Isoprenoids derive from two universal precursors: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP).[1] In mammals, these key intermediates are crucial for cell survival, e.g. in cholesterol biosynthesis, and are generated via the mevalonate pathway, while in most bacteria as well as in malaria parasites such as Plasmodium falciparum, the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway is used.[2] The DXP pathway is absent in humans and is, therefore, considered to be an important drug target against many infectious diseases.[3]
Scheme 1 shows the last step of the DXP pathway, the conversion of (E)-4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP, 1) to a mixture of IPP (2) and DMAPP (3). This reaction is catalyzed by HMBPP reductase (IspH), a monomeric enzyme that consists of three domains binding to a redox-active [Fe4S4] cluster in its central cavity (Figure 1a).[4] The cluster is linked to the protein by the side-chains of three cysteine residues, and one of the iron atoms possesses an unoccupied coordination site that has not been found to bind to any amino acid residue. Mechanistic studies with wild type Escherichia coli IspH as well as IspH mutants have revealed two different conformations of 1 inside the active site that are adopted in the catalytic cycle (Figure 1b and c): one in which O1 binds to the 4th iron atom, and a second in which it undergoes numerous hydrogen bond interactions with its diphosphate group and protein residues.[5]
Scheme 1.
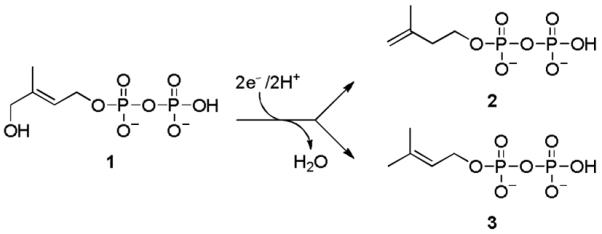
Reductive dehydration catalyzed by IspH.
Figure 1.
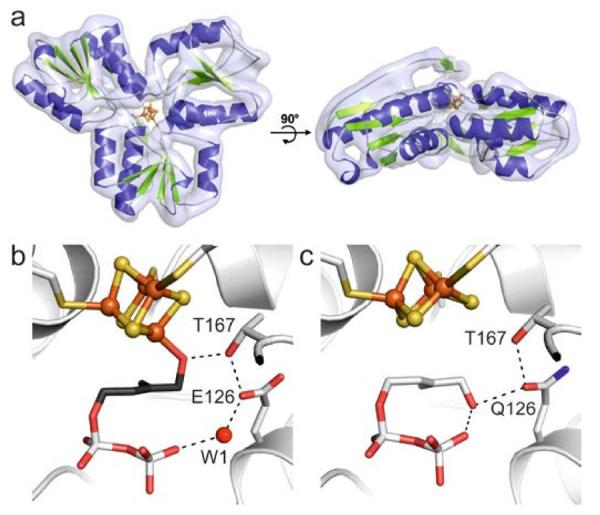
Crystal structure of IspH in complex with 1. a) Top and side view of the overall structure of E. coli IspH. Active site of b) wild type IspH forming an alkoxide complex with 1 and c) the IspH E126Q mutant bound to 1 in the cyclic conformation revealing a [Fe3S4] cluster.
The proposed mechanism for the IspH reaction is shown in Scheme 2 and involves four intermediate states that have been identified by crystallography, Mößbauer and electron paramagnetic resonance (EPR) spectroscopy.[5–6] The detailed structure of IspH in the absence of exogenous ligands is not known (state 0) but binding of 1 to oxidized IspH leads to formation of an alkoxide complex with weak pi interactions (state I; spin S=0). One-electron reduction of the cluster results in [Fe4S4]+ with spin S=1/2, and correlates with a rotation of the ligand's hydroxymethyl group away from the cluster to form a cyclic conformation (state II) which has essential impact on the stereochemical course of the IspH reaction.[7] The transfer of two electrons from the cofactor to the substrate produces a HiPIP-type [Fe4S4]3+ cluster and leads to C-O bond cleavage and water release. The allyl anion (state III) then abstracts a proton from the diphosphate group, either at the ligand's C2 or C4 atom, to form IPP and DMAPP, respectively.
Scheme 2.
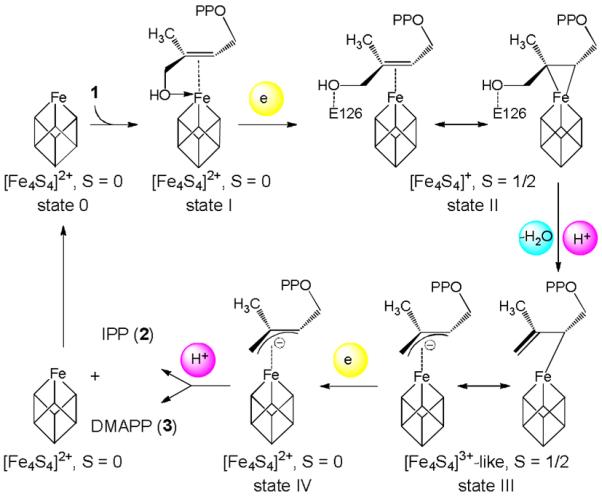
Proposed mechanism of IspH catalysis.
Besides the intensive investigation of the IspH reaction mechanism, a remarkable effort was put into the design and characterization of inhibitors.[8] Recently, synthesis and spectroscopic studies of three substrate analogs with the hydroxyl group in HMBPP replaced by fluoro (4)[9], amino (5)[10], or thiol (6) groups have been reported. Compound 4 is slowly converted by IspH, whereas 5 and 6 inhibit the enzyme. In order to analyze the structure-function relationship of these derivatives we synthesized 4[11], 5[12], and 6 (see SI), performed co-crystallization with E. coli IspH and determined the crystal structure of the complexes.
The X-ray structure of IspH in complex with the fluoro analog 4 was determined to 1.8 Å resolution [Rfree = 23.2%, Figure 2a, Protein Data Bank (PDB)[13] ID 4H4C] and reveals that 4 binds to the active site of IspH in a similar way as the substrate 1.[14] However, the C-F bond is rotated by 106° compared to the C-O bond in the IspH:1 complex (Figure 2b), the fluorine atom is thus located inside a hydrophobic pocket stabilized by van der Waals interactions with His74Cδ (3.6 Å), Ala73C (3.9 Å), and Ala73Cβ (3.9 Å). This unique conformation allows water molecules to occupy positions W1 and W2.[14] Although it displays an unusual orientation, 4 is converted to 2 or 3 by IspH, but with a decreased rate (kcat = 28 min−1) compared to 1 (kcat = 604 min−1). The differences in these reaction rates are likely due, at least in part, to the increased bond energies of C-F versus C-O.[15] Furthermore, the lack of a direct interaction with the apical iron atom leads to the high Km value of 4 (Km = 104 μM) compared to 1 (Km = 20 μM).
Figure 2.
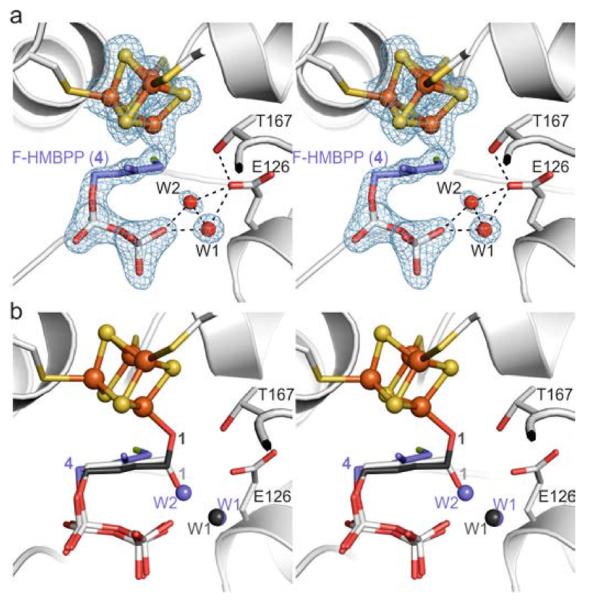
Complex structure of IspH bound to the fluorinated derivative 4. a) Active site of IspH showing the bound ligand and two water molecules. A 2FO-FC omit electron density map (blue mesh, contoured at 1.0 σ) is shown for the [Fe4S4] cluster, the ligand, and the solvent molecules in the first coordination sphere; dotted lines indicate hydrogen bonds. b) Structural superposition of IspH:4 with the alkoxide complex (IspH:1) and the cyclic intermediate (IspH E126Q:1).
Recent inhibition studies have shown that the amino and thiol substrate analogs 5 and 6 exhibit potent inhibition of IspH with IC50 values of 0.15 μM and 0.21 μM, respectively.[10] Additionally, Mößbauer spectroscopy has suggested that both ligands interact with the [Fe4S4] cluster. However, it is not immediately obvious that 5 binds to the 4th iron atom via its amino group, or whether it forms an alternative complex that allows a water molecule to coordinate to the 4th iron atom as previously observed with an acetylene inhibitor.[8c]
The structure of 5 in complex with IspH was determined to 1.35 Å resolution (Rfree = 21.0% Figure 3a, PDB[13] ID 4H4D) and clearly shows two ligand conformations within the same crystal[16]: (i) a ligand-cluster complex in which the amino group coordinates to the apical iron atom and (ii) a conformation in which the amino group is rotated by approximately 74° in the opposite direction to that observed with 4. The amino-iron complex is similar to that seen with the alkoxide-iron complex formed by 1 (Figure 3b), indicating that the affinity of the free amino group with the [Fe4S2]2+ cluster is comparable to that of the hydroxyl group. The second conformation observed in the crystal structure is stabilized by hydrogen bonding of the amino (or ammonium) group to one of the diphosphate oxygen atoms (2.8 Å), Glu126Oε (2.9 Å) and Thr167Oγ (3.1 Å), and a water molecule in the W1 position (3.1 Å).
Figure 3.
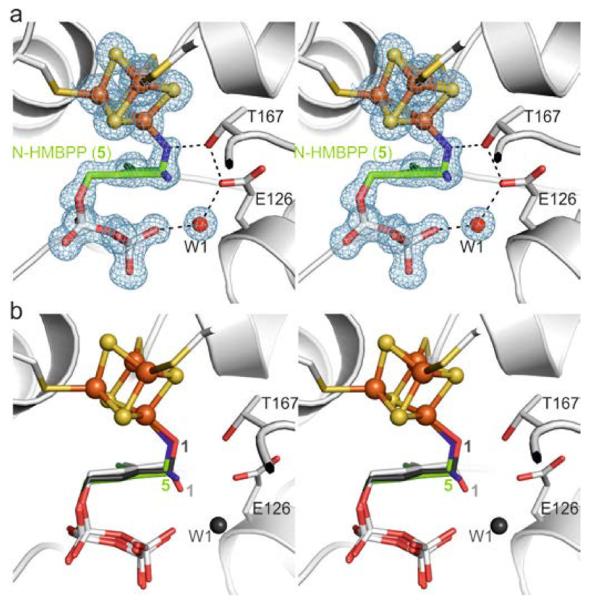
X-ray structure of IspH in complex with the amino derivative 5. a) Active site with the ligand in two orientations. The electron density map is displayed according to Figure 2a. b) Superposition of the IspH:5 complex structures with the alkoxide complex in IspH:1 and the cyclic intermediate in IspH E126Q:1.
The amino-iron complex is in good agreement with the Mößbauer spectroscopic data indicating a tetrahedral (3S/N) coordination sphere at the apical iron atom.[10] Thus, it seems likely that the alternative conformation may have arisen due to cluster reduction in the X-ray beam – a result that would be of particular interest in the context of inhibition of the IspH reaction which is, of course, carried out under reducing conditions.[17] However, unlike with 1, no turnover was observed with 5. One possible explanation for this is the formation of a stable ammonium-carboxylate ion pair that prevents the release of ammonia (Scheme 4).
Scheme 4.
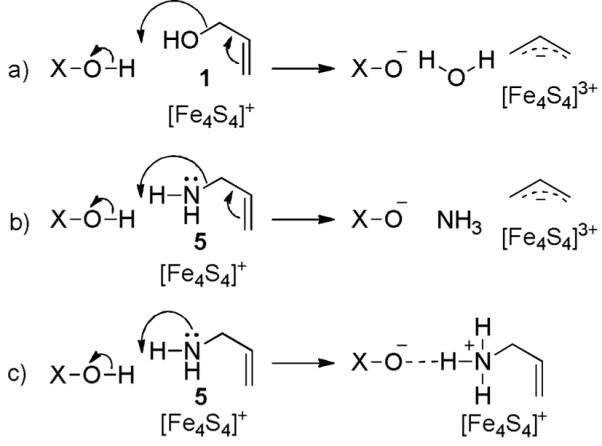
Mechanism of IspH catalysis with 1 and inhibition by 5. a) Dehydroxylation of 1, facilitated by an acidic proton donor (Glu126OH or the diphosphate) represented by X-OH. b) Corresponding hypothetical reaction with 5. c) Formation of a stable ammonium-carboxylate ion pair.
Next, we determined the crystal structure of IspH bound to 6 to 1.7 Å resolution (Rfree = 21.3%, Figure 4a, PDB[13] ID 4H4E), which displays complex formation between the unique iron atom of the cluster and the thiol group of 6. No solvent molecule is located in the active site, since the increased size of the bridging group leads to movement of the hydrocarbon chain of 6 towards the diphosphate moiety. Furthermore, the hydrogen bond network is modified, preventing the stabilization of the water molecules in the position as found in the IspH:1 complex (Figure 4b). The coordination by the ligand's thiol group generates a [Fe4S4] cluster that is coordinated by four sulfur ligands, similar to that found in most proteins catalyzing electron transfer reactions.[18]
Figure 4.
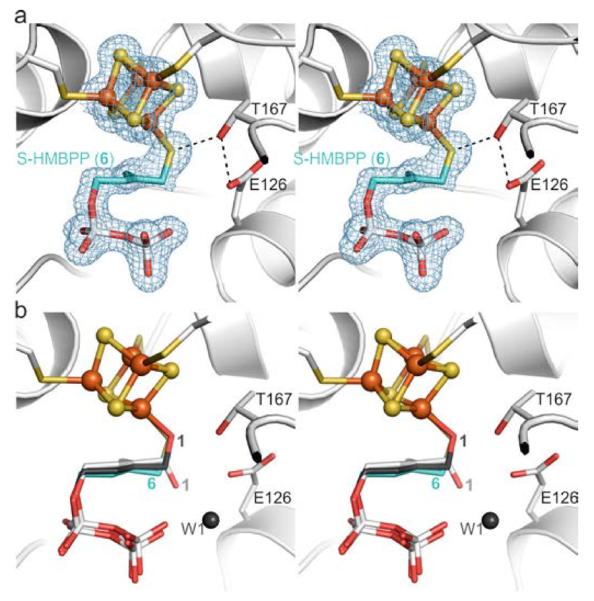
IspH bound to the thiol derivative 6. a) Active site with thiol-iron complex. The electron density map is displayed according to Figure 2a. b) Structural superposition of IspH:6 with the alkoxide complex in the IspH:1 structure and the cyclic ligand orientation in the IspH E126Q:1 complex.
In conclusion, the results presented here provide new insights into the mechanism of the IspH reaction with the artificial substrate 4, which involves a previously unknown intermediate that is not bound to the [Fe4S4] cluster. With the amino (5) and thiol (6) species, we find in both cases that the ligands bind to the 4th iron atom. However, it still remains to be determined whether IspH inhibition is due to bonding of these ligands to the oxidized protein, the reduced protein, or both. With 5, we see evidence for a second conformation in which the side-chain is not interacting with the cluster and propose formation of a non-reactive ion-pair complex with Glu126Oε and the diphosphate group (possibly in both oxidized and reduced IspH). As expected, the IspH:6 complex reveals that the ligand's sulfur atom binds to the 4th iron atom, and there is no alternative conformation, most likely due to the fact that the SH group is a poor base/hydrogen bond acceptor. The structures of these substrate analogs thus provide new mechanistic insights, as well as suggest novel strategies for the development of antibiotic and antimalarial drugs.
Supplementary Material
Scheme 3.
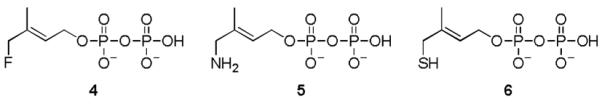
Analogs of the IspH substrate 1.
Footnotes
This work was supported by the Hans-Fischer Gesellschaft, DFG Grant GR1861/5–1, and NIH grant GM65307. We are grateful to Dr. Florian Kraus for his valuable contribution to the manuscript and David Hartmann for the synthesis of the fluoro-analog of HMBPP. We would like to thank the staff of the X06SA-beamline at the Paul Scherer Institute, Swiss Light Source, Villigen, Switzerland for help during data collection.
References
- [1].Ruzicka L. Cell. Mol. Life Sci. 1953;9:357–367. [Google Scholar]
- [2].a) Bloch K. Steroids. 1992;57:378–383. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]; b) Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Biochem. J. 1993;295(Pt 2):517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Diederich F. CHIMIA. 2008;62:226–230. [Google Scholar]
- [4].a) Rekittke I, Wiesner J, Rohrich R, Demmer U, Warkentin E, Xu W, Troschke K, Hintz M, No JH, Duin EC, Oldfield E, Jomaa H, Ermler U. J. Am. Chem. Soc. 2008;130:17206–17207. doi: 10.1021/ja806668q. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gräwert T, Rohdich F, Span I, Bacher A, Eisenreich W, Eppinger J, Groll M. Angew. Chem. Int. Ed. 2009;48:5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]; c) Xu W, Lees NS, Hall D, Welideniya D, Hoffman BM, Duin EC. Biochem. 2012;51:4835–4849. doi: 10.1021/bi3001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Gräwert T, Span I, Eisenreich W, Rohdich F, Eppinger J, Bacher A, Groll M. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1077–1081. doi: 10.1073/pnas.0913045107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Span I, Gräwert T, Bacher A, Eisenreich W, Groll M. J. Mol. Biol. 2012;416:1–9. doi: 10.1016/j.jmb.2011.11.033. [DOI] [PubMed] [Google Scholar]
- [6].Wang W, Wang K, Span I, Jauch J, Bacher A, Groll M, Oldfield E. J. Am. Chem. Soc. 2012;134:11225–11234. doi: 10.1021/ja303445z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Citron CA, Brock NL, Rabe P, Dickschat JS. Angew. Chem. Int. Ed. 2012;51:4053–4057. doi: 10.1002/anie.201201110. [DOI] [PubMed] [Google Scholar]
- [8].a) Wang K, Wang W, No JH, Zhang Y, Oldfield E. J. Am. Chem. Soc. 2010;132:6719–6727. doi: 10.1021/ja909664j. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang W, Wang K, Liu YL, No JH, Li J, Nilges MJ, Oldfield E. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4522–4527. doi: 10.1073/pnas.0911087107. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Span I, Wang K, Wang W, Zhang Y, Bacher A, Eisenreich W, Li K, Schulz C, Oldfield E, Groll M. Nat. Commun. 2012;3:1042. doi: 10.1038/ncomms2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Xiao Y, Zhao ZK, Liu P. J. Am. Chem. Soc. 2008;130:2164–2165. doi: 10.1021/ja710245d. [DOI] [PubMed] [Google Scholar]; b) Xiao Y, Chang WC, Liu HW, Liu P. Org. Lett. 2011;13:5912–5915. doi: 10.1021/ol202559r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ahrens-Botzong A, Janthawornpong K, Wolny JA, Tambou EN, Rohmer M, Krasutsky S, Poulter CD, Schünemann V, Seemann M. Angew. Chem. Int. Ed. 2011;50:11976–11979. doi: 10.1002/anie.201104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, Poulter CD. J. Org. Chem. 1986;51:4768–4779. [Google Scholar]
- [12].Das D, Tnimov Z, Nguyen U, Thimmaiah G, Lo H, Abankwa D, Wu Y, Goody R, Waldmann H, Alexandrov K. ChemBioChem. 2012;13:674–683. doi: 10.1002/cbic.201100733. [DOI] [PubMed] [Google Scholar]
- [13].Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr., Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Eur. J. Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- [14].The crystal structure of IspH bound to 4 reveals an excess (approximately 10%) electron density between the fluorine atom and the apical iron atom due to partial hydrolysis to 1 during crystallization, with 1 coordinating to the iron-sulfur cluster.
- [15].O'Hagan D. J. Org. Chem. 2012;77:3689–3699. doi: 10.1021/jo300044q. [DOI] [PubMed] [Google Scholar]
- [16].X-ray crystallography is an averaging method, which enables the determination of atomic coordinates of an ensemble of molecules. The IspH:5 complex structure displays two ligand conformations within the same crystal, which means that one part of the ensemble adopts one conformation, whereas the other part of the ensemble display another conformation.
- [17].We should mention that reduction of the [Fe4S4] cluster were previously observed with the IspH:1 complex at a temperature of 100 K.
- [18].a) Beinert H, Holm RH, Munck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]; b) Johnson DC, Dean DR, Smith AD, Johnson MK. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


