Abstract
Expression of the four transcription factors, that is, Oct4, Sox2, cMyc, and Klf4 has been shown to generate induced pluripotent stem cells (iPSCs) from many types of specialized differentiated somatic cells. It remains unclear, however, whether fully committed skeletal muscle progenitor cells (myoblasts) have the potency to undergo reprogramming to develop iPSCs in line with previously reported cases. To test this, we have isolated genetically marked myoblasts derived from satellite cell of adult mouse muscles using the Cre-loxP system (Pax7-CreER:R26R and Myf5-Cre:R26R). On infection with retroviral vectors expressing the four factors, these myoblasts gave rise to myogenic lineage tracer lacZ-positive embryonic stem cell (ESC)-like colonies. These cells expressed ESC-specific genes and were competent to differentiate into all three germ layers and germ cells, indicating the successful generation of myoblast-derived iPSCs. Continuous expression of the MyoD gene, a master transcription factor for skeletal muscle specification, inhibited this reprogramming process in myoblasts. In contrast, reprogramming myoblasts isolated from mice lacking the MyoD gene led to an increase in reprogramming efficiency. Our data also indicated that Oct4 acts as a transcriptional suppressor of MyoD gene expression through its interaction with the upstream enhancer region. Taken together, these results indicate that suppression of MyoD gene expression by Oct4 is required for the initial reprogramming step in the development of iPSCs from myoblasts. This data suggests that the skeletal muscle system provides a well-defined differentiation model to further elaborate on the effects of iPSC reprogramming in somatic cells.
Keywords: Induced pluripotent stem cell, Satellite cell, MyoD, Oct4, Skeletal muscle, Myoblast
Introduction
Induced pluripotent stem cells (iPSCs) have been generated from somatic cells reprogrammed by transduction of four transcription factors, that is, Oct4, Sox2, cMyc, and Klf4 [1]. These iPSCs have been derived from fibroblasts, B lymphocytes [2], natural killer T cells [3], stomach cells, liver cells [4], pancreatic β-cells [5], and neural stem cells [6]. The iPSCs generated from adult tissues have been thought to mimic ESCs and hold tremendous therapeutic promise due to their ability to proliferate, differentiate, and self-renew. However, many of the molecular mechanisms of the reprogramming process, particularly tissue-specific factors limiting pluripotency, remain undefined.
Along these lines, recent work demonstrated that the efficiency of iPSC reprogramming is dependent on the differentiation status of the cell source. For example, stem cells and progenitor cells are more readily reprogrammed to iPSCs compared with differentiated cells [7]. In addition, reprogramming mature B cells to iPSCs requires additional interruption of the transcriptional state that had maintained the cells’ identity [2]. These results suggest that the promotion of differentiation inhibits the reprogramming process. Furthermore, cells inherit a stable genetic program partly through various epigenetic markers such as DNA methylation and histone modifications. This cellular memory needs to be erased during genetic reprogramming for the cellular program to revert to that of an earlier developmental stage [8]. However, the relationship between the transcriptional activity of the four factors and the cellular memory status remains unclear.
Adult skeletal muscle possesses extraordinary regeneration capabilities. After exercise or muscle injury, large numbers of new muscle fibers are normally formed within a week due to the expansion and differentiation of muscle satellite cells [9]. Satellite cells are a small population of myogenic stem cells for muscle regeneration, which are normally mitotically quiescent [10]. Following injury, satellite cells initiate proliferation to produce myogenic precursor cells, or myoblasts, to mediate the regeneration of damaged muscle tissue [9, 11]. The myoblasts undergo multiple rounds of cell division prior to terminal differentiation and formation of multinucleated myotubes by cell fusion.
Our knowledge of the molecular mechanisms regulating skeletal muscle development and differentiation has been dramatically accelerated following the discovery of the MyoD-family of master transcription factors that can convert a broad range of cells to a myogenic lineage [12]. MyoD belongs to the myogenic regulatory factors, a group of skeletal muscle-specific basic helix-loop-helix transcription factors that play essential roles in myogenic specification and differentiation during embryogenesis and satellite cell development [13–15]. Satellite cell-derived primary myoblasts isolated from adult mice lacking the MyoD gene (MyoD−/−) display accelerated growth rates as well as delayed terminal differentiation and muscle regeneration [13, 16–19]. MyoD−/− myoblasts have been shown to display characteristics that are more primitive than wild-type myoblasts and may represent an intermediate stage between stem cells and myogenic precursor cells [13, 19].
To address whether myoblasts, or fully committed myogenic cells, can be reprogrammed to iPSCs and whether MyoD inhibits the reprogramming process of iPSC generation, we examined the induction of iPSCs from myoblasts isolated from both wild-type and MyoD−/− mice by infection with retroviral vectors containing the Oct4, Sox2, cMyc, and Klf4 genes.
Materials and Methods
Mice
MyoD−/− mice [20] were kindly provided by Dr. Michael A. Rudnicki. B6;129S4-Pou5f1tm2(EGFP)Jae/J (Oct4EGFP/+ or Oct4-GFP) mice [21] were purchased from Jackson Laboratory (Bar Harbor, ME, www.jax.org). MyoD−/−:Oct4-GFP mice were established by crossing MyoD−/− mice with Oct4-GFP mice. MyoD−/−:Oct4-GFP mice and their litter mates MyoD+/+:Oct4-GFP mice were used for isolation of MyoD−/− and wild-type primary myoblasts, respectively. Myf5B6; 129S4-Myf5tm3(cre)sar/J (Myf5Cre/+ or Myf5-Cre) mice [22] and 129S-Gt(ROSA)26-Sortm1Sor/J (ROSA26 reporter or R26R) mice [23] were also purchased from Jackson Laboratory. Pax7creERPp/+ (Pax7-CreER) mice were obtained from Dr. Charles Keller [24]. Myf5-Cre:R26R mice or Pax7-CreER:R26R mice were established by crossing Myf5-Cre or Pax7-CreER and R26R mice, respectively. Genotyping to detect the mutated alleles of MyoD−/−, Oct4-GFP, Myf5-Cre, and R26R mice was performed by polymerase chain reaction (PCR) using primers described on the website of Jackson Laboratory. Genotyping for Pax7-CreER mice was described previously [24]. B6;129-Gt(ROSA)26-Sortm1(rtTA*M2)Jae, Col1altm2(tetO–Pou5f1)Jae/J (iOct4) mice were also purchased from Jackson Laboratory [25]. Nod:Scid immunodeficient mice and ICR (CD1) mice were purchased from Charles River (Wilmington, MA, www.criver.com). The Institutional Animal Care and Use Committee for the University of Minnesota approved all experimental protocols. Care was taken to minimize the number of animals used, as well as any pain and suffering.
Myoblast Isolation and Culture
Quiescent satellite cells were isolated from the hind limb skeletal muscle of 1- to 2-month-old adult mice after muscle digestion with collagenase type B and dispase II (Roche Applied Science, Basel, Switzerland, www.roche-applied-science.com) [26–28]. Fluorescence-activated cell sorting (FACS) was performed on a FACS Aria sorter equipped with triple lasers (BD Biosciences, San Jose, CA, www.bdbiosciences.com) as described previously [28, 29]. The following antibodies were used for FACS sorting: Integrin α7 (MBL International, Woburn, MA, www.mblintl.com) with Alexa 488-labeled anti-mouse IgG, biotin-labeled Integrin β1 with allophycocyanin (APC)-labeled avidin, phycoerythrin (PE)-labeled CD45, PE-labeled CD31, and PE-labeled Sca-1 (all from BD Biosciences). Mouse and rat normal IgG (BD Biosciences) were used in the control experiment. Sorting gates were strictly defined based on single antibody-stained control cells, as well as the forward scatter (FSC) and side scatter (SSC) patterns of satellite cells. After the FSC/SSC gating, the triple-negative cells for CD45-PE, CD31-PE, and Sca-1-PE were gated out. Finally, double-positive cells for Integrin α7-Alexa 488 and Integrin β1-APC were sorted to enrich for satellite cells. Sorted satellite cells were cultured on collagen-coated dishes in myoblast growth medium consisting of HAM’s F-10 medium (Invitrogen, Carlsbad, CA, www.invitrogen.com) supplemented with 20% fetal bovine serum (FBS; Fisher Scientific, Pittsburgh, PA, www.fishersci.com), penicillin/streptomycin (50 µg/ml; Invitrogen), and 5 ng/ml basic fibroblast growth factor (R&D Systems, Minneapolis, MN, www.rndsystems.com). Differentiation medium, Dulbecco’s modified eagle medium (DMEM; Invitrogen) supplemented with 5% horse serum (Invitrogen), and penicillin/ streptomycin (50 µg/ml; Invitrogen), was used to induce myogenic differentiation. For genetic lineage tracing, Pax7-CreER:R26R mice were given intraperitoneal (I.P.) injections of tamoxifen (Sigma Aldrich, St. Louis, MO, www.sigmaaldrich. com; 300 μl, 10 mg/ml, diluted in corn oil [Sigma-Aldrich]) daily for 3 days. Two days after the third injection, hind limbs were harvested for myoblast isolation [30]. For inducible Oct4 expression, doxycycline (Sigma-Aldrich) was administrated at a concentration of 2 µg/ml. To ensure that the muscle cells retained physiological characteristics, all experiments were performed using cells that had been passaged between 2 and 4 times.
Preparation for Murine Embryonic Fibroblasts, Tail Tip Fibroblasts, and Muscle-Derived Fibroblasts
For murine embryonic fibroblast (MEF) isolation, uteri isolated from 13.5-day-pregnant mice were dissected as described previously [1] and γ-irradiated for feeder cell preparation. To establish tail tip fibroblasts (TTFs), the tails from adult mice were used as described previously [1]. For isolation of muscle-derived fibroblasts, hind limb skeletal muscle derived from adult mice was digested with collagenase type B and dispase II (Roche Applied Science, Basel, Switzerland, www.roche-applied-science.com) as described previously [31]. Dissociated cells were cultured on tissue culture plates (noncollagen coated) to obtain MEFs, TTFs, and muscle-derived fibroblasts and were maintained in DMEM supplemented with 10% FBS.
Generation and Culture of iPSCs
The pMX-based retroviral vectors encoding the mouse cDNAs of Oct4, Sox2, cMyc, and Klf4 [1] (Addgene, Cambridge, MA, www.addgene.org) were kindly provided by Dr. Toshio Kitamura [32]. The pMX-based retroviral vectors for MyoD expression were constructed by insertion of a mouse MyoD cDNA fragment into the EcoRI site of the pMX vectors. These pMX-based retroviral vectors were separately cotransfected by packaging defective helper plasmids into PlatE cells [32] using Lipofectamine 2000 Reagent (Invitrogen). Forty-eight hours after transfection, virus supernatants were harvested. Low-passage (2–3 passages) myoblasts or fibroblasts were seeded at a density of 2–5 × 105 cells per 6-cm plate 24 hours prior to viral infection. Myoblasts or fibroblasts were incubated with supernatants of retroviral vectors containing the four factors (Oct4, Sox2, cMyc, and Klf4) or five factors (Oct4, Sox2, cMyc, Klf4, and MyoD) with 10 µg/ml of polybrene (Millipore, Bedford, MA, www.millipore.com) for exactly 3 hours. Then, the medium was replaced with myoblast growth medium and cultured for 3 days. The infected cells were replated onto MEF feeder cells and cultured on tissue culture dishes (BD Biosciences, San Jose, CA, www.bdbiosciences.com) coated with 0.1% gelatin (Sigma-Aldrich) in ESC medium containing Knockout DMEM (Invitrogen) supplemented with 20% Knockout serum replacement (Invitrogen), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich), 0.1 mM nonessential amino acids (Invitrogen), penicillin/streptomycin (50 µg/ml; Invitrogen), and 5,000 U/ml leukemia inhibitory factor (Millipore). Two weeks after subculture, Oct4-GFP+ and ESC-like colonies were subcloned onto MEF feeder cells as iPSC colonies. Generated iPSCs were maintained on the MEF feeder cells in ES medium.
Histochemical Staining and Immunostaining
The cultured cells and embryos were stained for β-galactosidase expression as described previously [33, 34]. Alkaline phosphatase (AP) staining was performed as described previously [35]. For immunostaining, cells were fixed with 2% paraformaldehyde (PFA) and incubated with primary antibodies followed by secondary antibodies as described previously [36]. The following primary antibodies were used: anti-Nanog (Bethyl Laboratories, Montgomery, TX, www.bethyl.com), anti-Oct4 (Santa Cruz, Santa Cruz, CA, www.scbt.com), anti-Sox2 (R&D Systems), anti-cMyc (Abcam), anti-Klf4 (Santa Cruz), anti-stage specific embryonic antigen-1 (SSEA-1) (Developmental Studies Hybridoma Bank, Iowa City, IA, dshb.biology.uiowa.edu), anti-MyoD (BD Pharmingen, San Diego, CA, www.bdbiosciences.com), anti-sarcomeric myosin heavy chain (MHC; Developmental Studies Hybridoma Bank), and anti-Pax7 (Developmental Studies Hybridoma Bank). The following secondary antibodies were used: Alexa 488-conjugated antimouse IgG, Alexa 594-conjugated antimouse IgM, and Alexa 594-conjugated anti-rabbit IgG (all from Invitrogen). 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) was used for counter staining of nuclei. Fluorescence images were captured by a DP-1 digital camera attached to a CKX31 inverted florescence microscope with ×20 and ×40 LUCPlanFLN objectives and a BX51 fluorescence microscope with ×20 and ×40 UPlanFLN objectives (all from Olympus America, Center Valley, PA, www.olympusamerica.com). Photoshop CS2 (Adobe Systems, San Jose, CA, www.adobe.com) was used for image processing.
Genomic PCR and RT-PCR
Preparation of genomic DNA was carried out as reported previously [21]. Total RNA was isolated from cells by TRIZOL (Invitrogen). Purified total RNA was reverse-transcribed (Roche Applied Science: Transcriptor First Strand cDNA Synthesis Kit) and 20–35 cycles were performed (Eppendorf Thermal Cycler, Hauppauge, NY, www.eppendorf.com) using the gene-specific primer pairs described in Supporting Information Fig. S1. Optimal PCR cycles for each pair were determined by several different amplifications of the PCR products.
Embryoid Body Formation
The formation of embryoid bodies was carried out as described previously [37]. Briefly, undifferentiated iPSCs were cultured in hanging drops for 3 days at a density of 1,600 cells per 20 microliter of differentiation medium, which consisted of DMEM supplemented with penicillin/streptomycin (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), 0.1 mM 2-mercaptonethanol (Sigma-Aldrich), 5% horse serum (Invitrogen), and 10% FBS (Fisher Scientific). The embryoid bodies were transferred to suspension cultures for an additional 3 days. Finally, the embryoid bodies were plated into differentiation medium on 48-well plates coated with Matrigel (all from BD Biosciences). The medium was changed every 3 days.
Teratoma Formation
Cultured iPSCs (1 × 106 cells) on MEF feeder cells were subcutaneously injected into 2- to 3-month-old Nod:Scid immunodeficient mice. Three weeks after injection, teratomas were fixed with 4% PFA and frozen in O.C.T.-compound (Fisher Scientific). Cryosections were stained with H&E solution (Sigma-Aldrich).
Aggregation of iPSCs with Zona-Free Embryos
Eight-cell stage embryos were flushed out from 2.5 dpc pregnant CD1 female mice. Clumps of iPSCs (2–20 cells per clump) were selected after a brief period of trypsinization. These selected iPSC clumps and zona-free morula embryos were transferred into micro drops of KSOM medium (Millipore) supplemented with 10% FBS under mineral oil (Fisher Scientific). After overnight culture at 37°C in 5% CO2 incubator, these aggregated blastocysts were transferred into uterine horns of 2.5 dpc pseudopregnant recipient CD1 female mice or into oviducts of 0.5 dpc pseudopregnant recipient CD1 female mice [38].
Luciferase Reporter Assays
The firefly luciferase reporter genes 4R-SV-luciferase [39, 40], proximal regulatory region (PRR)-luciferase, distal regulatory region (DRR)-PRR-luciferase [33], and F3-2.5-luciferase [41], kindly provided by Dr. David J. Goldhamer, were used in this study. pRL-TK (Promega, Madison, WI, www.promega.com) was used as an internal control. Myoblasts were transfected with expression vectors for MyoD (pMX-MyoD), Oct4 (pMX-Oct4), empty vectors (pMXs), and the luciferase reporter genes using Lipofectamine 2000 (Invitrogen). Cells were harvested at 48 hours after transfection. Luciferase activity was measured with a plate reader (LD400; Beckman Coulter, Brea, CA, www.beck-mancoulter.com) using a dual luciferase reporter assay system (Promega).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was performed on iOct4 myoblast cultures using a ChIP assay kit (Millipore) and the manufacturer’s instructions [42]. Myoblasts infected with empty control pMXs or pMX-MyoD retrovirus vectors were harvested at 24 hours after treatment with or without doxycycline. Immunoprecipitation was done overnight at 4°C with control rabbit IgG (BD Biosciences), anti-MyoD (Santa Cruz), and anti-Oct4 antibodies (Santa Cruz). The PCR primers, core enhancer region (CER)-IP, DRR-IP, PRR-IP, and immunoglobulin heavy chain (IgH)-IP, used for this assay are described in Supporting Information Fig. S1.
Statistics
All data are expressed as means ± SEM. Statistical significance between groups was analyzed by Student’s t test. At least three independent experiments were performed and values denote mean ± SEM (n = 3). Asterisk and double asterisk indicate experimental pairs where differences between the compared values were statistically significant (*, p < .05 and **, p < .01, respectively).
Results
Generation of Myoblast-Derived iPSCs
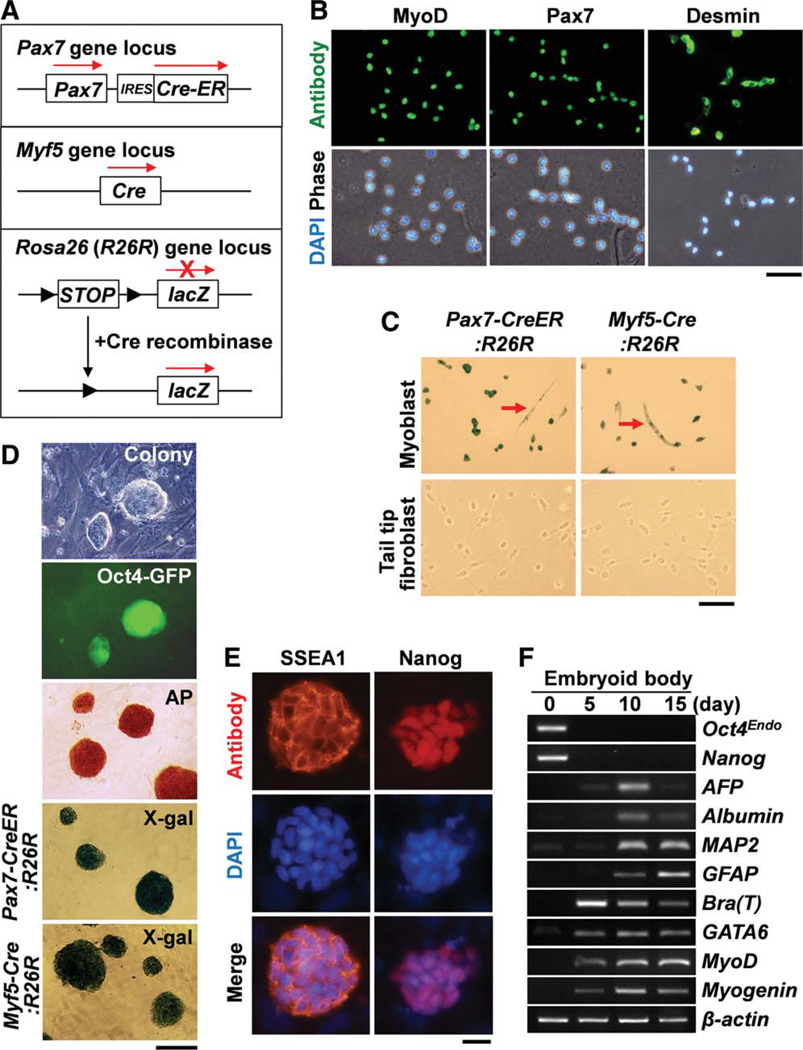
To address whether myoblasts or fully committed myogenic cells can be reprogrammed to iPSCs, we examined the induction of iPSCs from primary myoblasts isolated from adult mice. Initially, we isolated and expanded primary myoblasts from adult mice (1–2 months age) carrying Oct4-GFP transgene [21] by FACS and expanded them as described previously [43]. In addition, to confirm the myogenic origin of the myoblast-derived iPSCs, we used genetic myogenic lineage tracing. We crossed Pax7-CreER or Myf5-Cre mice with ROSA26R (R26R) reporter mice [23] to generate Pax7-CreER:R26R or Myf5-Cre:R26R compound mice (Fig. 1A). Pax7-CreER mice carry the CreER gene introduced into the Pax7 gene locus [24]. Myf5-Cre mice carry the Cre recombinase gene introduced into the Myf5 gene locus [22]. Tamoxifen was administered to induce gene recombination in Pax7-CreER:R26R mice. Following tamoxifen treatment Pax7-CreER:R26R mice and Myf5-Cre:R26R mice permanently express lacZ in Myf5+ and Pax7+ myogenic cells and their progeny [23, 44]. Immunofluorescent detection in the isolated myoblasts with anti-Pax7, anti-MyoD, and anti-desmin antibodies clearly showed that more than 98% of isolated cells were Pax7+, MyoD+, and/or desmin+ myoblasts (Fig. 1B). The myoblasts promptly differentiated into MHC+ multinuclear myotubes in high cell density or under differentiation medium (data not shown). Primary myoblasts and fibroblasts were purified from Pax7-CreER:R26R and Myf5-Cre:R26R mice. X-gal staining clearly showed lacZ expression in the myoblasts but not in the fibroblasts isolated from Pax7-CreER:R26R and Myf5-Cre:R26R mice (Fig. 1C).
Figure 1.
Generation of induced pluripotent stem cells (iPSCs) from primary myoblasts. (A): Schematic view of genetic lineage tracing strategy for Pax7-CreER:R26R and Myf5-Cre:R26R compound mice. (B): Panels show fluorescence views of myoblasts after antibody staining for MyoD, Pax7, and Desmin. Scale bar = 50 µm. (C): Culture of myoblasts from Pax7-CreER:R26R and Myf5-Cre:R26R compound mice shows lacZ+ permanently labeled cells including both myoblasts and multinucleated myotubes (arrows). By contrast, tail tip fibroblasts did not express lacZ. Scale bar = 100 µm. (D): ESC-like colonies were generated from myoblasts from Oct4-GFP mice. These colonies were positive for GFP and AP staining. Colonies generated from myoblasts isolated from Pax7-CreER:R26R and Myf5-Cre:R26R were positive for X-gal staining. Scale bar = 200 µm. (E): The myoblast-derived colonies expressed SSEA1 and Nanog. Scale bar = 20 µm. (F): Reverse transcription-polymerase chain reaction experiments show that embryonic bodies generated from myoblast-derived iPSCs expressed differentiation markers for three germ layers. AFP and Bra (T) denote α-fetoprotein and brachyury (T) genes, respectively. β-actin was monitored as a loading control. Nuclei were counterstained with DAPI (blue). Abbreviations: AP, alkaline phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; SSEA1, stage specific embryonic antigen-1. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Isolated Pax7-CreER:R26R, Myf5-Cre:R26R and Oct4-GFP [21] myoblasts were infected with pMX-retroviral vectors carrying four transcription factors (Oct4, Sox2, cMyc, and Klf4), as described previously [1]. As a control, musclederived fibroblasts and tail tip fibroblasts were also used for infection with retroviral vectors carrying these four transcription factors. ESC-like colonies were normally observed by day 10–14 after infection for these myoblasts (Fig. 1D) and fibroblasts (data not shown). ESC-like colonies were picked up and expanded to obtain myoblast-derived iPSCs. We observed that these myoblast-derived iPSC colonies generated from Oct4-GFP mice clearly expressed green fluorescent protein (GFP) (Fig. 1D). ESC-like iPSC colonies generated from both Pax7-CreER:R26R and Myf5-Cre:R26R myoblasts showed that myoblast-derived iPSC colonies from both compound mice expressed the lacZ lineage tracer (Fig. 1D), confirming that iPSCs generated from primary myoblasts had a myogenic origin. In agreement with previous findings [1], these iPSC colonies strongly expressed alkaline phosphatase and SSEA1 at the cell membrane and Nanog in the nucleus (Fig. 1D, 1E). In addition, RT-PCR experiments showed that all nine independent myoblast-derived iPSC colonies expressed ESC-specific genes such as endogenous Oct4 and Nanog at a similar level as ESCs (Supporting Information Figs. S2A and S3A). Furthermore, expression of the myogenic markers MyoD and Pax7 was lost, both of which were expressed in the parent myoblasts (Supporting Information Figs. S2A and S3A). Genomic PCR showed that these nine iPSC colonies retained infected retroviral vector DNA for Oct4, Sox2, cMyc, and Klf4 (Supporting Information Fig. S2B). By contrast, exogenous expression from these retroviral vectors was markedly reduced (Supporting Information Fig. S2C). Thus, retroviral vector DNA silencing occurred during iPSC generation, as reported previously [1]. The efficiency of iPSC generation from myoblasts was 0.0019% ± 0.0005% (n = 4). By contrast, the efficiency of iPSC generation from muscle-derived fibroblasts was 0.0047% ± 0.0009% (n = 3). These efficiencies are comparable with the efficiencies associated with iPSCs derived from other murine cell sources [1, 45, 46]. These results clearly indicated that myoblasts were successfully reprogrammed into iPSCs.
Multilineage Differentiation Potential of Myoblast-Derived iPSCs
To determine the differentiation potential of the myoblastderived iPSCs, we induced differentiation using embryoid body formation method over 15 days. RT-PCR for differentiation-specific gene expression showed that marker genes for endoderm, ectoderm, and mesoderm were detected during embryoid body formation (Fig. 1F).
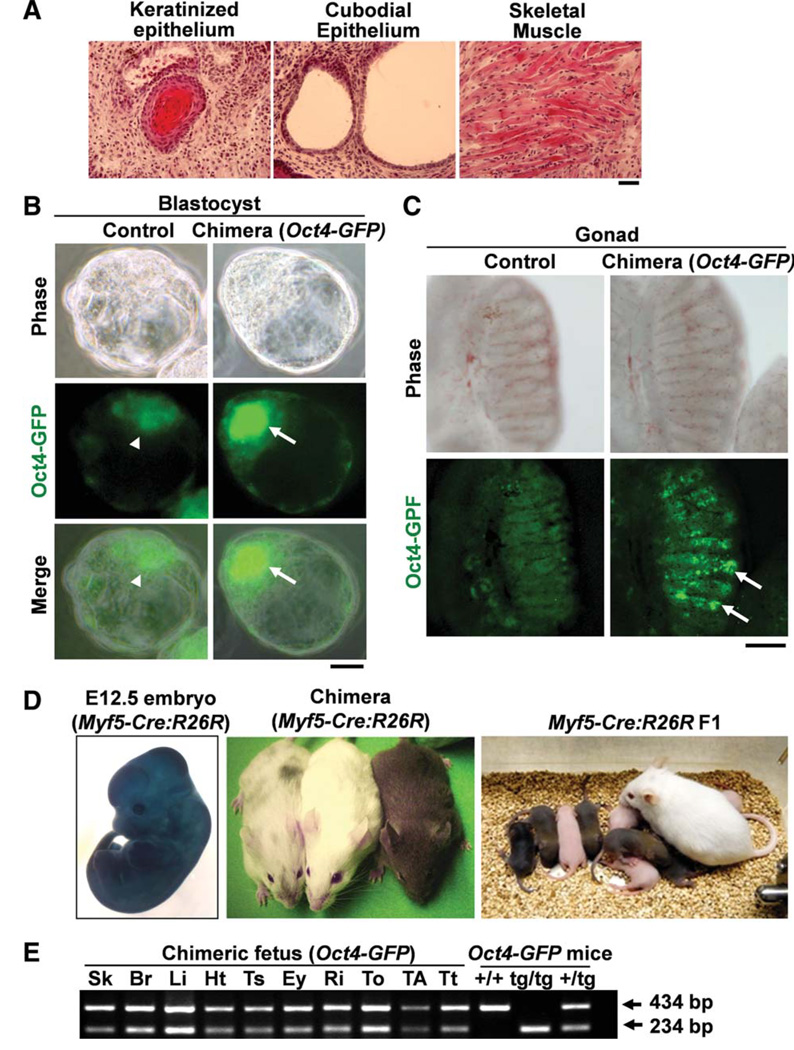
Next, to examine the developmental potential of the myoblast-derived iPSCs in vivo, we subcutaneously injected the iPSCs into Nod:Scid immunodeficient mice. Three weeks after injection, the iPSCs produced teratomas containing primitive tissues originating from all three germ layers including keratinized epithelium (ectoderm), cuboidal epithelium (endoderm), and skeletal muscle (mesoderm; Fig. 2A). Taken together, both in vitro embryoid body culture and in vivo teratoma formation indicated that myoblast-derived iPSCs are able to differentiate into all three germ layers.
Figure 2.
Developmental potential of myoblast-derived induced pluripotent stem cells (iPSCs). (A): Sections of teratoma formation after subcutaneous injection of myoblast-derived iPSCs into Nod:Scid mice. H&E staining shows iPSC contribution to all three germ layers, keratinized epithelium (ectoderm), cuboidal structures (endoderm), and skeletal muscle (mesoderm). Scale bar = 50 µm. (B): Myoblast-derived iPSCs generated from Oct4-GFP mice efficiently incorporated into GFP+ inner cell mass (arrows) of blastocysts after aggregation with CD1 mouse embryos. Non-aggregated blastocysts show auto fluorescence background (arrowheads). Scale bar = 25 µm. (C): Myoblast-derived iPSCs generated from Oct4-GFP mice contributed to the germ line cells (arrows), which expressed Oct4-GFP in the gonad of 13.5 dpc chimeric embryos. Control nonchimeric gonad shows no GFP staining. Scale bar = 100 µm. (D): X-gal staining for chimeric embryo (12.5 dpc) after aggregation with myoblastderived iPSCs generated from Myf5-Cre:R26R mice (left panel). Chimeric mice generated from Myf5-Cre:R26R myoblast-derived iPSCs showed mixed coat color at 3 weeks age (middle panel). The mice indicate medium contributed chimera (mixed color), CD1 nursing mother (white color), and high-contributed chimera (agouti color). The agouti color pups indicate germ line transmission of Myf5-Cre:R26R myoblast-derived iPSCs (right panel). (E): Genomic polymerase chain reaction (PCR) confirmed the chimerism of fetal mice generated from Oct4-GFP myoblastderived iPSCs. The chimeric fetal tissues contained both wild-type (434 bp) and Oct4-GFP transgene (234 bp) alleles: Sk, Br, Li, Ht, Ts, Ey, Ri, To, TA, Tt. PCR results for control DNAs prepared from tail tips of wild-type (þ/þ), homozygous (tg/tg), and heterozygous (+/tg) Oct4-GFP mice are also shown. Abbreviations: Br, brain; Ey, eye; GFP, green fluorescent protein; Ht, heart; Li, liver; Ri, rib; Sk, skin; TA, tibialis anterior muscle; Ts, testis; To, tongue muscle; Tt, tail tip. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Developmental Potential of Myoblast-Derived iPSCs
To examine pluripotency of the myoblast-derived iPSCs, we aggregated the iPSCs with mouse embryos (CD1 strain) at the eight-cell stage. iPSCs derived from Oct4-GFP myoblasts were able to integrate into the GFP+ inner cell mass of the blastocysts (Fig. 2B). In addition, the presence of GFP+ cells in the gonads of 13.5 dpc embryos (Fig. 2C) indicated that myoblast-derived iPSCs contributed to germ cells, demonstrating their germ cell differentiation potential. We also aggregated iPSCs derived from Myf5-Cre:R26R myoblasts with CD1 embryos. We detected widespread lacZ expression in the whole body of 12.5 dpc chimeric embryos (Fig. 2D). In addition, genomic DNA PCR confirmed that these iPSCs contributed to a wide variety of fetal tissues originating from all three germ layers (Fig. 2E). We also noticed that the iPSCs generated from both Oct4-GFP (data not shown) and Myf5-Cre:R26R (Fig. 2D) myoblasts were capable of contributing to the formation of chimeric mice and germ line transmitted offspring. These data strongly indicate that the iPSCs reprogrammed from myoblasts possess phenotypical and functional characteristics of pluripotent stem cells.
MyoD Suppresses Reprogramming of Myoblasts
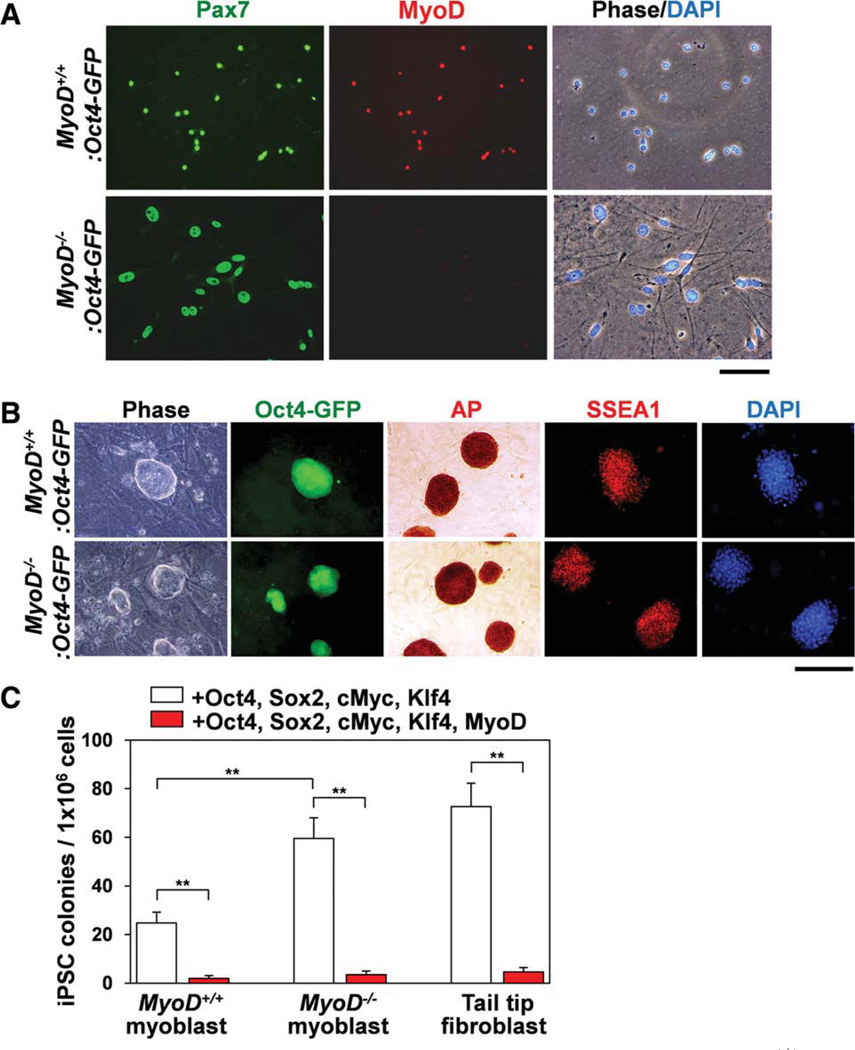
As described earlier, the efficiency of myoblast reprogramming to iPSCs was relatively low compared with the iPSCs generated from fibroblasts. We hypothesized that myogenic differentiation status may inhibit the molecular process of iPSC reprogramming by the four factors. Previous studies indicated that MyoD is a potent myogenic master transcription factor that converts many nonmyogenic cells to a myogenic lineage [12, 47]. To examine whether MyoD expression affected the reprogramming efficiency of myoblasts, we created MyoD+/−:Oct4-GFP mice by crossing mice carrying MyoD null alleles (MyoD−/−) and Oct4-GFP mice. Next, MyoD+/−:Oct4-GFP mice were crossed with each other to generate MyoD null (MyoD :Oct4-GFP) and the control MyoD+/+:Oct4-GFP mice. Isolated MyoD−/−:Oct4-GFP myoblasts displayed elongated shapes compared with the rounded control MyoD+/+:Oct4-GFP myoblasts (Fig. 3A) [13]. These MyoD−/−:Oct4-GFP myoblasts clearly expressed the myogenic marker Pax7 but not MyoD. Both MyoD+/+:Oct4-GFP and MyoD :Oct4-GFP myoblasts were used to generate iPSCs by infection with retroviral vectors (Oct4, Sox2, cMyc, and Klf4). Two weeks after infection, both MyoD+/+:Oct4-GFP and MyoD−/−:Oct4-GFP myoblasts generated ESC-like colonies that expressed GFP, alkaline phosphatase, and SSEA1 proteins, along with ESC-specific proteins such as endogenous Oct4 and Nanog (Fig. 3B and Supporting Information Fig. S3B). Interestingly, the efficiency of iPSC generation from MyoD :Oct4-GFP myoblasts was significantly higher than that of MyoD+/+:Oct4-GFP myoblasts (0.0060% ± 0.0009% [n = 4] vs. 0.0025% ± 0.0004% [n = 5], p < .01) (Fig. 3C). The efficiency for the MyoD :Oct4-GFP myoblast-derived iPSCs was comparable with that of MyoD+/+:Oct4-GFP fibroblasts (0.0060% ± 0.0009% [n = 4] vs. 0.0073% ± 0.0010% [n = 3]). The infection efficiencies of retroviruses were not different between these three cell types (Supporting Information Fig. S4A). These results suggested that MyoD could be a negative regulator of iPSC generation.
Figure 3.
MyoD suppresses myoblast-derived iPSC formation. (A): Cultured myoblasts isolated from control (MyoD+/+:Oct4-GFP) and MyoD null (MyoD :Oct4-GFP) mice were immunostained for Pax7 and MyoD. Scale bar = 100 µm. (B): iPSC colonies were generated from myoblasts isolated from control MyoD+/+:Oct4-GFP and MyoD :Oct4-GFP mice. These iPSC colonies were positive for Oct4-GFP, AP staining, and SSEA1. Scale bar = 200 µm. (C): The number of generated iPSC colonies was counted after infection with four factors (Oct4, Sox2, cMyc, and Klf4) or five factors (Oct4, Sox2, cMyc, Klf4, and MyoD). iPSC colonies generated from MyoD−/−:Oct4-GFP myoblasts or MyoD+/+:Oct4-GFP tail tip fibroblasts show higher numbers compared with iPSC colonies generated from the control MyoD+/+:Oct4-GFP myoblasts. Forced expression of MyoD significantly diminished iPSC colony formation for all three cell types. Nuclei were counterstained with DAPI (blue) in (A) and (B). Abbreviations: AP, alkaline phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; iPSC, induced pluripotent stem cell; SSEA1, stage specific embryonic antigen-1. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
To investigate whether forced expression of MyoD can inhibit iPSC generation, we infected myoblasts with a retroviral vector for MyoD expression together with the four factors (Oct4, Sox2, cMyc, and Klf4). Two weeks after infection, we counted the number of GFP+ ESC-like colonies generated from control MyoD+/+:Oct4-GFP myoblasts, MyoD :Oct4-GFP myoblasts, and MyoD+/+:Oct4-GFP fibroblasts following infection with or without MyoD retroviral vector (Fig. 3C). Clearly, forced expression of MyoD significantly reduced efficiency of iPSC generation in these cell types. These results strongly suggest that MyoD expression inhibits the myoblast reprogramming process and the downregulation or inactivation of MyoD may be essential for the initial step in reprogramming myoblasts to iPSCs.
Oct4 Suppresses MyoD Gene Expression in Myoblasts
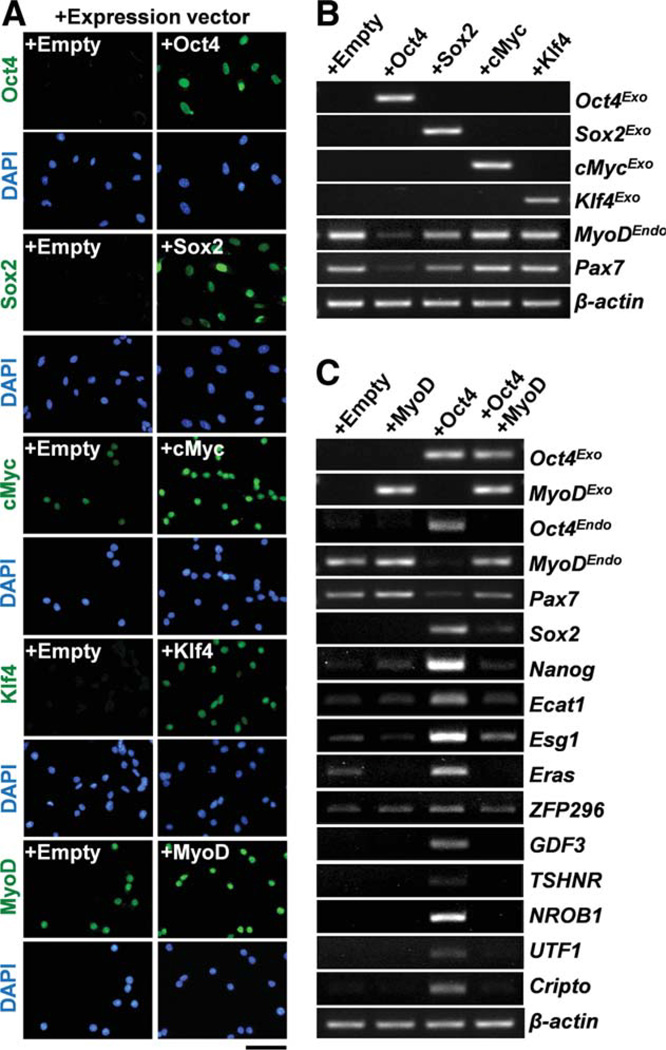
To examine which of the four factors may alter MyoD gene expression in myoblasts, we infected wild-type myoblasts with a single retroviral vector expressing Oct4, Sox2, cMyc, or Klf4, instead of all four factors together. Two days after infection, immunostaining, and RT-PCR experiments clearly showed that most of the myoblasts expressed the transgenes and their proteins in their nuclei (Fig. 4A, 4B). Interestingly, ectopic expression of Oct4 significantly reduced MyoD and Pax7 gene expression in the myoblasts (Fig. 4B). Ectopic expression of Sox2 also reduced the expression of MyoD and Pax7 to some extent. cMyc or Klf4 did not downregulate MyoD or Pax7. These results strongly suggest that Oct4 is the primary factor in the downregulation of MyoD gene expression during iPSC generation.
Figure 4.
Expression of Oct4 suppresses MyoD expression and induces ESC-specific gene expression. (A): Retroviruses expressing Oct4, Sox2, cMyc, Klf4, or MyoD were infected to primary myoblasts. Trans-gene expression was detected by antibodies for Oct4, Sox2, cMyc, Klf4, and MyoD (second column). Empty denotes myoblasts infected with an empty expression vector as a control (first column). Scale bar = 50 µm. Nuclei were counterstained with DAPI (blue). (B): Reverse transcription-polymerase chain reaction (RT-PCR) was performed for RNA from myoblasts at 48 hours after infection with retroviral expression vectors for Oct4, Sox2, cMyc, or Klf4. Exogenous Oct4 (Oct4Exo, Sox2 (Sox2-Exo, cMyc (cMycExo, or Klf4 (Klf4Exo) expression was detected. Ectopic expression of Oct4 or Sox2 but not cMyc or Klf4 suppressed MyoDEndo and Pax7 expressions. (C): RT-PCR was performed for RNA from myoblasts at 48 hours after infection with retroviral expression vectors for Oct4 or MyoD, MyoD+Oct4. Ectopic expression of Oct4 induced many ESC-specific genes such as endogenous Oct4 (Oct4Endo ; third lane) compared with the control empty vector (first lane) or MyoD (second lane). These Oct4-induced gene expressions were abrogated by forced expression of MyoD (fourth lane), β-actin was monitored as a loading control in (B) and (C). Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Therefore, we further characterized Oct4-expressing myoblasts by RT-PCR. Importantly, ectopic expression of Oct4 upregulated many ESC-specific genes including endogenous Oct4, Sox2, and Nanog (Fig. 4C), suggesting that Oct4 alone can partially reprogram myoblasts. Similar upregulation of ESC-specific genes was observed in Oct4-expressing fibro-blasts (Supporting Information Fig. S4B). However, infection of retroviral Oct4 expression vector alone could not induce iPSCs from myoblasts and fibroblasts (unpublished observation). Forced expression of MyoD led to the abrogation of the ESC-specific gene expression that was induced by ectopic expression of Oct4. By contrast, myogenic marker genes such as endogenous MyoD and Pax7 were maintained in these cells (Fig. 4C), indicating that MyoD dominantly regulates myogenic status even though Oct4 is expressed.
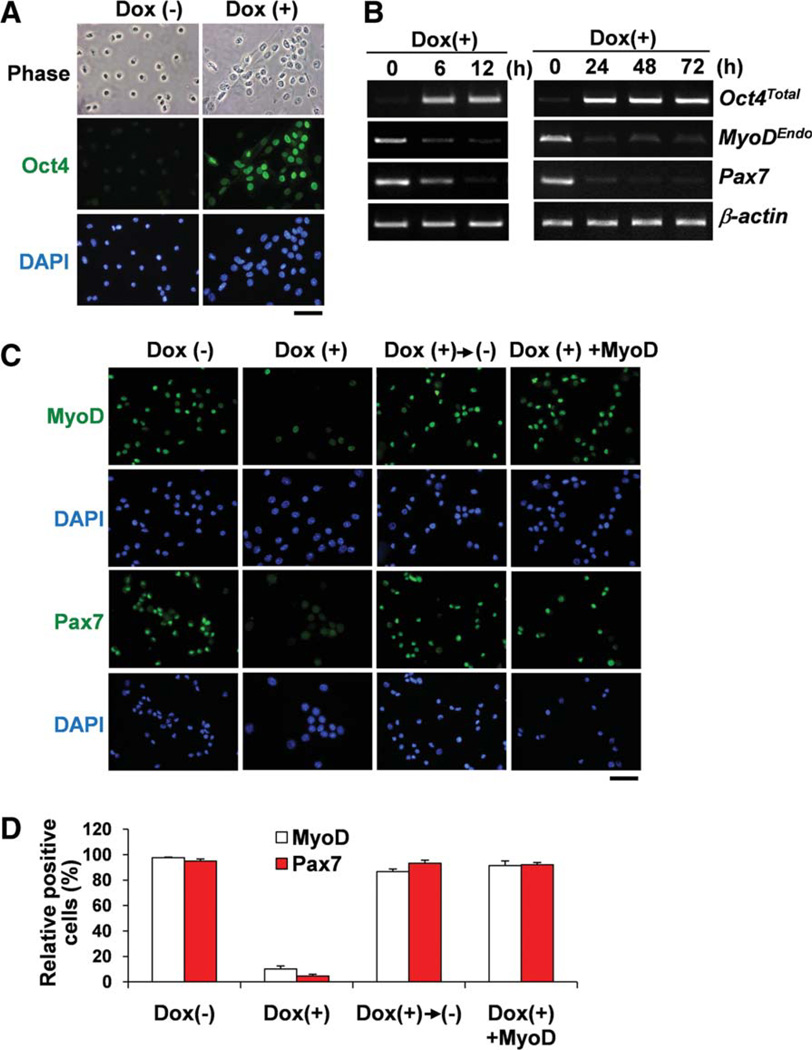
To further characterize the effects of ectopic expression of Oct4 in myoblasts, we used a doxycycline-dependent Oct4 gene expression system (Tet-ON system) [25]. Primary myoblasts and tail tip fibroblasts were isolated from adult induced Oct4 (iOct4) mice. Immunostaining and RT-PCR experiments clearly showed that the Oct4 expression was quickly induced by doxycycline administration within 6 hours in iOct4 myoblasts (Fig. 5A, 5B). Similar to myoblasts and fibroblasts infected with retrovirus Oct4 expression vector, both iOct4 myoblasts and iOct4 fibroblasts also showed downregulation of MyoD gene expression and upregulation of ESC-specific gene expression within 48 hours after treatment with doxycycline (Supporting Information Fig. S4B). Interestingly, MyoD gene expression was quickly downregulated by doxycycline-induced Oct4 expression within 6 hours (Fig. 5B), indicating that MyoD gene expression may be directly suppressed by Oct4. Oct4 also suppressed Pax7 gene expression but the downregulation was not as drastic compared with MyoD. Immunostaining also showed that both MyoD and Pax7 proteins were downregulated by ectopic expression of Oct4 (Fig. 5C, 5D). However, removal of doxycycline reversed this downregulation leading to a quick recovery of MyoD and Pax7 expression. Forced expression of MyoD could overcome Oct4-dependent downregulation of MyoD and Pax7 (Fig. 5C, 5D).
Figure 5.
Inducible Oct4 suppresses MyoD expression in myoblasts. (A): Myoblasts isolated from iOct4 mice carry inducible Oct4 gene expression system shows Oct4 induction by 24 hours after treatment with doxycycline. No Oct4 expression was detected in control myoblasts without doxycycline. Scale bar = 50 µm. (B): Reverse transcription-polymerase chain reaction was performed for myoblasts after treatment with doxycycline for 72 hours. MyoDEndo downregulation occurred within 6 hours along with Oct4 induction after treatment with doxycycline. Pax7 downregulation was slightly slower than MyoD. β-actin was monitored as a loading control. (C): Immunostaining confirmed downregulation of MyoD and Pax7 in myoblasts by 24 hours after treatment with doxycycline (second column). Without doxycycline, myoblasts expressed MyoD and Pax7 (first column). Third column shows that doxycycline-treated myoblasts were incubated in culture medium without doxycycline for an additional 24 hours. These myoblasts reactivated MyoD and Pax7 expressions. Fourth column shows that forced MyoD-expressing myoblasts were treated with doxycycline. Pax7 expression was not downregulated in the myoblasts. Scale bar = 100 µm. (D): Relative MyoD or Pax7+ myoblasts shown in (C). Nuclei were counterstained with DAPI (blue) in (A) and (C). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; Dox(+), treatment with doxycycline; Dox(−), treatment without doxycycline. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Ectopic Expression of Oct4 Downregulated MyoD Expression in iOct4 Myoblasts
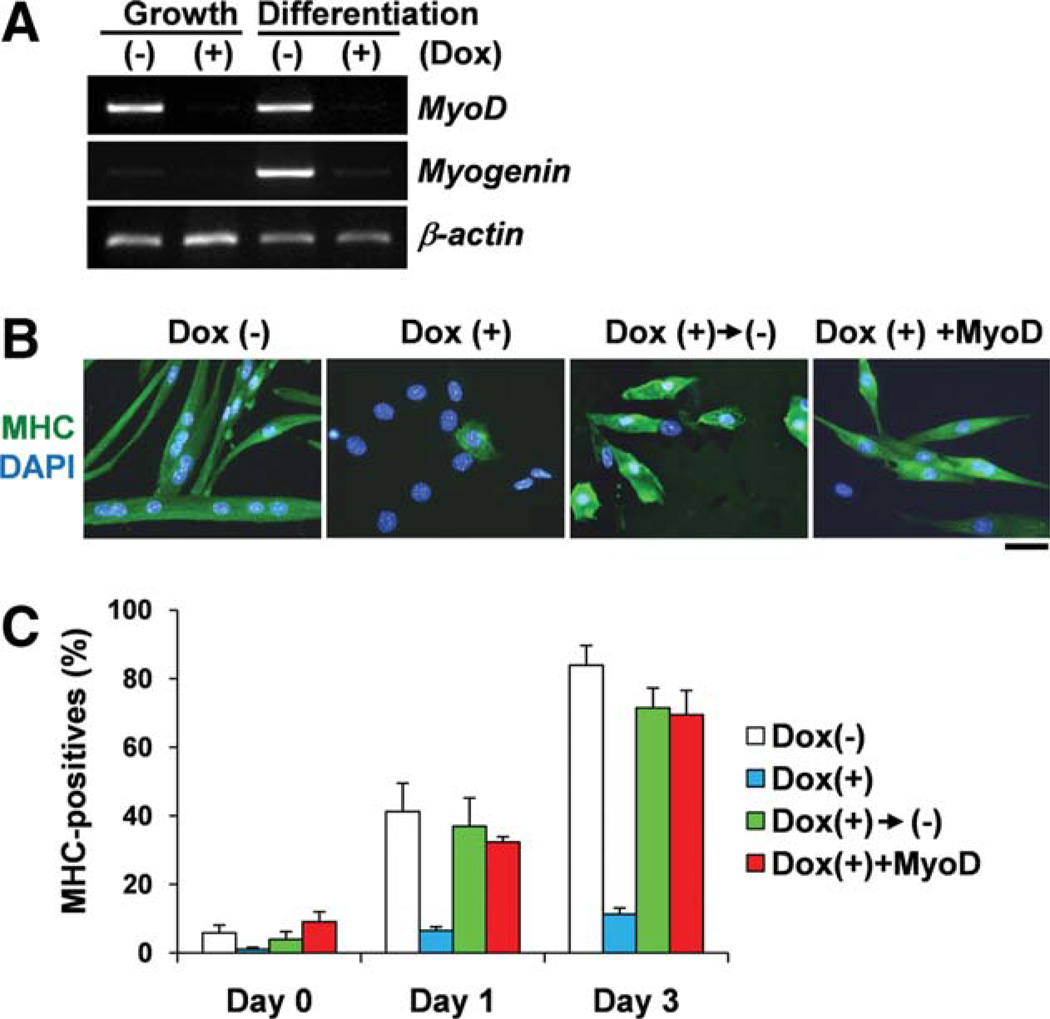
Therefore, we examined terminal differentiation of Oct4-expressing myoblasts after serum withdrawal. Clearly, MyoD and myogenin expression was downregulated by ectopic expression of Oct4 in differentiated muscle cells. In addition, MHC+ myogenic differentiation was markedly suppressed by ectopic expression of Oct4 (Fig. 6A–6C). When Oct4 expression was eliminated, myogenic differentiation was recovered. In addition, forced expression of MyoD could induce myogenic differentiation even though Oct4 was expressed (Fig. 6A–6C). Taken together, these results strongly suggest that Oct4 is a suppressor for MyoD gene expression and myogenic differentiation. In addition, forced expression of MyoD can overcome Oct4-mediated inhibition of myogenic differentiation.
Figure 6.
Expression of Oct4 suppresses myogenic differentiation. (A): Reverse transcription-polymerase chain reaction for iOct4 myoblast culture demonstrates that treatment with doxycycline suppressed MyoD gene expression in growth medium and both MyoD and myogenin gene expression at day 3 differentiation medium (first column) compared with the control cells. (B): Treatment with doxycycline suppressed MHC+ myogenic differentiation at day 3 differentiation medium (first column) compared with the control cells, which shows MHC+ multinucleated myotubes (second column). Doxycycline-treated myoblasts were incubated in differentiation medium without doxycycline for an additional 3 days. These cells underwent differentiation to MHC+ myocytes (third column). Forced MyoD-expressing myoblasts underwent differentiation to MHC+ myocytes even after treatment with doxycycline (fourth column). Scale bar = 50 µm. Nuclei were counterstained with DAPI (blue). (C): Relative MHC+ myocytes and myotubes shown in (B). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; Dox(+), treatment with doxycycline; Dox(−), treatment without doxycycline; MHC, myosin heavy chain. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
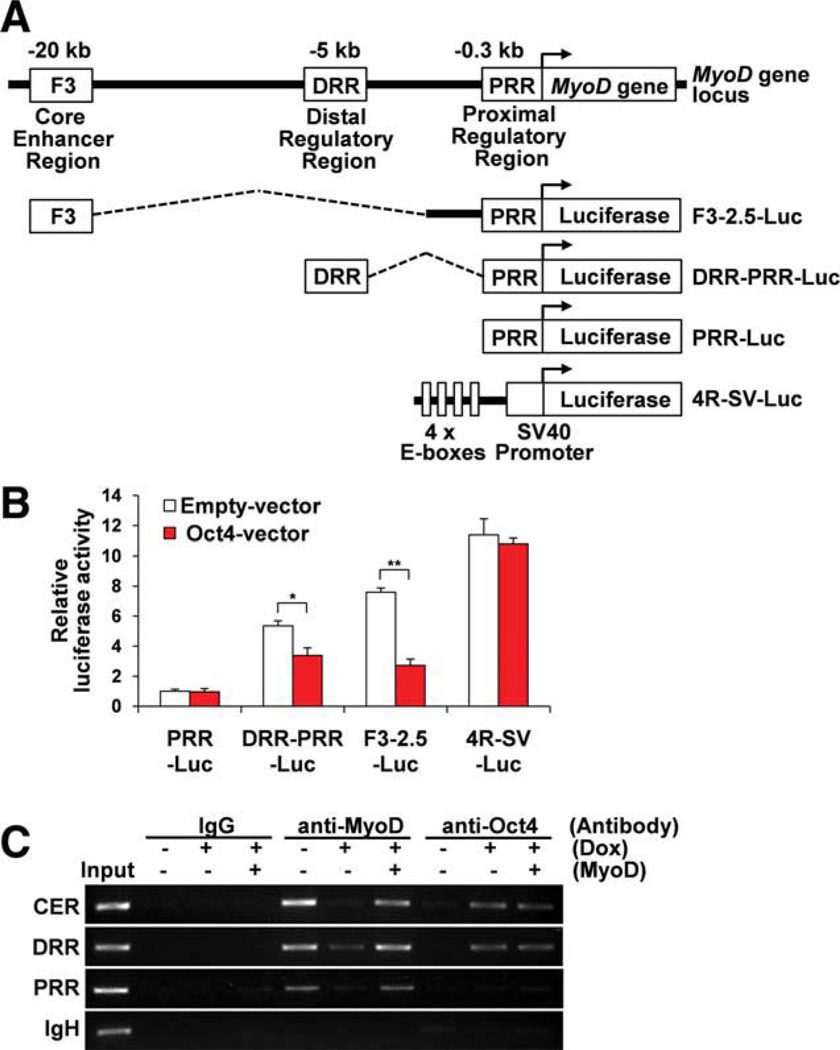
Next, to determine whether Oct4 directly downregulates MyoD gene expression through suppression of MyoD gene enhancer activities, we tested the luciferase activities of MyoD enhancer-driven luciferase and 4R-SV-luciferase reporter genes. MyoD contains three transcriptional regulatory elements located upstream of the MyoD gene, termed the PRR, the DRR [33], and the F3 region containing the CER (Fig. 7A) [41]. These three regions are essential for proper MyoD gene expression during muscle development [33, 41, 48, 49]. The 4R-SV-luciferase reporter gene contains 4 E-boxes, MyoD-binding sites, derived from the muscle creatine kinase (MCK) gene enhancer region [39, 40]. Cotransfection experiments demonstrated that Oct4 suppressed DRR-PRR-luciferase and F3-2.5-luciferase activities but did not suppress PRR-luciferase or 4R-SV-luciferase activities in myoblasts (Fig. 7B). The failed suppression of 4R-SV-luciferase activity by Oct4 indicated that Oct4 does not inhibit the MyoD-mediated transcriptional regulation by direct suppression of MyoD protein function or competition for the target DNA binding. Taken together, Oct4 is able to downregulate MyoD gene expression by suppression of MyoD enhancer (DRR and CER) activities possibly by directly binding to these regions [50]. Next, we examined whether Oct4 binds directly to the upstream enhancer region sites of MyoD. A ChIP assay clearly demonstrated that Oct4 binds to the upstream enhancer region sites containing the CER or DRR but not to the sites containing the PRR. MyoD binding to the upstream region sites containing the CER, DRR, or PRR was also detected as described in previous work using ChIP-Seq [50]. MyoD binding to the upstream region sites was abolished by Oct4 expression due to downregulation of MyoD expression (Fig. 7C). However, in forced MyoD-expressing myoblasts, MyoD was still able to bind to the upstream region sites even when ectopic Oct4 expression was induced, indicating that Oct4 did not prevent the binding of MyoD to the upstream region sites. Thus, Oct4 may act as a repressor for MyoD gene expression through direct binding to the MyoD upstream enhancer region sites, possibly interacting with other negative regulator(s). Therefore, during myoblast-derived iPSC generation, MyoD gene expression may be directly suppressed by Oct4 as an initial step of the reprogramming process.
Figure 7.
Oct4 suppresses MyoD gene transcription through binding to MyoD upstream enhancer region sites. (A): Schematic view for MyoD gene enhancer regions, MyoD-luciferase reporter genes and 4R-SV-lucif-erase reporter gene. (B): Luciferase activity was assessed after cotransfection with MyoD-luciferase or 4R-SV-luciferase reporter genes and empty vector or Oct4 expression vector into myoblasts. Oct4 expression suppressed DRR-PRR-luciferase and F3-2.5-luciferase activities but not suppressed PRR-luciferase and 4R-SV-luciferase activities. (C): Chromatin immunoprecipitation assay with antibodies against anti-MyoD and anti-Oct4 on MyoD upstream enhancer regions containing CER, DRR, and PRR or immunoglobulin heavy chain (IgH) enhancer region was performed for DNAs isolated from iOct4 myoblasts treated with or without doxycycline. MyoD retrovirus was also infected to doxycycline-treated myoblasts. Normal rabbit IgG was used for the negative control experiments. Input denotes each PCR product from naked myoblast genomic DNA. Abbreviations: CER, core enhancer region; Dox, doxycycline; DRR, distal regulatory region; PRR, proximal regulatory region. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
The first important finding in this study was that iPSCs can be successfully derived from MyoD-positive primary myoblasts, fully committed myogenic cells, by retroviral infection of four factors (Oct4, Sox2, cMyc, and Klf4). These myoblast-derived iPSCs display similar characteristics to ESCs. We detected ESC-specific gene expression and proteins in myoblast-derived iPSCs. In addition, myoblast-derived iPSCs formed embryoid bodies and teratomas, and contributed to the production of chimeric mice and the offspring, demonstrating their potential to develop into all three germ layers (ectoderm, endoderm, and mesoderm) as well as into germ cells. These results strongly indicated that the myoblast-derived iPSCs possess pluripotent differentiation potential, consistent with previous studies that cell reprogramming is not restricted to specific cell types [1, 2, 4, 5, 51, 52]. Recent work demonstrates that freshly isolated skeletal muscle precursors (SMPs) can be reprogrammed to iPSCs using doxycycline-inducible lentiviral vectors expressing Oct4, Sox2, cMyc, and Klf4 [53]. SMPs are a muscle stem cell population expressing Pax7 but negative for MyoD [54]. Our genetic lineage tracing studies using Cre recombinase Pax7-CreER:R26R and Myf5-Cre:R26R mice demonstrated that the iPSCs generated from primary myoblasts are truly myogenic in origin as they express myogenic markers Pax7 and Myf5 prior to reprogramming. Therefore, to our knowledge, this study is the first showing that fully committed myogenic cells can be reprogrammed to iPSCs.
The second important finding in this study was that MyoD expression prevents the reprogramming of myoblasts into iPSCs. MyoD is a myogenic master transcription factor whose expression alone can program many nonmuscle cells to myogenic lineage [12, 47]. Infection with MyoD expression vector significantly reduced the efficiency of iPSC generation in myoblasts and fibroblasts. In contrast, the efficiency of iPSC production is higher in myoblasts derived from MyoD−/− mice than myoblasts derived from wild-type mice. These results strongly indicate that suppression of MyoD gene expression in myoblasts is required for the initial step of reprogramming myoblasts to iPSCs. Similar results were obtained from B cellderived iPSC reprogramming in which B cell-specific transcription factor Pax5 inhibits reprogramming [2].
We also investigated the mechanism of MyoD gene suppression in myoblasts following retroviral infection of the four factors. Among the four factors, we demonstrated that Oct4 is the primary inhibitor of MyoD gene expression. Ectopic expression of Oct4 in myoblasts immediately shut down MyoD gene expression within 6 hours, possibly through direct transcriptional repression of MyoD enhancers. Our ChIP assay demonstrated that Oct4 is able to bind to the upstream enhancer region sites of the MyoD gene. However, Oct4 did not prevent MyoD from binding to the upstream enhancer region sites of the MyoD gene. Thus, Oct4 may function as a repressor for MyoD gene expression through direct binding to the upstream enhancer region sites, possibly interacting with other negative regulator(s). Recent work demonstrates that ectopic expression of Oct4 suppresses Pax7 gene expression by binding to the upstream site of the Pax7 gene in C2C12 myoblasts [55]. In addition, Oct4 can bind to the upstream sites of Cdx2 and Cldn4, trophectoderm-specific genes, and suppress their expression [56]. More recently, Dax1, a molecule interacting with Oct4, has been identified as a negative regulator of Oct4 in ESCs [57]. Therefore, Oct4-interacting factors such as Dax1 may negatively regulate MyoD gene expression. During myogenesis, Oct4 expression has never been detected by immunostaining or Oct4-GFP transgene expression (unpublished observation). Recent high-throughput DNA sequencing (ChIP-Seq) demonstrates that binding scores for Oct4 at the MyoD or Pax7 locus in ESCs are very low or absent [58]. In contrast, Oct4 binds at the transcriptionally inactive Myf5 locus in ESCs [59]. Currently, there is no data for ChIP-mediated Oct4-binding profiles in muscle cells. One possibility is that Oct4-related transcription factor(s) containing a POU domain and/or a homeodomain such as Hoxa-11 and Hoxa-13 [60], which are expressed in myogenic lineages can negatively regulate MyoD gene expression by binding to these enhancer regions during muscle development and regeneration.
Recent work demonstrated that the origin of iPSCs affects DNA methylation profiles, thereby altering gene expression and differentiation capacity of the iPSCs [53, 61]. For example, iPSCs derived from blood cells maintain epigenetic memory as residual DNA methylation signatures that are characteristic of their somatic tissue of origin [62]. In addition, these bloodderived iPSCs possess a higher hematopoietic cell differentiation efficiency while restricting alternative cell fates. Therefore, the differentiation potential to specific cell types may differ broadly among iPSCs generated from different sources. In addition, our data showed that MyoD expression is easily suppressed by Oct4 in myoblasts. Nevertheless, MyoD−/− myoblasts display higher efficiency for iPSC generation than wildtype myoblasts, indicating that wild-type myoblasts resist iPSC reprogramming. This discrepancy may be due to differences in gene expression profiles [19] or epigenetic status, including methylation profiles, between wild-type and of MyoD−/− myoblasts. Currently, we are investigating whether myoblast-derived iPSCs possess any epigenetic memory of myogenic status which might contribute to a higher myogenic differentiation potential than that of nonmuscle cell-derived iPSCs.
Muscle-derived stem cell transplantation is a potential therapeutic application for muscle degenerative diseases including muscular dystrophy [63]. Myoblasts have been used for this purpose clinically and experimentally [19, 64]. A current limitation of myoblast transplantation is that myoblasts expanded ex vivo lose self-renewal activity and do not efficiently contribute to the muscle stem cell pool in the recipient muscle [65, 66]. Factors that reprogram myoblasts to a more dedifferentiated stem cell-like population might maintain self-renewal activity and efficiently contribute to the stem cell pool after cell transplantation [19, 67]. Therefore, understanding the molecular mechanisms underlying the production of myoblast-derived iPSCs may lead to new avenues for the efficient stem cell transplantation of genetically modified myoblasts. Importantly, efficiency of direct skeletal muscle differentiation of ESCs is still very poor [68]. The possibility exists that myoblast-derived iPSCs may maintain epigenetic memory of myogenic status, which can contribute to higher myogenic differentiation potential. Therefore, the use of myoblast-derived iPSCs can be regarded as a potential cell therapy for muscular dystrophy.
Conclusion
We successfully generated iPSCs from myoblasts and proved that these myoblast-derived iPSCs are truly myogenic in origin. Our findings suggest that suppression of MyoD gene expression by Oct4 is required for the initial reprogramming step in the induction of pluripotent stem cells from myoblasts. In addition, the skeletal muscle system is a well-established differentiation model, allowing a more detailed study of somatic cell iPSC reprogramming.
Acknowledgments
We thank Drs. Michael A. Rudnicki for providing MyoD mutant mice, David J. Goldhamer for providing the luciferase reporter genes, Toshio Kitamura for providing the pMXs vector and PlatE cells, Nobuaki Kikyo for providing MEFs and ESCs, Genya Gekker for technical assistance in the Stem Cell Institute FACS facility, Jesse L. Mull for critical reading of the manuscript, Lucas Greder and Tetsuya Tani for technical assistance. This work was supported by grants from the Muscular Dystrophy Association (MDA), the Nash Avery Foundation, and Gregory Marzolf Jr. Award.
Footnotes
Author contributions: S.W.: collection and/or assembly of data, manuscript writing; H.H.: collection and/or assembly of data; Y.A.: collection and/or assembly of data; C.T.: collection and/or assembly of data; M.V.: collection and/or assembly of data; C.K.: provision of study material or patients; J.R.D.: provision of study material or patients; A.A.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. S.W. and H.H. contributed equally to this work.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Hanna J, Markoulaki S, Schorderet P, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watarai H, Fujii S, Yamada D, et al. Murine induced pluripotent stem cells can be derived from and differentiate into natural killer T cells. J Clin Invest. 2010;120:2610–2618. doi: 10.1172/JCI42027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoi T, Yae K, Nakagawa M, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 5.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JB, Zaehres H, Wu G, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 7.Eminli S, Foudi A, Stadtfeld M, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 9.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 10.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins CA. Satellite cell self-renewal. Curr Opin Pharmacol. 2006;6:301–306. doi: 10.1016/j.coph.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Weintraub H, Davis R, Tapscott S, et al. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 13.Sabourin LA, Girgis-Gabardo A, Seale P, et al. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 15.Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Megeney LA, Kablar B, Garrett K, et al. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 17.Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol. 2000;224:122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- 18.White JD, Scaffidi A, Davies M, et al. Myotube formation is delayed but not prevented in MyoD-deficient skeletal muscle: Studies in regenerating whole muscle grafts of adult mice. J Histochem Cytochem. 2000;48:1531–1544. doi: 10.1177/002215540004801110. [DOI] [PubMed] [Google Scholar]
- 19.Asakura A, Hirai H, Kablar B, et al. Increased survival of muscle stem cells lacking the MyoD gene after transplantation into regenerating skeletal muscle. Proc Natl Acad Sci USA. 2007;104:16552–16557. doi: 10.1073/pnas.0708145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudnicki MA, Braun T, Hinuma S, et al. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 21.Lengner CJ, Camargo FD, Hochedlinger K, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tallquist MD, Weismann KE, Hellstrom M, et al. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 23.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 24.Nishijo K, Hosoyama T, Bjornson CR, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochedlinger K, Yamada Y, Beard C, et al. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Asakura A, Seale P, Girgis-Gabardo A, et al. Myogenic specification of side population cells in skeletal muscle. J Cell Biol. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuang S, Kuroda K, Le Grand F, et al. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirai H, Romanova L, Kellner S, et al. Post-mitotic role of nucleostemin as a promoter of skeletal muscle cell differentiation. Biochem Biophys Res Commun. 2010;391:299–304. doi: 10.1016/j.bbrc.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma M, Asakura Y, Hirai H, et al. Flt-1 haploinsufficiency ameliorates muscular dystrophy phenotype by developmentally increased vasculature in mdx mice. Hum Mol Genet. 2010;19:4145–4159. doi: 10.1093/hmg/ddq334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shea KL, Xiang W, LaPorta VS, et al. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2009;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldring K, Jones GE, Thiagarajah R, et al. The effect of galectin-1 on the differentiation of fibroblasts and myoblasts in vitro. J Cell Sci. 2002;115:355–366. doi: 10.1242/jcs.115.2.355. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura T, Koshino Y, Shibata F, et al. Retrovirus-mediated gene transfer and expression cloning: Powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 33.Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: Conserved elements mediate expression in embryonic axial muscle. Dev Biol. 1995;171:386–398. doi: 10.1006/dbio.1995.1290. [DOI] [PubMed] [Google Scholar]
- 34.Verma M, Asakura A. Efficient single muscle fiber isolation from alcohol fixed adult muscle following {beta}-galactosidase staining for satellite cell detection. J Histochem Cytochem. 2011;59:60–67. doi: 10.1369/jhc.2010.956730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe S, Kondo S, Hayasaka M, et al. Functional analysis of homeodomain-containing transcription factor Lbx1 in satellite cells of mouse skeletal muscle. J Cell Sci. 2007;120:4178–4187. doi: 10.1242/jcs.011668. [DOI] [PubMed] [Google Scholar]
- 37.Chang H, Yoshimoto M, Umeda K, et al. Generation of transplantable, functional satellite-like cells from mouse embryonic stem cells. FASEB J. 2009;23:1907–1919. doi: 10.1096/fj.08-123661. [DOI] [PubMed] [Google Scholar]
- 38.Eakin GS, Hadjantonakis AK. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Nat Protoc. 2006;1:1145–1153. doi: 10.1038/nprot.2006.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weintraub H, Davis R, Lockshon D, et al. MyoD binds cooperatively to two sites in a target enhancer sequence: Occupancy of two sites is required for activation. Proc Natl Acad Sci USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker MH, Perry RL, Fauteux MC, et al. MyoD synergizes with the E-protein HEB beta to induce myogenic differentiation. Mol Cell Biol. 2006;26:5771–5783. doi: 10.1128/MCB.02404-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldhamer DJ, Brunk BP, Faerman A, et al. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- 42.Filippova GN, Thienes CP, Penn BH, et al. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 43.Hirai H, Verma M, Watanabe S, et al. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brack AS, Conboy MJ, Roy S, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 45.Eminli S, Utikal J, Arnold K, et al. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 46.Kunisato A, Wakatsuki M, Kodama Y, et al. Generation of induced pluripotent stem cells by efficient reprogramming of adult bone marrow cells. Stem Cells Dev. 2010;19:229–238. doi: 10.1089/scd.2009.0149. [DOI] [PubMed] [Google Scholar]
- 47.Weintraub H, Tapscott SJ, Davis RL, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen JC, Ramachandran R, Goldhamer DJ. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev Biol. 2002;245:213–223. doi: 10.1006/dbio.2002.0638. [DOI] [PubMed] [Google Scholar]
- 49.Chen JC, Goldhamer DJ. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev Biol. 2004;265:502–512. doi: 10.1016/j.ydbio.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Cao Y, Yao Z, Sarkar D, et al. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JB, Zaehres H, Arauzo-Bravo MJ, et al. Generation of induced pluripotent stem cells from neural stem cells. Nat Protoc. 2009;4:1464–1470. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- 52.Do JT, Joo JY, Han DW, et al. Generation of parthenogenetic induced pluripotent stem cells from parthenogenetic neural stem cells. Stem Cells. 2009;27:2962–2968. doi: 10.1002/stem.233. [DOI] [PubMed] [Google Scholar]
- 53.Polo JM, Liu S, Figueroa ME, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerletti M, Jurga S, Witczak CA, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang KC, Lin IH, Teng HF, et al. Simultaneous overexpression of Oct4 and Nanog abrogates terminal myogenesis. Am J Physiol Cell Physiol. 2009;297:C43–C54. doi: 10.1152/ajpcell.00468.2008. [DOI] [PubMed] [Google Scholar]
- 56.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 57.Sun C, Nakatake Y, Akagi T, et al. Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol Cell Biol. 2009;29:4574–4583. doi: 10.1128/MCB.01863-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 59.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto M, Kuroiwa A. Hoxa-11 and Hoxa-13 are involved in repression of MyoD during limb muscle development. Dev Growth Differ. 2003;45:485–498. doi: 10.1111/j.1440-169x.2003.00715.x. [DOI] [PubMed] [Google Scholar]
- 61.Colman A, Dreesen O. Induced pluripotent stem cells and the stability of the differentiated state. EMBO Rep. 2009;10:714–721. doi: 10.1038/embor.2009.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tedesco FS, Dellavalle A, Diaz-Manera J, et al. Repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blau HM. Cell therapies for muscular dystrophy. N Engl J Med. 2008;359:1403–1405. doi: 10.1056/NEJMcibr0805708. [DOI] [PubMed] [Google Scholar]
- 65.Montarras D, Morgan J, Collins C, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 66.Collins CA, Olsen I, Zammit PS, et al. Stem cell function, selfrenewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Pajcini KV, Corbel SY, Sage J, et al. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darabi R, Gehlbach K, Bachoo RM, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]