Abstract
Terahertz (THz) radiation lies between the infrared and microwave regions of the electromagnetic spectrum and is non-ionizing. We show that exposure of artificial human skin tissue to intense, picosecond-duration THz pulses affects expression levels of numerous genes associated with non-melanoma skin cancers, psoriasis and atopic dermatitis. Genes affected by intense THz pulses include nearly half of the epidermal differentiation complex (EDC) members. EDC genes, which are mapped to the chromosomal human region 1q21, encode for proteins that partake in epidermal differentiation and are often overexpressed in conditions such as psoriasis and skin cancer. In nearly all the genes differentially expressed by exposure to intense THz pulses, the induced changes in transcription levels are opposite to disease-related changes. The ability of intense THz pulses to cause concerted favorable changes in the expression of multiple genes implicated in inflammatory skin diseases and skin cancers suggests potential therapeutic applications of intense THz pulses.
Recent progress in the development of broadband terahertz (THz) pulse sources based on ultrafast lasers1 has led to applications of THz pulses as a non-invasive tool in cancer diagnosis2,3,4,5,6, intra-operative tumor margin identification7, assessment of burns8 and in vivo skin9 and cornea10 hydration sensing. At the same time, we are only starting to uncover how THz radiation, and especially intense THz pulses with duration on the order of 1 ps and peak electric fields on the order of 100 kV/cm and higher, interacts with biological tissue11,12,13,14,15,16,17,18,19. With photon energies typically in the range from 0.5–15 meV, THz radiation is non-ionizing, and the mechanisms by which it interacts with cells and tissues are fundamentally different from those involved in interactions of living matter with high-energy ionizing radiation (i.e., UVB, UVC, X-rays, gamma rays) that cause damage by directly breaking covalent bonds in DNA and other biomolecules.
Initially, it was assumed that the THz-induced effects are exclusively thermal in nature2,11,20,21. While heating due to strong absorption of THz radiation by water in biological materials may be significant in the case of continuous-wave excitation16, average thermal effects are minimal for picosecond-duration THz pulse sources with low repetition rates20. Theoretical modelling as well as experiments have shown that many important cellular biomolecules, including DNA and proteins, have intrinsic vibrational resonances in the THz range22,23,24,25,26,27. Resonant coupling of THz radiation to those vibrational modes may therefore affect conformation states and dynamics of various cellular biomolecules, and thus impact cellular functions15,27.
Recently, we demonstrated that intense, picosecond THz pulses significantly induce phosphorylation of H2AX in human skin tissue, indicative of DNA damage18. We also observed THz-pulse-induced increases in the levels of multiple cell cycle regulatory and tumor suppressor proteins, such as p53, p21, p16, Ku70, and EGR1, suggesting that cellular DNA repair machinery is activated in response to THz-pulse-induced DNA damage18. Furthermore, recent experiments have demonstrated that exposure to broadband, picosecond-duration THz pulses results in gene expression changes that depend on the type of exposed cells or tissues and are non-thermal in nature. For example, broadband, picosecond-duration THz pulses influenced differentiation of mammalian stem cells and elicited parameter-specific changes in the expression levels of genes that were transcriptionally active at a given differentiation time point14,15. In human skin tissue, it was observed that intense THz pulses caused changes in expression levels of several cancer-related genes19.
Since the penetration of THz radiation into the human body is limited to the sub-mm range28, its effects on skin tissue are most relevant for potential clinical applications. In our preliminary study of the effects of intense THz pulses on human skin tissue, we analyzed gene expression in the dissected portions of the tissue containing both directly exposed as well as neighboring unexposed portions, and noted changes in gene expression19. Yet, it is important to precisely delineate the changes that occur exclusively in THz-exposed tissue. Here we report on the impact of intense THz pulses on the global gene expression profile in human skin tissue. We find that exposure to intense THz pulses results in coordinated changes in expression of multiple genes involved in epidermal differentiation. Many of the affected genes are implicated in inflammatory skin diseases such as psoriasis and atopic dermatitis and in non-melanoma skin cancer, suggesting the possibility for novel treatment modalities based on intense THz pulses.
Results
We have exposed artificial human skin tissues to picosecond-duration (Fig. 1a) broadband (0.2–2.5 THz, Fig. 1b) THz pulses with 1 kHz repetition rate, 1/e2 spot-size diameter of 1.5 mm (Fig. 1c) and pulse energies of either 1.0 μJ or 0.1 μJ. For comparison, we have exposed skin tissues to UVA pulses (400 nm, 0.1 ps, 0.024 μJ). While UVA is not directly absorbed by DNA, its cytotoxicity and genotoxicity are mediated by the generation of reactive oxygen species which in turn damage DNA and other cellular components29.
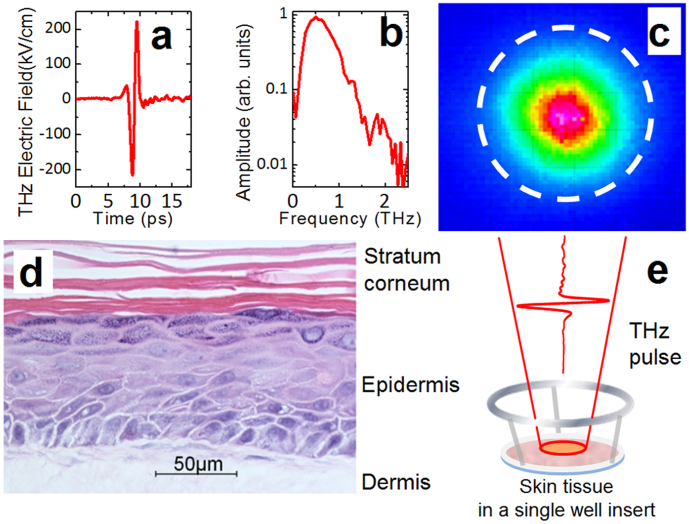
Figure 1. Intense THz pulses and skin tissue samples used in this study.
(a) Electric field wave form of the THz pulse. (b) Spectral bandwidth of the THz pulse shown in (a). (c) THz spot profile at the focus. The 1/e2 diameter of the THz beam is 1.5 mm. The 2 mm-diameter dashed circle shows the portion of the tissue that was excised for gene profiling. (d) Histology of an EpiDermFT tissue section (400X image, courtesy of MatTek). (e) Schematic illustration of the THz exposure procedure. The EpiDermFT tissue is in a single well insert placed at the focus of THz beam.
The full thickness human normal skin tissue used, EpiDermFT, is a multilayered, highly differentiated model of dermis and epidermis (Fig. 1d). It is mitotically and metabolically active, preserves the arrangement and communication of cells present in tissues in vivo30, and thus provides an excellent platform for assessing the effects of exposure to intense THz pulses18,19. Tissue samples were placed at the focus of the pulsed THz beam for 10 minutes (Fig. 1e) or at the focus of the pulsed 400 nm beam for two minutes. Global gene expression in excised 2 mm-diameter exposed portions of the tissues (as illustrated schematically by a dashed circle in Fig. 1c) was analyzed using an Illumina HumanHT-12 v4 Expression BeadChip 30 minutes after irradiation.
We found that intense THz pulses profoundly affect gene expression in directly exposed human skin. Exposure to THz pulses with an energy of 1.0 μJ for 10 minutes altered the levels of 442 genes (Supplementary Table S1), while irradiation by pulses with 10 times lower energy (0.1 μJ) for the same time duration affected the expression of 397 genes as compared to unexposed controls (Supplementary Table S2). Interestingly, exposure to UVA pulses caused changes in the expression of only 290 genes (Supplementary Table S3). The observed changes in gene expression profile were not caused by THz-induced tissue heating. THz exposures were carried out at ambient temperature (21°C), and the time-averaged power (1 mW) was low. Applying a theoretical formalism developed in ref. 20, we estimate that the temperature increase due to THz exposure was less than 0.7°C. Furthermore, none of the heat shock protein encoding genes were differentially expressed in THz- or UVA-exposed tissues.
Analysis of the overlaps in the lists of genes differentially expressed upon exposure to intense pulsed THz and UV radiation revealed interesting trends (Fig. 2). While there is a significant overlap between genes affected by both THz pulse energies, little overlap exists between THz- and UVA-induced gene expression profiles, which is not surprising since there is a fundamental difference in how low photon energy (~4 meV), non-ionizing THz radiation and high photon energy (3.1 eV) UVA radiation interact with living cells.
Figure 2. Intense THz-pulse-induced gene expression in human skin.
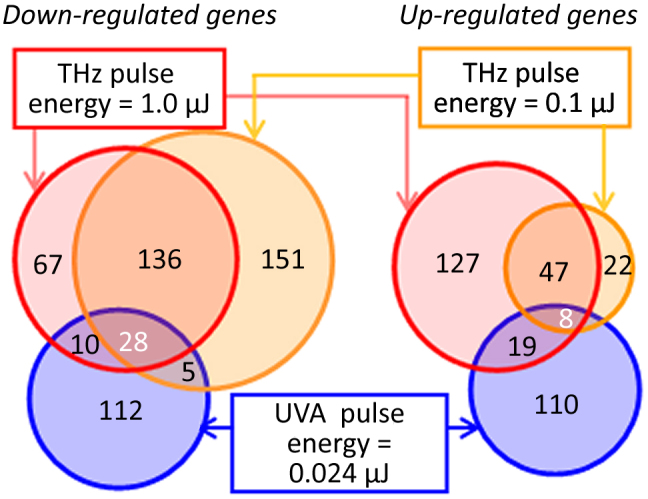
Venn diagrams summarizing differentially-expressed genes in EpiDermFT tissues exposed to either 1.0 μJ or 0.1 μJ THz or UVA (400 nm, 0.024 μJ) pulses, as described in Materials and Methods. Genes with a False Discovery Rate (FDR)-adjusted p-value < 0.05 and log2 fold change > 0.6 (1.5× change) were considered differentially expressed. Left diagram: down-regulated genes. Right diagram: up-regulated genes. The complete lists of the affected genes, the fold change and p-values are given in Supplementary Tables S1, S2 and S3.
Differentially-expressed genes that were common for both THz pulse exposure regimes are of particular interest as potential THz-pulse-exposure biomarkers that hold the key to understanding cellular effects of intense THz pulses. We identified 219 such genes, of which 164 are down-regulated and 55 are up-regulated as a result of THz pulse exposure. Among them are genes involved in molecular etiology of dermatological diseases (psoriasis, atopic dermatitis, epidermal hyperplasia, and dermatitis), cancer, and genes with important functions in apoptotic signaling pathways.
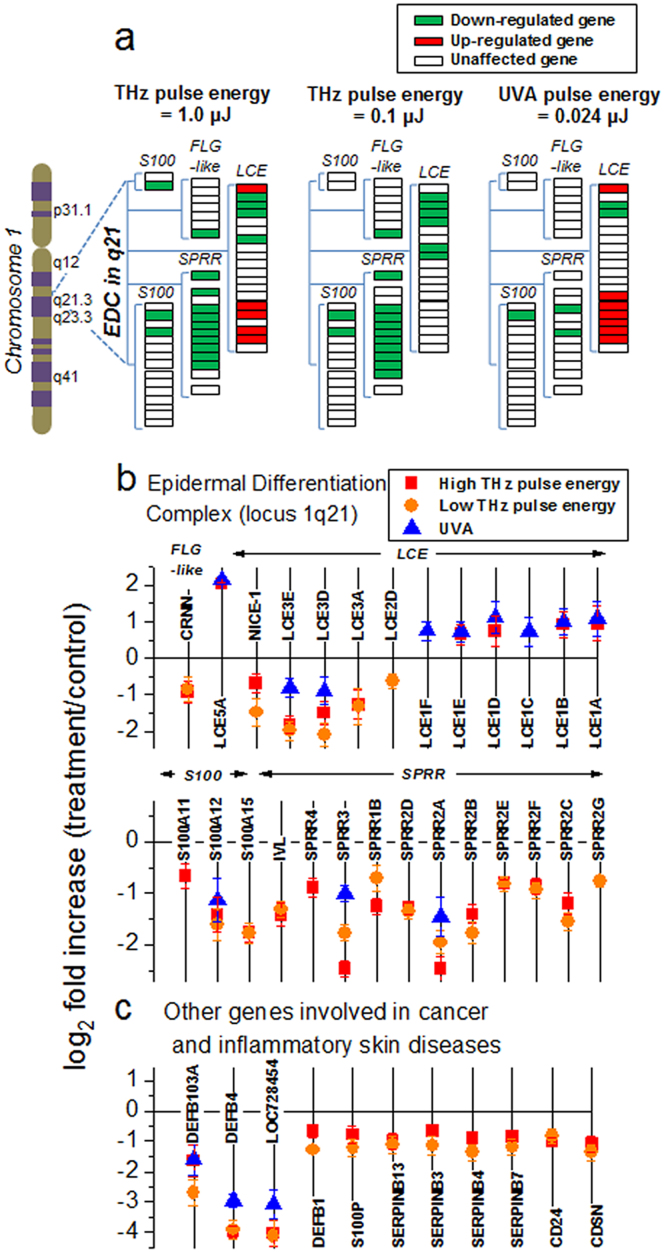
We also find that the affected genes are not uniformly distributed throughout the genome. The most striking changes were observed in the expression of epidermal differentiation complex (EDC) genes. EDC describes the 1.6-Mb locus on human chromosome 1q21 that contains 57 genes encoding S100 proteins as well as structural proteins of epidermal cornification (Fig. 3a)31,32,33. Of them, 16 were markedly down-regulated by exposure to intense THz pulses in both intensity regimes, compared to only 5 affected by UVA pulses (Fig. 3b). THz-pulse-affected genes include members of the Small Proline Rich Region (SPRR), involucrin (IVL), Late Cornified Envelope (LCE) and the S100 families, as well as NICE-1 and cornulin (CRNN). In addition to EDC genes, exposure to intense THz pulses also altered expression levels of many other genes implicated in cancer and dermatological diseases (Fig. 3c).
Figure 3. Differentially expressed Epidermal Differentiation Complex (EDC) genes and selected other genes associated with non-melanoma skin cancer or inflammatory skin diseases.
(a) EDC genes that were either up-regulated (green rectangles) or down-regulated (red rectangle) after exposure of skin tissues to either 1.0 μJ or 0.1 μJ THz pulse energies, or UVA pulses. Unaffected genes are depicted as white rectangles. (b) log2 fold changes for differentially-expressed genes belonging to four families of EDC genes (FLG-like, LCE, S100, and SPRR). (c) Other genes (selected) involved in dermatological diseases and cancer, whose expression levels were altered by exposure to both THz pulse energy regimes. In (b) and (c), error bars indicate corresponding standard deviations, and in some cases are smaller than the symbol size. Mean values of three biological or technical replicates, log2 fold changes relative to control mean values, and FDR-adjusted p-values for genes in (b) and (c) are given in Supplementary Table S4. Individual replicates, mean values and corresponding standard deviations are given in Supplementary Table S5.
Discussion
EDC complex genes are responsible for terminal differentiation of keratinocytes and regulation of epidermal barrier function, as well as for skin immune and inflammation responses. Up-regulation of EDC genes leads to increased proliferation and differentiation of keratinocytes commonly observed in inflammatory skin disorders such as psoriasis and atopic dermatitis32,34,35, as well as in skin cancers36. The 1q21 locus has been identified as one of the key psoriasis susceptibility loci (PSORS4) in clinical genome-wide linkage studies34. Of the EDC genes that were down-regulated by intense THz pulses (Fig. 3b), many, including S100A1537, S100A1237,38, SPRR2A, SPRR3, SPRR2B36, LCE3 genes39, are up-regulated in psoriasis34,36.
Enhanced expression of multiple EDC genes, including members of the SPRR family, S100 genes that encode for calcium-binding proteins, and multiple cornified envelope genes, has been identified as a hallmark of cutaneous squamous cell carcinomas (SCC)36,40. Of these SCC-associated EDC genes, eight, including S100A11, S100A12, SPRR1B, SPRR2B, SPRR2C, SPRR3, involucrin (IVL), and LCE3D, are down-regulated by intense THz pulse exposure (Fig. 3b). Down regulation of these genes by intense THz pulses may open new avenues for targeted treatments for psoriasis and SCC.
Besides EDC genes, THz pulses affected numerous other genes involved in psoriasis, atopic dermatitis and other inflammatory skin diseases, as well as in many cancer types, such as aggressive oral squamous cell carcinoma (OSCC) and non-melanoma skin cancer (SCC and basal cell carcinoma (BCC)) (Fig.3c). We observe down regulation of corneodesmosin (CDSN), a member of the PSORS1 psoriasis susceptibility locus41, and CD24, a key pro-inflammatory gene, both of which are usually over-expressed in psoriatic skin. The CD24 gene encodes for a mucin-like adhesion molecule, and its over expression has been also repeatedly associated with aggressive tumor progression, metastasis and poor prognosis in various cancers42,43. It is also up-regulated in SCC36. Additionally, THz exposure significantly lowered mRNA expression of human β-defensins (DEFB103A, DEFB4, LOC728454, and DEFB1). Interestingly, the impact of intense THz pulses on the expression levels of β-defensins was similar to that of UVA pulses. Enhanced expression of β-defensins is associated with non-melanoma skin cancer (basal cell carcinoma (BCC) and SCC)36,40,44,45,46, and OSCC47, as well as psoriasis34.
Exposure to intense THz pulses also suppressed levels of four serine proteinase inhibitor (serpin) B genes that map to the serpin superfamily locus at chromosome 18q21.348, once again demonstrating a non-uniform distribution of intense-THz-pulse-affected genes throughout the genome. Two of them, serpinB3 and serpinB4, are also known as squamous cell carcinoma antigens 1 and 2, respectively. Their elevated expression is associated with aggressive SCCs49,50 and psoriasis36. SerpinB13, or hurpin, is also over-expressed in SCC, as well as in psoriatic lesions51. Another down-regulated member of the serpin family is serpinB7, or megsin. Finally, intense THz pulses (but not UVA pulses) down-regulated the S100P gene which is related in function to the S100A family of genes of EDC and encodes for a member of the Ca2+-binding proteins family. Over-expression of S100P has been shown to act as a proliferative and anti-apoptotic factor in many tumor types52,53,54. Moreover, targeted disruption of S100P was recently shown to suppress the growth of hepatocellullar carcinoma cells52. While the exact role of serpins as well as S100P in skin cancer and inflammatory skin diseases is yet to be elucidated, their concerted down-regulation by intense THz pulses might have clinical relevance and should therefore be further investigated.
Furthermore, the mechanisms by which intense THz pulses influence gene expression are not yet known. Based on mesoscopic modelling of DNA breathing dynamics in a THz field, it has been suggested that THz radiation may amplify existing (or create new) open states in the double helix, thereby affecting transcription initiation or binding of transcription factors15,55. Alternatively, the changes in gene expression may constitute a cellular response to intense-THz-pulse-induced damage to DNA18 or intracellular proteins, which initiates multiple cellular-damage-repair pathways. In the future, a detailed analysis of the impact of intense THz pulses on the activity of various transcription factors common to affected genes is needed to elucidate the exact cellular and molecular effects of intense THz pulses.
Additionally, analysis of transcriptional changes seen in skin tissue that is directly exposed to intense THz radiation and in the mixture of directly exposed and neighboring naïve tissue19 indicates that exposure to intense THz pulses can induce out-of-field bystander effects. While bystander effects are accepted to occur after exposure to ionizing radiation56,57, the existence, extent and mechanisms of THz-pulse-induced bystander effects need to be further analysed.
While a number of earlier studies investigated the biological effects of continuous-wave THz radiation and found that the majority of the observed effects are thermal in nature, the present study is one of the very few to date that have explored genotoxic, cytotoxic and epigenetic effects of picosecond broadband THz pulses. The low average power of THz pulse sources results in biologically insignificant temperature increases of only fractions of a degree at most. However, the high energy density per pulse produces peak powers and corresponding electric fields that can be extremely high. It is likely that these high peak electric fields are responsible for the observed THz-pulse-driven cellular effects.
In 2003, Clothier and Bourne58 reported that picosecond-duration, 0.2–3.0 THz bandwidth THz pulses had no discernible effect on the differentiation, activity or viability of primary human keratinocytes in vitro. Based on the experimental details provided in the paper, we estimate that the energy density per pulse was ~3 pJ/cm2 (3 × 10−6 μJ/cm2). In a recent work, Williams et al17. explored the influence of more intense THz pulses produced by the ALICE (Daresbury Laboratory, UK) synchrotron source, with energy densities per pulse reaching 10 nJ/cm2 (0.01 μJ/cm2), on the attachment, morphology, proliferation and differentiation of human epithelial and embryonic stem cells. Like Clothier and Bourne58, they reported no changes in cell properties. On the other hand, a series of experiments investigating the effects of broadband (~1–15 THz, centered at 10 THz), subpicosend THz pulses on mouse mesenchymal stem cells (Bock et al14., Alexandrov et al15.) revealed THz-induced changes in the expression of the genes transcriptionally active at a given differentiation stage of stem cells. Furthermore, the same studies found that THz pulse exposure accelerated cell differentiation toward adipose phenotype. In these experiments, the energy density per pulse was significantly higher at 1 μJ/cm2, which may suggest the existence of a THz pulse energy density (and corresponding peak THz electric field) threshold for the induction of changes in cellular function. Finally, in our work, the energy density per pulse was ~6 μJ/cm2 for 0.1 μJ THz pulses and ~60 μJ/cm2 for 1 μJ THz pulses, resulting in specific gene expression changes in human skin tissue. Clearly, more extensive studies are needed to determine pulse energy density thresholds and other exposure parameters for the onset of THz-pulse-induced changes to gene expression and cellular function.
In summary, we have shown that intense THz pulses have profound impact on global gene expression in human skin. Furthermore, we observed that the distribution of intense-THz-pulse-sensitive genes is not uniform across the genome, with a significant number of affected genes belonging to the epidermal differentiation complex in the 1q21 locus. Finally, the observed THz-induced changes in expression of EDC genes and other genes implicated in non-melanoma skin cancers and inflammatory skin disorders such as psoriasis, suggests the potential application of intense THz pulses for treatment aimed at normalizing the expression of these disease-related genes. Picosecond duration, intense THz pulses are non-ionizing, do not cause tissue overheating and can be focused with ~1 mm precision for delivering safe, non-invasive treatment. Of course, since the present study concentrated on analyzing changes in gene expression in artificial human skin tissue 30 minutes after 10-minute-long exposure to intense THz pulses, it essentially provides a snapshot of THz-pulse-induced effects. Extensive large-scale studies tracking the THz-induced-changes in gene expression and the resulting changes in protein expression over a period of time following the exposure of healthy as well as diseased tissues will have to be carried out to assess the full potential of intense THz pulses for therapeutic applications. We envision the pilot study presented here as an essential roadmap for this future work.
Methods
Generation and characterization of intense THz pulses
We have exposed human skin tissues to THz pulses with 1 kHz repetition rate and pulse energies of either 1.0 μJ or 0.1 μJ. The THz pulses were generated by optical rectification of tilted-pulse-front 800 nm pulses from an amplified Ti:sapphire laser source in a LiNbO3 crystal59,60. A black polyethylene sheet placed after the LiNbO3 crystal filtered out the remaining 800 nm beam while letting the THz pulses through with minimal attenuation. The energy of the THz pulses was monitored by a pyroelectric detector (Spectrum Detector, Inc.) with an active area of 6 × 7 mm2. Wire grid polarizers were used to attenuate the THz pulse energy. The THz pulse waveform (Fig. 1a) was measured by electro-optic sampling in 0.5 mm-thick [110] ZnTe crystal. The THz spot size at the focus was determined by imaging the THz beam on a pyroelectric infrared camera. The peak electric field was calculated as discussed in Ref. 19. The peak THz electric field was 220 kV/cm for 1.0 μJ and 70 kV/cm for 0.1 μJ energy THz pulses. Exposure to 400 nm pulses for two minutes was also used. The UVA pulses were generated by second harmonic generation in a BBO crystal. A shortpass filter placed after the BBO crystal blocked any remaining 800 nm beam. The UVA spot size at the tissue surface was 2.7 mm diameter.
Tissue models and handling
We used the in vivo full thickness artificial human skin tissues model, EpiDermFT by MatTek, to study the effects of intense THz pulses on gene expression. EpiDermFT recreates normal skin tissue structure with differentiated dermis and epidermis. It consists of human-derived epidermal keratinocytes and dermal fibroblasts that are mitotically and metabolically active31. The tissues were cultured according to the manufacturer's protocol, using an air-liquid interface tissue culture technique. Unexposed tissue samples served as controls. Skin tissues in single well plate inserts were placed at the focus of the THz beam for 10 minutes, or at the focus of the UVA beam for 2 minutes.
Four tissue samples were used for each of the four experimental conditions (control, 1.0 μJ THz and 0.1 μJ THz pulses, and UVA). Following exposure, the tissues were incubated at 37°C in 5% CO2 atmosphere for 30 minutes in a multiwell dish filled with fresh medium, followed by a snap freeze in liquid nitrogen. Control tissues underwent the same procedure other than being irradiated. The central region (2 mm in diameter, as shown in Fig. 1c) containing the exposed portion of the frozen tissues was cut out and used for gene expression analysis.
Whole-genome gene expression profiling
RNA isolation
Total RNA was isolated using the Illustra RNAspin mini kit (GE Healthcare Life Sciences, Buckinghamshire, UK) were processed following the manufacturer's instructions. Samples were eluted in Ultrapure DNase/RNase-free distilled water, which was provided in the kit. RNA samples were quantified using ultraviolet spectroscopy (NanoDrop, Wilmington, DE) and were further assessed for RNA integrity (RIN) on the Agilent 2100 Bioanalyzer (Santa Clara, CA) using the RNA Nano-chip Kit. RNA samples with RIN values of seven or better were used for the further analysis.
Library preparation
CRNA was created using the Ambion's Illumina TotalPrep RNA Amplification Kit (Applied Biosystems, Carlsbad, CA) with an input of 400 ng of total RNA per sample. Briefly, oligo-dT primers were used to synthesize first strand cDNA containing a phage T7 promoter sequence. Single-stranded cDNA was converted into a double-stranded DNA template via DNA polymerase. RNase H simultaneously acted to degrade the RNA. Samples of cDNA were purified in filter cartridges to remove excess RNA, primers, enzymes and salts. The recovered cDNA was subjected to in vitro transcription using biotinylated UTPs. This step created, labeled and amplified cRNA. A final purification step removed unincorporated NTPs, salts, inorganic phosphates and enzymes, which prepared the samples for hybridization.
Hybridization and detection
Illumina's direct hybridization assay kit was used to process samples according to the manufacturer's protocol (Illumina, San Diego, CA). Overnight, 750 ng from each cRNA sample was hybridized into the Illumina HumanHT-12_v4 Whole Genome Expression BeadChip arrays. Afterward, a 10-min incubation with a supplied wash buffer at 55°C preceded a 5-min room temperature wash. The arrays were incubated in 100% ethanol for 10 min. A second room temperature wash lasted 2 min with gentle shaking, which completed this high stringency wash step. The arrays were blocked with a buffer for 10 min and washed before a 10-min steptavidin-Cy3 (1:1,000) probing. After a 5-min wash at room temperature, the BeadChips were dried and imaged. Six controls were also built into the Whole-Genome Gene Expression Direct Hybridization Assay system to cover aspects of the array experiments, including controls for: the biological specimen (14 probes for housekeeping controls), 3 controls for hybridization (6 probes for Cy3-labeled hybridization, 4 probes for low stringency hybridization, and 1 probe for high stringency hybridization), signal generation (2 probes for biotin control), and approximately 800 probes for negative controls on an 8-sample BeadChip. The arrays were scanned on the iScan platform (Illumina), and data were normalized and scrutinized using Illumina BeadStudio Software.
BeadChip statistical analysis and data processing
The false discovery rate (FDR) was controlled using the Benjamini- Hochberg method. The Illumina Custom Model took the FDR into account and was used to analyze the data. Differential gene expression (at least a 2-fold change) from sham-treated tissues was determined to be statistically significant if the p-value after the Benjamini-Hochberg method adjustment was lower than 0.05. The values were transformed to show a log2 scale. Lists of regulated transcripts were inserted into the web-based DAVID Bioinformatics Resources 6.7 (NIAID/NIH) Functional Annotation Tool.
Accession numbers
The raw and processed gene expression data for this project have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (GSE48586).
Author Contributions
L.V.T., O.K. and F.A.H. conceived the concept. L.V.T. and O.K. wrote the manuscript with contributions of all authors. A.K.A. built the intense THz pulse source under supervision of F.A.H. THz pulse and UVA exposure experiments were carried out by L.V.T., A.K.A., F.A.H. and O.K. Gene expression analysis was done by O.K., R.R.-J.and A.G. Statistical analysis was performed by A.G. and R.W. All authors reviewed the manuscript and contributed to discussions.
Supplementary Material
Supplementary information - tables
Acknowledgments
We wish to acknowledge financial support from the Alberta Cancer Foundation and the Alberta Innovates – Health Solutions (AIHS), Natural Sciences and Engineering Research Council of Canada (NSERC), Canada Foundation for Innovation (CFI), and the Alberta Science and Research Investments Program (ASRIP). We are also grateful for technical assistance from Don Mullin, Greg Popowich, Rommy Rodriguez-Juarez and Jody Filkowski.
References
- Tonouchi M. Cutting-edge terahertz technology. Nat. Photonics 1, 97–105 (2007). [Google Scholar]
- Fitzgerald A. J. et al. An introduction to medical imaging with coherent terahertz frequency radiation. Phys. Med. Biol. 47, R67–84 (2002). [DOI] [PubMed] [Google Scholar]
- Ashworth P. C. et al. Terahertz pulsed spectroscopy of freshly excised human breast cancer. Opt. Express 17, 12444–54 (2009). [DOI] [PubMed] [Google Scholar]
- Woodward R. M., Wallace V. P., Arnone D. D., Linfield E. H. & Pepper M. Terahertz pulsed imaging of skin cancer in the time and frequency domain. J. Biol. Phys. 29, 257–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Fan S., Sun Y. & Pickwell-Macpherson E. The potential of terahertz imaging for cancer diagnosis: A review of investigations to date. Quant. Imaging Med. Surg. 2, 33–45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph C. S., Yaroslavsky A. N., Neel V. A., Goyette T. M. & Giles R. H. Continuous wave terahertz transmission imaging of nonmelanoma skin cancers. Lasers Surg. Med. 43, 457–62 (2011). [DOI] [PubMed] [Google Scholar]
- Ashworth P. C. et al. An intra-operative THz probe for use during the surgical removal of breast tumors. Infrared, Millimeter and Terahertz Waves (2008). dx.doi.org/10.1109/ICIMW.2008.4665810. [Google Scholar]
- Arbab M. H. et al. Terahertz reflectometry of burn wounds in a rat model. Biomed. Opt. Express 2, 2339–47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen J. Y. et al. Towards medical terahertz sensing of skin hydration. Stud. Health Technol. Inform. 142, 364–8 (2009). [PubMed] [Google Scholar]
- Bennett D. et al. Assessment of corneal hydration sensing in the terahertz band: in vivo results at 100 GHz. J. Biomed. Opt. 17, 97008–1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmink G. J. & Grundt J. E. Current State of Research on Biological Effects of Terahertz Radiation. J. Infrared, Millimeter, Terahertz Waves 32, 1074–1122 (2011). [Google Scholar]
- Korenstein-Ilan A. et al. Terahertz radiation increases genomic instability in human lymphocytes. Radiat. Res. 170, 224–34 (2008). [DOI] [PubMed] [Google Scholar]
- Hintzsche H. et al. Terahertz radiation induces spindle disturbances in human-hamster hybrid cells. Radiat. Res. 175, 569–74 (2011). [DOI] [PubMed] [Google Scholar]
- Bock J. et al. Mammalian stem cells reprogramming in response to terahertz radiation. PLoS One 5, e15806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov B. S. et al. Specificity and heterogeneity of terahertz radiation effect on gene expression in mouse mesenchymal stem cells. Sci. Rep. 3, 1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmink G. J. et al. In vitro investigation of the biological effects associated with human dermal fibroblasts exposed to 2.52 THz radiation. Lasers Surg. Med. 43, 152–63 (2011). [DOI] [PubMed] [Google Scholar]
- Williams R. et al. The influence of high intensity terahertz radiation on mammalian cell adhesion, proliferation and differentiation. Phys. Med. Biol. 58, 373–91 (2013). [DOI] [PubMed] [Google Scholar]
- Titova L. V. et al. Intense THz pulses cause H2AX phosphorylation and activate DNA damage response in human skin tissue. Biomed. Opt. Express 4, 559–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titova L. V. et al. Intense picosecond THz pulses alter gene expression in human skin tissue in vivo. Proc. SPIE 8585, 85850Q (2013). [Google Scholar]
- Kristensen T. T., Withayachumnankul W., Jepsen P. U. & Abbott D. Modeling terahertz heating effects on water. Opt. Express 18, 4727–39 (2010). [DOI] [PubMed] [Google Scholar]
- Berry E. et al. Do in vivo terahertz imaging systems comply with safety guidelines? J. Laser Appl. 15, 192–198 (2003). [Google Scholar]
- Prohofsky E. W., Lu K. C., Van Zandt L. L. & Putnam B. F. Breathing modes and induced resonant melting of the double helix. Phys. Lett. A 70, 492–494 (1979). [Google Scholar]
- Chitanvis S. M. Can low-power electromagnetic radiation disrupt hydrogen bonds in dsDNA? J. Polym. Sci. Part B: Polym. Phys. 44, 2740–2747 (2006). [Google Scholar]
- Fischer B. M., Walther M. & Jepsen P. U. Far-infrared vibrational modes of DNA components studied by terahertz time-domain spectroscopy. Phys. Med. Biol. 47, 3807–14 (2002). [DOI] [PubMed] [Google Scholar]
- Kim S. J., Born B., Havenith M. & Gruebele M. Real-time detection of protein-water dynamics upon protein folding by terahertz absorption spectroscopy. Angew. Chem. Int. Ed. 47, 6486–9 (2008). [DOI] [PubMed] [Google Scholar]
- Markelz A., Whitmire S., Hillebrecht J. & Birge R. THz time domain spectroscopy of biomolecular conformational modes. Phys. Med. Biol. 47, 3797–805 (2002). [DOI] [PubMed] [Google Scholar]
- Cherkasova O. P., Fedorov V. I., Nemova E. F. & Pogodin A. S. Influence of Terahertz Laser Radiation on the Spectral Characteristics and Functional Properties of Albumin. Opt. Spectrosc. 107, 534–7 (2009). [Google Scholar]
- Siegel P. H. Terahertz technology in biology and medicine. IEEE Trans. Microwave Theory Tech. 52, 2438–47 (2004). [Google Scholar]
- Cadet J., Sage E. & Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat. Res. 571, 3–17 (2005). [DOI] [PubMed] [Google Scholar]
- Sedelnikova O. A. et al. DNA double-strand breaks form in bystander cells after microbeam irradiation of three-dimensional human tissue models. Cancer Res. 67, 4295–302 (2007). [DOI] [PubMed] [Google Scholar]
- Mischke D., Korge B. P., Marenholz I., Volz A. & Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J. Invest. Dermatol. 106, 989–92 (1996). [DOI] [PubMed] [Google Scholar]
- de Guzman Strong C. et al. A milieu of regulatory elements in the epidermal differentiation complex syntenic block: implications for atopic dermatitis and psoriasis. Hum. Mol. Genet. 19, 1453–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypriotou M., Huber M. & Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 21, 643–9 (2010). [DOI] [PubMed] [Google Scholar]
- Roberson E. D. & Bowcock A. M. Psoriasis genetics: breaking the barrier. Trends Genet. 26, 415–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffjan S. & Stemmler S. On the role of the epidermal differentiation complex in ichthyosis vulgaris, atopic dermatitis and psoriasis. Br. J. Dermatol. 157, 441–9 (2007). [DOI] [PubMed] [Google Scholar]
- Haider A. S. et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J. Invest. Dermatol. 126, 869–81 (2006). [DOI] [PubMed] [Google Scholar]
- Eckert R. L. et al. S100 proteins in the epidermis. J. Invest. Dermatol. 123, 23–33 (2004). [DOI] [PubMed] [Google Scholar]
- Semprini S. et al. Evidence for differential S100 gene over-expression in psoriatic patients from genetically heterogeneous pedigrees. Hum. Genet. 111, 310–3 (2002). [DOI] [PubMed] [Google Scholar]
- Bergboer J. G. et al. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am. J. Pathol. 178, 1470–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L. G. et al. Microarray analysis of cutaneous squamous cell carcinomas reveals enhanced expression of epidermal differentiation complex genes. Mol. Carcinog. 49, 619–29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. et al. Corneodesmosin expression in psoriasis vulgaris differs from normal skin and other inflammatory skin disorders. Lab. Invest. 81, 969–76 (2001). [DOI] [PubMed] [Google Scholar]
- Baumann P. et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 65, 10783–93 (2005). [DOI] [PubMed] [Google Scholar]
- Lee J. H., Kim S. H., Lee E. S. & Kim Y. S. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol. Rep. 22, 1149–56 (2009). [DOI] [PubMed] [Google Scholar]
- Muehleisen B. et al. Distinct Innate Immune Gene Expression Profiles in Non- Melanoma Skin Cancer of Immunocompetent and Immunosuppressed Patients. PLOS One 7, e40754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambichler T. et al. Pattern of mRNA expression of beta-defensins in basal cell carcinoma. BMC Cancer 6, 163 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu Y. K., Abe K., Ferris L. K., Sarkar S. N. & Ferris R. L. Human β-defensin 3 promotes NF-κB-mediated CCR7 expression and anti-apoptotic signals in squamous cell carcinoma of the head and neck. Carcinogenesis 32, 168–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaki K. et al.. High concentration of beta-defensin-2 in oral squamous cell carcinoma. Anticancer Res. 22, 2103–7 (2002). [PubMed] [Google Scholar]
- Schneider S. S. et al. A serine proteinase inhibitor locus at 18q21.3 contains a tandem duplication of the human squamous cell carcinoma antigen gene. Proc. Natl. Acad. Sci. USA 92, 3147–51 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A. et al. Tumor-related protein, the squamous cell carcinoma antigen binds to the intracellular protein carbonyl reductase. Int. J. Oncol. 36, 1395–400 (2010). [DOI] [PubMed] [Google Scholar]
- Catanzaro J. M. et al. Elevated expression of squamous cell carcinoma antigen (SCCA) is associated with human breast carcinoma. PLoS One 6, e19096 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussali H. et al. Expression of hurpin, a serine proteinase inhibitor, in normal and pathological skin: overexpression and redistribution in psoriasis and cutaneous carcinomas. Exp. Dermatol. 14, 420–8 (2005). [DOI] [PubMed] [Google Scholar]
- Kim J. K. et al. Targeted disruption of S100P suppresses tumor cell growth by down-regulation of cyclin D1 and CDK2 in human hepatocellular carcinoma. Int. J. Oncol. 35, 1257–64 (2009). [PubMed] [Google Scholar]
- Arumugam T., Simeone D. M., Van Golen K. & Logsdon C. D. S100P promotes pancreatic cancer growth, survival, and invasion. Clin. Cancer. Res. 11, 5356–64 (2005). [DOI] [PubMed] [Google Scholar]
- Schor A. P., Carvalho F. M., Kemp C., Silva I. D. & Russo J. S100P calcium-binding protein expression is associated with high-risk proliferative lesions of the breast. Oncol. Rep. 15, 3–6 (2006). [PubMed] [Google Scholar]
- Alexandrov B. S., Gelev V., Bishop A. R., Usheva A. & Ramussen K. Ø. DNA breathing dynamics in the presence of a terahertz field. Phys. Lett. A 374, 1214–1217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk O. et al. microRNAome changes in bystander three-dimensional human tissue models suggest priming of apoptotic pathways. Carcinogenesis 31, 1882–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothersill C. & Seymour C. B. Radiation-induced bystander effects--implications for cancer. Nat. Rev. Cancer 4,158–64 (2004). [DOI] [PubMed] [Google Scholar]
- Clothier R. H. & Bourne N. Effects of THz exposure on human primary keratinocyte differentiation and viability., J. Biol. Phys. 29, 179–85 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard F. et al. Generation of intense terahertz radiation via optical methods. IEEE J. Sel. Top. Quantum Electron. 17, 5–16 (2011). [Google Scholar]
- Hoffmann M. C. & Fülöp J. A. Intense ultrashort terahertz pulses: generation and applications. J. Phys. D: Appl. Phys. 44, 083001 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information - tables