minutes) because the buffer concentration was increased as the function of evaporation time increased. Therefore, detection period should be done within 15 minutes before the background increases significantly. After patterning the etching area by standard photolithography, the locally roughened surface was fabricated using inductively coupled plasma (ICP) ion reactive etching. The microelectrodes were patterned by standard photolithography techniques and metal etching. The reaction chamber was made by PDMS (curing condition: 75°C for an hour) at a 10:1 ration of base-to-curing agents.
Author Contributions
I.-F.C. conceived, designed and performed the experiments. H.-C.C. and T.-Y.C. provided reagents, blood and bacteria. I-F.C. and C.M.H. wrote the paper. F.-L.Y. supervised this study. All authors reviewed the manuscript.
Acknowledgments
This work was supported by the National Science Council of Taiwan (NSC 100-2221-E-006-026-MY3 and NSC 101-2218-E-492 -002). We thank Professor T-C Chang for providing the cultured bacteria and Prof. Yi-Je Juang's Lab for supporting SEM techniques. We also thank the National Nano Device Laboratory for supplying the micro-fabrication equipment.
References
- Hage D. S. Immunoassays. Anal. Chem. 67, 455–462 (1995). [DOI] [PubMed] [Google Scholar]
- Lequin R. M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clinical Chemistry 51, 2415–2418(2005). [DOI] [PubMed] [Google Scholar]
- Heller M. J. DNA Microarray Technologu: Devices, Systems, and Applications. Annu. Rev. Biomed. Eng. 4, 129–153 (2002). [DOI] [PubMed] [Google Scholar]
- Pechorsky A., Nitzan Y. & Lazarovitch T. Identification of pathogenic bacteria in blood cultures: Comparison between conventional and PCR methods. J. Microbiol. Methods 78, 325–330 (2009). [DOI] [PubMed] [Google Scholar]
- Tu Q. & Chang C. Diagnostic applications of Raman spectroscopy. Nanomed Nanotechnol Biol Med. 8, 545–558 (2012). [DOI] [PubMed] [Google Scholar]
- Harz M. et al. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. J. Biophoton. 2, 70–80 (2009). [DOI] [PubMed] [Google Scholar]
- Campion A. & Kambhampati P. Surface-enhanced Raman scattering. Chem. Soc. Rev. 27, 241 (1998). [Google Scholar]
- Jarvis R. M. & Goodacre R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 76, 40– (2004). [DOI] [PubMed] [Google Scholar]
- Kim K. B., Han J. -H., Choi H., Kim H. C. & Chung T. D. Dynamic Preconcentration of Gold Nanoparticles for Surface-Enhanced Raman Scattering in a Microfluidic System. Small 8, 378–383 (2012). [DOI] [PubMed] [Google Scholar]
- Quang L.-X. et al. A portable surface-enhanced Raman scattering sensor integrated with a lab-on-a-chip for field analysis. Lab Chip 8, 2214–2219 (2008). [DOI] [PubMed] [Google Scholar]
- Kahl M., Voges E., Kostrewa S., Viets C. & Hill W. Periodically structured metallic substrates for SERS. Sens. Actuators B 51, 285 (1998). [Google Scholar]
- Gamby J. et al. Polycarbonate microchannel network with carpet of Gold NanoWires as SERS-active device. Lab Chip 9, 1806–1808 (2009). [DOI] [PubMed] [Google Scholar]
- Huang X., El-Sayed I. H., Qian W. & El-Sayed M. A. Cancer cells assemble and align gold nanorods conjugated to antibodies to produce highly enhanced, sharp, and polarized surface Raman spectra: a potential cancer diagnostic marker. Nano Lett. 7, 1591–1597 (2007). [DOI] [PubMed] [Google Scholar]
- Jarvis R. M., Brooker A. & Goodacre R. Surface-enhanced Raman scattering for the rapid discrimination of bacteria. Faraday Discuss. 132, 281–292 (2006). [DOI] [PubMed] [Google Scholar]
- Cheng I-F., Lin C.-C., Lin D.-Y. & Chang H.-C. A dielectrophoretic chip with a roughened metal surface for on-chip Surface Raman Enhanced Scattering analysis of bacteria. Biomicrofluidics 4, 034104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis R. M. & Goodacre R. Characterisation and identification of bacteria using SERS. Chem. Soc. Rev. 37, 931–936 (2008). [DOI] [PubMed] [Google Scholar]
- Driskell J. D. et al. Rapid and Sensitive Detection of Rotavirus Molecular Signatures Using Surface Enhanced Raman Spectroscopy. PLOS One 5, e10222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhoumi A., Zhang D., Tam F. & Halas N. J. Surface-Enhanced Raman Spectroscopy of DNA. J. Am. Chem. Soc. 130, 5523–5529 (2008). [DOI] [PubMed] [Google Scholar]
- Filik J. & Stone N. Drop coating deposition Raman spectroscopy of protein mixtures. Analyst 132, 544–550 (2007). [DOI] [PubMed] [Google Scholar]
- Yazdi S. H. & White I. M. A nanoporous optofluidic microsystem for highly sensitive and repeatable surface enhanced Raman spectroscopy detection. Biomicrofluidics 6, 014105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan R. D. et al. Capillary flow as the cause of ring stains from dried liquid drops. Nature 389, 827–829 (1997). [Google Scholar]
- Keskin S. & Culha M. Label-free detection of proteins from dried-suspended droplets using surface enhanced Raman scattering. Analyst 137, 2651–2657 (2012). [DOI] [PubMed] [Google Scholar]
- Jarvis R. M. & Goodacre R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 76, 40– (2004). [DOI] [PubMed] [Google Scholar]
- Wellinghausen N. et al. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J. Clin. Microbiol. 47, 2759–2765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-Y. et al. Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood. Nat. Commun. 2, 538 (2011). [DOI] [PubMed] [Google Scholar]
- Cheng I-F., Han H. -W. & Chang H. -C. Dielectrophoresis and shear enhanced DNA hybridization for rapid discrimination of Candida species. Biosens. Bioelectron. 33, 36–43 (2012). [DOI] [PubMed] [Google Scholar]
- Chung C.-C. et al. Screening of the antibiotic susceptibility to β-lactam-induced elongation of Gram-negative bacteria based on dielectrophoresis. Anal. Chem. 84, 3347–3354 (2012). [DOI] [PubMed] [Google Scholar]
- Bharti B., Findenegg G. H. & Velev O. D. Co-Assembly of Oppositely Charged Particles into Linear Clusters and Chains of Controllable Length. Sci. Rep. 2, 1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. et al. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 16, 541–546 (1998). [DOI] [PubMed] [Google Scholar]
- Krishnan R. et al. Interaction of Nanoparticles at the DEP Microelectrode Interface under High Conductance Conditions. Electrochem commun. 11, 1661–1666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R. & Heller M. J. An AC electrokinetic method for enhanced detection of DNA nanoparticles. J. Biophoton. 2, 253–261 (2009). [DOI] [PubMed] [Google Scholar]
- Sonnenberg A., Marciniak J. Y., Krishnan R. & Heller M. J. Dielectrophoretic isolation of DNA and nanoparticles from blood. Electrophoresis 33, 2482–2490 (2012). [DOI] [PubMed] [Google Scholar]
- Deng Y.-L. & Juang Y.-J. Electrokinetic trapping and surface enhanced Raman scattering detection of biomolecules using optofluidic device integrated with a microneedles array. Biomicrofluidics 7, 014111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Siqueira e Oliveira F. S., Giana H. E. & Silveira L. Jr Discrimination of selected species of pathogenic bacteria using near-infrared Raman spectroscopy and principal components analysis. J Biomed Opt. 17, 107004 (2012). [DOI] [PubMed] [Google Scholar]
- Zeiri L. & Efrima S. Surface-enhanced Raman spectroscopy of bacteria: the effect of excitation wavelength and chemical modification of the colloidal milieu. J. Raman Spectrosc. 36, 667–675 (2005). [Google Scholar]
- Hamasha K. et al. Sensitive and specific discrimination of pathogenic and nonpathogenic Escherichia coli using Raman spectroscopy—a comparison of two multivariate analysis techniques. Biomed Opt Express 4, 481–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Díez E. C., Winder C. L., Ashton L., Currie F. & Goodacre R. Monitoring the mode of action of antibiotics using Raman spectroscopy: investigating subinhibitory effects of Amikacin on Pseudomonas aeruginosa. Anal Chem. 77, 2901–2906 (2005). [DOI] [PubMed] [Google Scholar]
- Menachery A. et al. Counterflow Dielectrophoresis for Trypanosome Enrichment and Detection in Blood. Sci. Rep. 2, 775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer L. G., Wilson M. L. & Weinstein M. P. Update on Detection of Bacteremia and Fungemia. Clin. Microbiol. Rev. 10, 444–465 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagupsky P. & Nolte F. S. Quantitative aspects of septicemia. Clin. Microbiol. Rev. 3, 269–279 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N. G., Ramos A., González A., Morgan H. & Castellanos A. Fluid flow induced by nonuniform ac electric fields in electrolytes on microelectrodes. I. Experimental measurements. Phys. Rev. E 61, 4011–4018 (2000). [DOI] [PubMed] [Google Scholar]
- Wu J. Biased AC Electro-Osmosis for On-Chip bioparticle processing. IEEE Transactions on Nanotechnology 5, 84–89 (2006). [Google Scholar]
- Gagnon Z. R. & Chang H.-C. Electrothermal ac electro-osmosis. Appl. Phys. Lett. 94, 024101 (2009). [Google Scholar]
- Lian M. & Wu J. Ultrafast micropumping by biased alternating current electrokinetics. Appl. Phys. Lett. 94, 064101 (2009). [Google Scholar]