Abstract
An analytical approach was developed to study the incorporation of selenium (Se), an important trace element involved in the protection of cells from oxidative stress, into the well-known probiotic Lactobacillus reuteri Lb2 BM-DSM 16143. The analyses revealed that about half of the internalized Se was covalently incorporated into soluble proteins. Se-enriched proteins were detected in 2D gels by laser ablation inductively coupled plasma mass spectrometry imaging (LA-ICP MSI) and identified by capillary HPLC with the parallel ICP MS (78Se) and electrospray Orbitrap MS/MS detection. On the basis of the identification of 10 richest in selenium proteins, it was demonstrated that selenium was incorporated by the strain exclusively as selenocysteine. Also, the exact location of selenocysteine within the primary sequence was determined. This finding is in a striking contrast to another common nutraceutical, Se-enriched yeast, which incorporates Se principally as selenomethionine.
In recent years selenium (Se)1 has received considerable attention as an essential element for human health. Severe Se deficiency is linked to oxidative stress and aging (1), elevated mortality with HIV (2), and irreversible brain injury (seizures, Parkinson's disease) (3). Se occurs in nature principally in four inorganic chemical forms: the highly toxic selenide (Se2−−) (4), the moderately toxic selenate (SeO42−−) and selenite (SeO32−−), and elemental selenium (Se0) which is essentially nontoxic and can be stored by several bacterial species as nanoparticles on the cell surface (5, 6, 7). Inorganic selenium can be converted by biological systems (microorganisms, plants, and mammals) into seleno-amino acids, which are then incorporated into proteins. The two most common seleno-amino acids are selenomethionine (SeMet) and selenocysteine (SeCys). The former is synthesized via a route similar to the sulfur metabolic pathway in which selenium substitutes sulfur with no alteration of the protein structure (8, 9). The insertion of SeCys is genetically encoded by the UGA (TGA) codon and requires a SECIS element downstream of such a codon, a specific tRNA[Ser]Sec and accessory proteins (10).
Selenoproteins containing genetically encoded SeCys are known to be synthesized by several bacteria. Among Gram-negative ones, E. coli produces three forms of selenated formate dehydrogenase (FdhN, FdhO, FdhH) (11). Among Gram-positive bacteria, all the selenoproteins experimentally known were found exclusively in anaerobic bacteria belonging to the clostridial clade. Examples include glycine reductase from Clostridium sticklandii (12) and Eubacterium acidaminophilum (13), proline reductase in C. sticklandii (14), xanthine dehydrogenase in C. acidiurici (15), and several antioxidant defense proteins (16). Enterococcus faecalis is the only member of the Firmicutes/Lactobacillales subdivision containing a SeCys-decoding trait (SelD) (17).
Lactobacillus reuteri species has been widely described as a probiotic: it produces antimicrobial compounds, such as reuterin, with a broad spectrum of action (18), it is effective against diarrhea in children (19) and possesses immunomodulatory (potent TNF-inhibitory activity) effects in humans (20). Because L. reuteri species are native inhabitants of human microbiota, the association of the probiotic feature L. reuteri Lb2 BM-DSM 16143 with its ability to fix selenium into proteins, offers an innovative approach to combat human selenium deficiency.
The objectives of this study were to investigate the ability of Firmicutes/Lactobacillales subdivision, Lactobacillus reuteri Lb2 BM-DSM 16143to incorporate selenium into proteins, and to investigate, for the first time, its speciation in order to identify the pathway(s) of this process (SeMet or SeCys). For this purpose an analytical approach based on laser ablation inductively coupled plasma mass spectrometry imaging (LA-ICP MSI) of Se-containing proteins in 2D gel electrophoresis, followed by their identification by capillary HPLC - electrospray Orbitrap MS/MS assisted by the quantitative control of selenium elution by ICP MS, was developed.
EXPERIMENTAL PROCEDURES
Culture Conditions
L. reuteri Lb2 BM-DSM 16143 was grown in MRS medium (Difco) supplemented with 4.38 mg L−1 sodium selenite corresponding to 2 mg L−1 selenium. One milliliter of precultured bacteria was inoculated in closed 250-ml screw-cap bottles, at 37 °C without shaking. Two biological replicates were performed.
Reagents and Chemicals
All the reagents were of analytical grade from Sigma - Aldrich unless stated otherwise. Water obtained from a Milli-Q purification system was used throughout.
Microwave-assisted Digestion and ICP-MS Analysis
Three types of samples were collected at different times during the cell growth: 1) pellet (to quantify cell incorporation of Se): an aliquot of culture containing 0.14 mg bacteria (dry weight), corresponding to 0.5 OD, was taken and centrifuged (10,000 × g, 4 °C, 15 min); 2) supernatant (to evaluate the amount of Se which was not taken up): 1 ml of the culture broth was centrifuged at 10,000 × g, 4 °C, 15 min to separate the biomass and to recover the supernatant, and 3) wash supernatant (to recover surface-associated Se(0) deposits (5): the harvested biomass was washed with 1 ml 50 mm EDTA, and centrifuged at 10,000 × g (4 °C, 15 min).
Each sample (1) pellet resuspended in 1 ml water; 2) 1 ml of supernatant; 3) 1 ml of EDTA wash), respectively, was microwave (Milestone Ethos 900-Mega II) digested in a Teflon vessel with a mixture of 6 ml conc HNO3 and 2 ml conc H2O2 (both Super Purity Solvent grade from Romil, Cambridge, UK). Mineralization was achieved with the following oven program: 20 min to reach 220 °C at 1400 W; 15 min at 220 °C and 1400 W; cooling for 30 min. The digested samples were then made up to a volume of 10 ml with water and stored at 4 °C before inductively coupled plasma mass spectrometry (ICP-MS) analysis. Samples were analyzed after 10-fold dilution. The analyses were carried out in biological duplicate and technical triplicate on a 7700 ICP-MS (Agilent, Hachi-oji, Japan), equipped with a frequency-matching RF generator and the third generation Octopole Reaction System (ORS3), operating with helium as cell gas. The parameters were set as follows: radiofrequency power 1550 W, plasma gas flow 14 L min−1; carrier gas flow 1.0 L/min; He gas flow 4.6 ml/min. 78Se was used to quantify selenium using a 4-point calibration curve (1, 10, 50, and 100 μg L−1) prepared in 5% HNO3. An internal standard (103Rh) was added to all the samples and calibration points.
Extraction of Proteins
After 6 h of growth a 50 mg amount of biomass was collected by centrifugation and washed in 50 ml 0.85% NaCl. The obtained pellet was resuspended in 3 ml 50 mm Tris-HCl buffer (pH 7.3) containing EDTA-free protease inhibitor (Complete, Roche), sonicated and recentrifuged (4000 × g, 20 min, 4 °C). To recover the largest amount of proteins, the pellet was resuspended again, sonicated and centrifuged and the two supernatants were combined. Samples were supplemented with 15 μl ml−1 of Nuclease Mix (GE Healthcare) and centrifuged (100,000 × g, 1 h, 4 °C) in a Beckman L8–60 M Ultracentrifuge (Type 60 rotor). The supernatants were dialyzed against four volumes of water. Protein extracts were quantified using the QuantiProTM BCA Assay Kit (Sigma-Aldrich).
Gel Electrophoretic Separations
For 1D-SDS-PAGE different amounts of soluble protein extract (20, 40, 80, and 100 μg) were separated on a 12% T SDS-PAGE after protein precipitation in acetone and resuspension in Laemmli loading dye (21). For 2D-SDS-PAGE aliquots of 100 μg and 400 μg for each of the samples were precipitated using the 2D Clean-Up Kit (GE Healthcare). After precipitation the samples were resuspended in 340 μl of rehydration buffer (GE Healthcare) supplemented with 1.7 μl of IPG buffer. Isoelectrofocusing was carried out using four to seven 18-cm Immobiline™ DryStrip gels strips (GE Healthcare Bio-Sciences, Uppsala, Sweden) in a IEF-SYS (Biostep, Jahnsdorf, Germany) under a constant current of 3 mA. The strips were then focused according to the following electrical conditions: 500 V for 1 h, 1000 V for 1 h, 5000 V for 3 h, held at 5000 V, 6000 V for 3 h, and held at 6000 V for 5 h until a total of 15000 V was reached. After focusing, IPG strips were reduced in an equilibration buffer (6 m urea, 30% (V/V) glycerol, 2% (w/V) SDS, 0.05 m Tris-HCl, pH 6.8) 1% (w/v) with dithiotreitol, and subsequently alkylated with 2.5% (w/v) iodoacetamide. SDS-PAGE was done using an electrophoresis unit TV100 (Biostep, Jahnsdorf, Germany) cell. Tris glycine was used as a running buffer. Migration was performed at 120 V and 160 V for 1D and 2D respectively. After electrophoresis gels were stained with Coomassie blue; after staining, gels were washed with a destaining solution containing 10% acetic acid and 10% methanol, then with water, and put on Whatman 3MM Chromatography paper, covered with Saran film. Gels were dried for 1h at 80 °C using a vacuum dryer Hoefer Slab Gel Dryer GD 2000 (Amersham Biosciences). Strips scanned by laser ablation (LA) - ICP MS were kept in fixing buffer (30% ethanol, 10% glycerol) for 30 min and were dried under laminar flow for 4 h.
Laser Ablation ICP-MS Analyses
NewWave Research (Freemont, CA) UP-213 laser coupled with Agilent 7500cs ICP-MS (Agilent, Hachi-Oji, Japan). The laser was operated in a focused spot mode at the repetition rate of 20 Hz, fluence of 3.80 J/cm2, spot size of 250 μm and scan speed 50 and 100 μm/s for gels and strips, respectively. Ablation was carried out with a He gas flow of 500 ml/min. The ablated aerosol was mixed in a T-connector with 2% nitric acid solution aerosol obtained using a Micromist nebulizer and a double pass Scott spray chamber.
Three Se isotopes (77Se, 78Se, and 80Se) were monitored in the collision cell mode using 3.6 ml/min of H2 as the collision/reaction gas. 78Se (23.78%) provided the highest signal-to-noise (S/N) ratio and was used for analysis. All the parameters were optimized using as standard bovine glutathione peroxidase 1 (GPx1) from Sigma-Aldrich. To obtain two-dimensional (2D) images of selenium distribution, 2D gel was systematically screened (line by line), with a distance between lines of 0.80 mm. The number and the length of lines depended on the dimension of the interesting gel area. LA-ICP MS data files for each analysis were converted into Excel files used to produce both electropherograms and 2D gel images. Finally, the images were plotted using programming a script in MATLAB 7.9.0 computing software. The absolute LOD for Se detection in PAGE gels was 15 pg. The effects of the instrumental parameters and the analytical performance of the method were discussed in detail elsewhere (22, 23).
In-gel Protein Hydrolysis
Areas corresponding to the 10 spots most abundant in terms of selenium concentration were excised from a gel obtained in the identical conditions in parallel and digested with trypsin. Before adding the enzyme, the excised pieces of gel were washed twice with 200 μl 200 mm ammonium bicarbonate in 40% acetonitrile at 37 °C. The washing solution was discarded and the pieces of gel were dried under a nitrogen flow. Protein digestion was performed overnight with 20 μl of 20 μg/ml solution of trypsin (Proteomics Grade from Sigma-Aldrich) and 50 μl of 40 mm ammonium bicarbonate in 9% acetonitrile at 37 °C. Then, samples were filtered using a 10 kDa cutoff Vivacon 500 filter (Sartorius, Gottingen, Germany) by centrifugation at 7000 × g for 20 min.
Capillary HPLC with the Parallel ICP-MS and Electrospray LTQ Orbitrap MS/MS
An Agilent 1260 Infinity 2D HPLC system consisting of an isocratic binary and a capillary pump was used (24). The mobile phases A and B were 0.1% formic acid and 0.1% formic acid in acetonitrile, respectively. After loading, the peptide mixture (8 μl) was first concentrated on a ZORBAX 300SB C18 (5 μm 35 × 0.5 mm) enrichment column (Agilent), by using the isocratic pump. Then, the analytes were eluted in back flush by the capillary pump and separated on a ZORBAX 300SB C18 (3.5 μm 100 × 0.3 mm) column at flow rate of 4 μl/min. Peptides were eluted using a gradient: 0–2 min 2% B linear, 2–5 min 2–10% B linear, 5–35 min 10–25% B linear, 35–40 min 25–40% B linear, 40–45 min 40–97% B linear, 45–50 min 97% B isocratic, and 50–55 min 97–2% linear.
The same chromatographic system was coupled alternatively to ICP MS and Orbitrap MS. The retention time correction (e.g. for the different tubing length) was performed by overlapping the UV chromatograms from the different runs.
The ICP MS instrument was Agilent 7700cs ICP-MS (Agilent, Tokyo, Japan) connected to the capillary column via a total consumption micronebulizer (24). Five percent of O2 was added to the plasma gas. ICP-MS was used in the collision cell mode using 10 ml/min of He as the collision/reaction gas. A tryptic digest of a bovine glutathione peroxidase 1 (GPx1) was used to check the performance of the system. The LOD for the capHPLC-ICP MS was 0.3 pg (referred to 40% acetonitrile).
The electrospray ionization (ESI) mass spectrometer was Velos Orbitrap (Thermo Electron, Bremen, Germany) operated in positive ion mode with the following parameters: heater temperature 50 °C, sheath gas pressure 5 psi, spray voltage 3.80 kV, capillary temperature 280 °C and S-Lens RF 67%. Data were acquired in both MS and MS/MS mode in the m/z range 300–1200 at a rate of 3 spectra/s. All samples were measured in a data dependent acquisition mode. In addition, selected precursor ions were fragmented in an independent HPLC run. The peptide masses were measured with a resolution of 60,000. Double and triple charged peptide ions were fragmented by high-collision dissociation (HCD) with a resolution of 30,000 and a normalized fragmentation energy of 40%.
Protein identification
The raw data were processed using the Xcalibur software version 2.1.0. MassMatrix file conversion tool version 3.0 was used to convert the raw data in common spectral file formats (.mgf mascot generic file). MASCOT software (www.matrixscience.com) version 2.4.0 was used for the protein identification against NCBInr database (NCBInr_20120920.fasta; 21582400 sequences; 7401135489 residues), with the taxonomy restriction to Other Firmicutes (2926062 sequences). The Mascot search parameters were: “trypsin” as enzyme allowing up to 3 missed cleavages, carbamidomethyl and selenocysteine on cysteine residues, oxidation of methionine, and formation of pyroGlu N-term on glutamine were selected as variable modifications. The parent peptide mass accuracy was set at 10 ppm and for MS/MS fragments obtained by HCD; 0.6 Da tolerance was allowed. Peptide score threshold provided from MASCOT software to evaluate quality of matches for MS/MS data was 41. No single peptide identification, even if unique, with an ion score lower than 41, was accepted.
RESULTS
Selenium Uptake by L. reuteri Lb2 BM-DSM 16143 During Growth
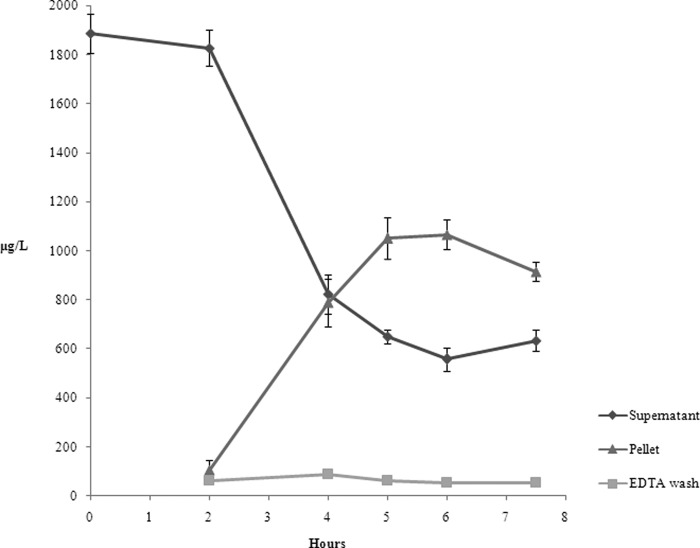
The ICP-MS analyses were performed on the pellet, supernatant and EDTA wash recovered from cultures of L. reuteri grown as described in the Experimental section. Because preliminary results (data not shown) indicated that there was no uptake of Se from the medium during the lag phase, attention was focused on the exponential growth phase. Fig. 1 reports the Se concentration (expressed in μg L−1) for each sample during L. reuteri growth. It reveals that all the selenium added to the medium before the inoculum (2 mg L−1) was completely dissolved and therefore bio-available for the strain. After 2 h growth a slight decrease of Se in the medium was observed, even if a significant uptake occurred only 4 h after the inoculum. The maximal internalized Se concentration was about 1150 μg L−1 after 5 h growth (middle exponential phase), whereas about 600 μg L−1 Se was not internalized. The increase of Se concentration in the pellet was consistent with the progressive decrease in the supernatant. Virtually no Se was measured in the EDTA wash during L. reuteri growth. The analytical protocol was validated by the mass balance of selenium. The sum of the Se amounts determined in the three analyzed samples equaled the concentration added to the medium. There was no Se loss during the experimental steps.
Fig. 1.
Results of the ICP-MS analyses of pellet, supernatant, and EDTA wash samples collected at different times during the exponential growth phase of L. reuteri Lb2 BM (lag phase is not shown). The Se concentration, expressed in μg/L, refers to the solution after digestion.
Detection of Selenium-containing Proteins in the Gels by Laser Ablation - ICP MS
To locate rapidly and precisely the selenium-containing proteins in large 2D electrophoresis gels, a sample was divided in four aliquots. Two of them were analyzed by 1D SDS-PAGE - LA-ICP-MS and 1D IEF - LA-ICP-MS, and the two others by 2D IEF-SDS-PAGE electrophoresis. Different selenium isotopes (77Se 7.63%, 78Se 23.78%, and 80Se 49.61%) were monitored. The 80Se isotope gives the highest signal but the background was also high and difficult to control. Therefore, 78Se was chosen for analysis.
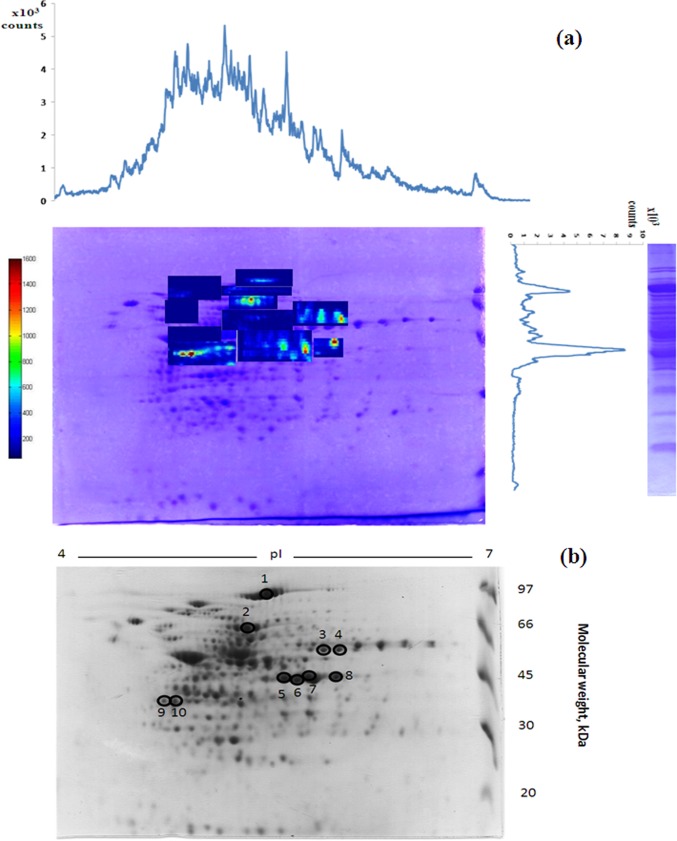
The LA- ICP MS electropherograms of 1D gels allowed the identification of the areas in the 2D gel supposed to contain proteins having accumulated the highest amount in Se. The most intense signals in the IEF were in the range of 4.5–6 pI and SDS -PAGE indicated the presence of major peaks in the 45–97 kDa mass range. The areas corresponding to these characteristics in the 2D gel were submitted to LA - ICP MS imaging analysis to localize precisely the position of the Se-bearing proteins. The approach is illustrated in Fig. 2A. The subsequent analysis focused on the 10 proteins which were the richest in terms of the accumulated selenium content.
Fig. 2.
A, Imaging of Se-containing protein spots in a 2D SDS-PAGE gel of a L. reuteri Lb2 BM soluble extract by Laser Ablation ICP MS. The to be imaged areas were defined on the basis of 1D (IEF or SDS) LA - ICP MS electropherograms. B, A parallel 2D SDS-PAGE of L. reuteri Lb2 BM soluble extract prepared for proteomics analysis. Circles and numbers refer to spots which were excised and further analyzed. (cf. Table I).
Identification of the Selenium-containing Proteins
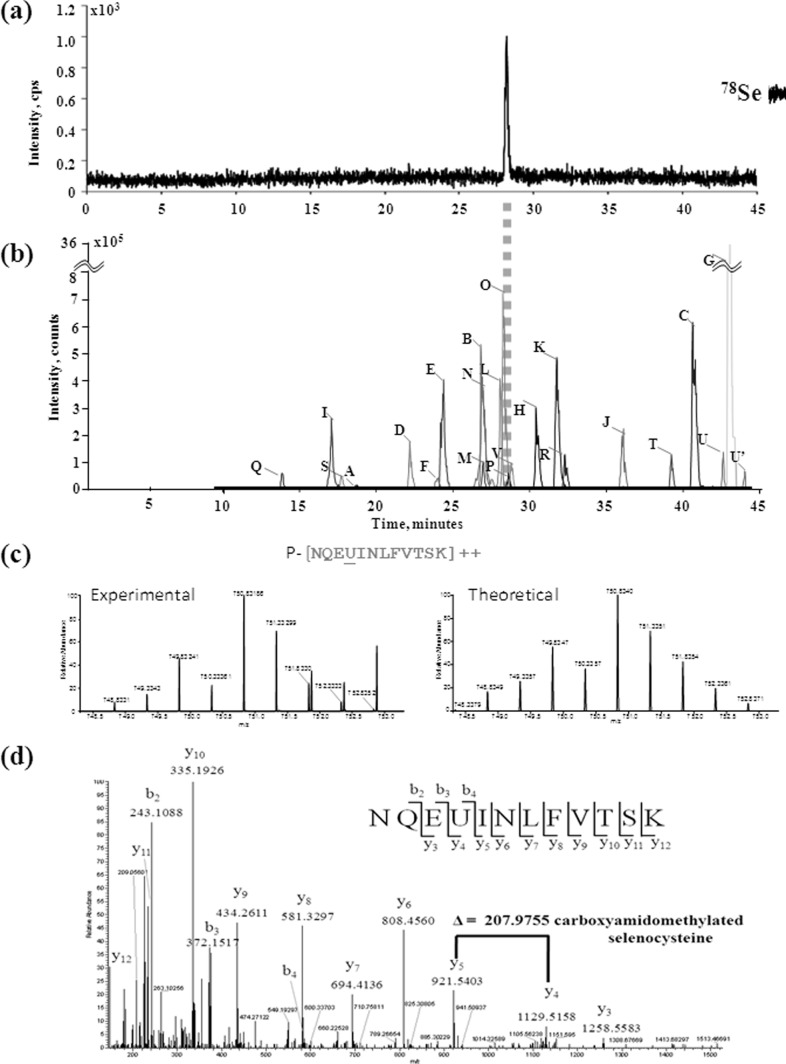
The areas corresponding to the 10 most intense Se spots were excised from the 2D electrophoretic gel, which had not undergone the LA-ICP MS imaging (Fig. 2B), de-stained and digested with trypsin. The identification procedure schematically illustrated in Fig. 3 targets in particular the Se-containing peptides which are detected after capillary HPLC separation in parallel by ICP MS and electrospray Orbitrap MS. The 78Se isotope intensity was measured on-line by ICP MS in order to detect selenium-containing peptides (Fig. 3A). The ICP MS detection is essential to guarantee that no Se-bearing species escapes ESI MS detection, and thus to conclude on the speciation of all the selenium in the proteins under study.
Fig. 3.
Example set of results from the analytical workflow (spot 1). A, Detection of selenocysteine-containing peptide NQEUINLFVTSK by capHPLC-ICPMS; B, XIC chromatograms of other tryptic peptides of the Se-containing protein (phosphoketolase). Note the correspondence of the retention times of the selenocysteine-containing peptide NQEUINLFVTSK using ICP MS and electrospray LTQ Orbitrap. The complete fingerprint of identified protein (all peptides from letter A to V) is given in a supplemental Table S1); C, comparison between the observed isotopic pattern and theoretical prediction of selenocysteine-containing peptide NQEUINLFVTSK; D, unambiguous identification of the Se cysteine-containing peptide NQEUINLFVTSK after interpretation of MS/MS spectrum of the double charged ion at m/z 750.8326.
The HPLC - Orbitrap ESI MS/MS analysis in the same chromatographic conditions allowed the identification of the Se-containing peptide(s) (Figs. 3C, 3D) and a number of other peptides (Fig. 3B) allowing the identification of the protein as in a canonical bottom up approach. As an example Fig. 3C shows the MS/MS spectrum of the selenocysteine-containing peptide from spot 1. From the interpretation of the MS/MS spectrum it was possible to localize the modification site at level of Cys 151 within the sequence of the phosphoketolase enzyme. The comparison between the retention time of putative selenocysteine-containing peptide from capillary HPLC with parallel detection ICP MS (Fig. 3A) and electrospray LTQ Orbitrap (Fig. 3D) is reported, showing a perfect correspondence of the peaks related to peptides containing selenocysteine. Similar results were obtained for all the putative selenocysteine-containing peptides detected in different protein spots.
The raw data from mass spectrometry were used for the identification of protein through the MASCOT online software, obtaining good results in terms of total protein score and sequence coverage the identified proteins and all the selenopeptides are reported in Table I.
Table I. List of the identified protein. For GAPDH and RihC in table are reported the results obtained from spot 8 and 10 respectively. Number of peptides, Peptide sequences and Individual ion scores are reported in supplementary Table S1.
| Spot | Protein | NCBI ID | MW (Da) | pI | Score | Seq. cov. (%) | Peptide sequence | Obs. mass | Theo. mass | Δm (ppm) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Phosphoketolase | gi 148544892 | 91346 | 5.09 | 2888 | 39 | NQEUINLFVTSK | 750,8326 | 750,8334 | 1,0655 |
| 2 | Pyruvate kinase | gi 148543982 | 51793 | 5.06 | 4077 | 72 | ELVUHVLNHGVLGS | 869,4182 | 869,4185 | 0,3451 |
| UNELGKPVITATQMLDSMQENPRPTR | 759,3545 | 759,3552 | 0,9218 | |||||||
| 3 | Arginine deiminase | gi 148543680 | 46214 | 5.39 | 1803 | 73 | UMoxSCPIVR | 543,7015 | 543,7023 | 1,4714 |
| UMSCPIVR | 535,7056 | 535,7049 | −1,3067 | |||||||
| DQQAUIGDGITINHMTFK | 699,6362 | 699,6373 | 1,5722 | |||||||
| 4 | 6-Phosphogluconate dehydrogenase | gi 184154309 | 53498 | 5.56 | 2005 | 56 | AGUIIR | 369,1643 | 369,1640 | −0,8126 |
| AEEDGKPUVAYIGPNGAGHYVK | 794,0203 | 794,0206 | 0,3778 | |||||||
| 5,7,8 | Glyceraldehyde 3-phosphate dehydrogenase | gi 184153036 | 35971 | 5.41 | 2913 | 71 | DDIIVSAGSUTTSCLAPMAK | 1072,9598 | 1072,9589 | −0,8388 |
| NDGVDFVLEUTGFYTSAEK | 1100,4534 | 1100,4528 | −0,5452 | |||||||
| 6 | Ornithine carbamoyltransferase | gi 148543661 | 37536 | 5.21 | 1791 | 58 | USFEVGAK | 473,1824 | 473,1826 | 0,4227 |
| USFEVGAKDEGAHVTYLGPSGSHIGHK | 963,4326 | 963,4337 | 1,1417 | |||||||
| ATENPNVLFEHULPAFHNLDTEVGK | 725,8341 | 725,8334 | −0,9644 | |||||||
| 9,10 | Ribonucleoside hydrolase RihC | gi 148543341 | 32552 | 4.70 | 1211 | 60 | VULDIDAEYFNK | 767,8191 | 767,8200 | 1,1721 |
Speciation of Selenium: SeCys is the Only Form Detected
The incorporation of selenium into amino acids was evaluated by analyzing all the methionine- and cysteine-containing peptides, searching for characteristic mass shift on scan spectra in the 300–1200 m/z range. Selenium isotopic pattern was observed for all reported peptides, and it was correlated with the theoretical prediction; moreover high accuracy (< 2ppm) was achieved owing to the high resolution (60, 000) (Fig. 3B).
The fine sequencing of the selenopeptides was reached by manual interpretation of the fragmentation spectra obtained in both the automatic and manual mode. For some peptides it was not possible to obtain adequate information from MS/MS spectra obtained in the data dependent acquisition mode because of the presence of interfering ions or due to very low relative abundance of the selenopeptides. The search for selenocysteines and selenomethionines was processed manually, by adding the specific mass shift regarding the substitution Se/S (+23.97223 and +15.98149 for double and triple charged peptides).
It is worth noting that in some cases the fragmentation spectra of SeCys-containing peptides were obtained by further SIM experiments because of the poor relative intensity of m/z signals in the Orbitrap TIC. However, when the fragmentation spectra of selenocysteine-containing peptides were acquired in automatically LC-MS/MS, the addition of the specific modification in Mascot, allowed the identification of SeCys as a modification, as reported in the supplemental Table S1.
The retention-times of all the SeCys-containing peptides were matched by the elution of Se observed in HPLC-ICP MS and all the peaks in HPLC-ICP MS chromatograms could be matched by an MS/MS identification. It can therefore be concluded that all the Se present in the 10 richest in terms of the Se-content soluble proteins was in the form of selenocysteine. No selenomethionine was detected.
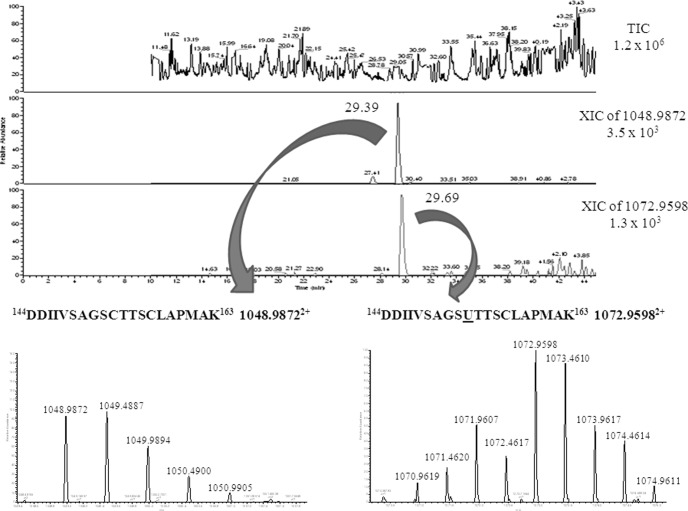
Sulfur-containing homologies of all the Se-containing peptides were found at concentrations exceeding a factor of 5. The difference of the retention time between the selenopeptides and the corresponding sulfur peptides was ca. 15s (Fig. 4).
Fig. 4.
Comparison between the chromatographic behavior of a Se-containing peptide (144DDIIVSAGSUTTSCLAPMAK163 1072. 95982+) and its sulfur analog (144DDIIVSAGSCTTSCLAPMAK163 1048.98732+) (spot 8).
The substitution Se/S seems to occur in specific cysteine residues within protein sequence. An explicative example is represented by GAPDH (spots 5, 7, 8) and arginine deiminase (spot 3) where only two of the three cysteine residues were substituted.
DISCUSSION
Experiments on selenium uptake by L. reuteri Lb2 BM-DSM 16143 during growth demonstrated the ability of the strain to recover selenium from the medium. Not all the Se added was internalized; moreover, the Se concentration used in this study negatively affected L. reuteri growth (5). Se concentration and culture conditions can probably be optimized in order to reduce selenium-induced stress on bacteria, especially for nutraceutical applications. It was previously demonstrated that the exceeding Se is partly released by L. reuteri in the form of surface-associated Se(0) particles (5). However, in this study no detectable increase in Se concentration was observed in the pellet wash, suggesting that the internalized selenium was metabolized mainly in a different way from the detoxification mechanism, and inserted into proteins. Moreover, the decrease of Se concentration in the biomass and the corresponding increase in the medium after 6 h growth suggests that internalized selenium is released into the external environment, probably incorporated into proteins.
Size-exclusion LC fractionation of the soluble protein extract using ICP MS detection indicated (data not shown) that about half of the internalized Se was covalently incorporated into proteins. An in depth investigation of the Se-incorporation in protein by 2D GE - LA-ICPMS, demonstrated that in this L. reuteri strain all the Se present in the proteins is incorporated as selenocysteine (SeCys), whereas methionine/selenomethionine substitution, which is typical for other microorganisms, such as yeast (25), did not occur. Note also that, in contrast to literature (26), the method developed here allows the preservation of the SeCys moiety during analysis Indeed, no dehydroalanine residues were detected.
The results corroborate data obtained by autoradiography using 75Se and 14C-carboxymethylseleno-cysteine indicating that lactic acid bacteria are able to incorporate selenium into intracellular proteins only as SeCys (27). SeCys2 (seleno-cystine, containing a di-selenide bridge) and MeSeCys (methyl-selenocysteine) were also the predominant organic forms of selenium detected in the Se-enriched yogurt containing Lactobacillus species (28, 29).
Our data determine the exact position within the primary sequence of proteins in which SeCys is inserted. SeCys was found in two glycolytic enzymes (glyceraldehyde 3-phosphate dehydrogenase, spots 5, 7, 8; pyruvate kinase, spot 2), two pentose phosphate pathway enzymes (phosphoketolase, spot 1; 6-phosphogluconate dehydrogenase, spot 4), two ADI pathway enzymes (arginine deiminase, spot 3; ornithine carbamoyltransferase, spot 6) and a ribonucleoside hydrolase RihC (spots 9, 10) in this L. reuteri strain. In some of these enzymes (PKP, PK, 6PGD, OTCase and RihC) all the cysteine residues reported in the sequence were modified into selenocysteine. In contrast to that, GAPDH and ADI behave in a different way: although they have in the primary sequence three Cys residues but in both enzymes only two of them are replaced by SeCys. In both cases two cysteine residues were present in the same peptide, 144DDIIVSAGSCTTSCLAPMAK163 for GAPDH and 398CMSCPIVR405 for ADI, both belonging to the active site, but only one of the two Cys (the underlined one) is replaced by SeCys (30, 31). Curiously, in each enzyme it was the catalytic Cys to be replaced (153Cys/153SeCys in GAPDH and 398Cys/398SeCys in ADI). Therefore, the catalytic Cys proved to be the most affected by selenization, suggesting that not all the Cys possess the same tendency to be converted into SeCys. It is worth noting that the presence of a SeCys residue in these sites could modify the catalytic properties of the enzymes, because the pka of SeCys (5.2) is much lower than that of Cys (8.3) (32). The presence of Se as SeCys in GAPDH was previously reported by Lacourciere et al. which demonstrated that selenium was retained by GAPDH after denaturation, suggesting its role as a selenium delivery protein (10).
Our data demonstrate that in this L. reuteri strain Se insertion into proteins exclusively occurs as selenocysteine, although the insertion mechanism still remains to be determined. Because in GAPDH and ADI there is a specific substitution of only two of the three Cys residues, it can be suggested that the insertion of SeCys can be genetically encoded, according to the mechanism used by other bacteria (4, 16). This hypothesis should be verified by analyzing gene sequences of the identified proteins searching for UGA codon and putative SECIS elements. However, the prediction of UGA codon and SECIS elements in prokaryotic cells is difficult, because most of the predictive tools are set for eukaryotic systems. An attempt to find a specific SeCys insertion sequence using the bSECISearch tool developed by Zhang and Gladyshev (33) carried out here did not give any positive result.
In this article, it was demonstrated that L. reuteri Lb2 BM-DSM 16143 is able to uptake inorganic Se from the medium and to metabolize it into an organic form incorporated into proteins that are crucial for the bacterial energy metabolism. The peculiarity of this strain is its ability to exclusively insert Se into selenocysteine. To the best of our knowledge, this is the first study that describes this event in a probiotic lactic acid bacterium, specifically identifying the sites in which cysteine/selenocysteine substitution occurs. L. reuteri Lb2 BM-DSM 16143 can therefore become a viable alternative to Se-rich yeast as a food supplement in Se-deficient subjects.
Supplementary Material
Footnotes
* This work was supported by Programma Operativo Nazionale “Ricerca e Competitività 2007–2013” PON01_01802” and PON01_00117.
 This article contains supplemental Table S1.
This article contains supplemental Table S1.
1 The abbreviations used are:
- Se
- selenium
- ICP-MS
- inductively coupled plasma MS
- LA
- laser ablation
- 2D
- two dimensional
- ESI
- electrospray ionization
- HCD
- high collisional dissociation.
REFERENCES
- 1. Burk R. F. (2002) Selenium, an antioxidant nutrient. Nutr. Clin. Care 5, 75–79 [DOI] [PubMed] [Google Scholar]
- 2. Singhal N., Austin J. (2002) A clinical review of micronutrients in HIV infection. J Int Assoc Physicians AIDS Care 1, 63–75 [DOI] [PubMed] [Google Scholar]
- 3. Rayman M. P. (2012) Selenium and human health. Lancet 379, 1256–1268 [DOI] [PubMed] [Google Scholar]
- 4. Turner R. J., Weiner J. H., Taylor D. E. (1998) Selenium metabolism in Escherichia coli. Biometals 11, 223–227 [DOI] [PubMed] [Google Scholar]
- 5. Lamberti C., Mangiapane E., Pessione A., Mazzoli R., Giunta C., Pessione E. (2011) Proteomic characterization of a selenium-metabolizing probiotic Lactobacillus reuteri Lb2 BM for nutraceutical applications. Proteomics 11, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 6. Andreoni V., Luischi M. M., Cavalca M. L., Erba D., Ciappellano S. (2000) Selenite tolerance and accumulation in the Lactobacillus species. Ann. Microbiol. 50, 77–88 [Google Scholar]
- 7. Dobias J., Suvorova E. I., Bernier-Latmani R. (2011) Role of proteins in controlling selenium nanoparticle size. Nanotechnology 22, 1–9 [DOI] [PubMed] [Google Scholar]
- 8. Schrauzer G. N. (2000) Selenomethionine: A Review of Its Nutritional Significance, Metabolism and Toxicity. J. Nutr. 130, 1653–1656 [DOI] [PubMed] [Google Scholar]
- 9. McSheehy S., Kelly J., Tessier L., Mester Z. (2005) Identification of selenomethionine in selenized yeast using twodimensional liquid chromatography-mass spectrometry based proteomic analysis. Analyst 130, 35–37 [DOI] [PubMed] [Google Scholar]
- 10. Lacourciere G. M., Levine R. L., Stadtman T. C. (2002) Direct detection of potential selenium delivery proteins by using an Escherichia coli strain unable to incorporate selenium from selenite into proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 9150–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stolz J. F., Basu P., Santini J. M., Oremland R. S. (2006) Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130 [DOI] [PubMed] [Google Scholar]
- 12. Cone J. E., Del Rio R. M, Davis J. N., Stadtman T. C. (1976) Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc. Natl. Acad. Sci. U.S.A. 73, 2659–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wagner M., Sonntag D., Grimm R., Pich A., Eckerskorn C., Söhling B., Andreesen J. R. (1999) Substrate-specific selenoprotein B of glycine reductase from Eubacterium acidaminophilum. Biochemical and molecular analysis. Eur. J. Biochem. 260, 38–49 [DOI] [PubMed] [Google Scholar]
- 14. Kabisch U. C., Gräntzdörffer A., Schierhorn A., Rücknagel K. P., Andreesen J. R., Pich A. (1999) Identification of D-proline reductase from Clostridium sticklandii as a selenoenzyme and indications for a catalytically active pyruvoyl group derived from a cysteine residue by cleavage of a proprotein. J. Biol. Chem. 274, 8445–8454 [DOI] [PubMed] [Google Scholar]
- 15. Wagner R., Cammack R., Andreesen J. R. (1984) Purification and characterization of xanthine dehydrogenase from Clostridium acidiurici grown in the presence of selenium. Biochim. Biophys. Acta. 791, 63–74 [Google Scholar]
- 16. Stock T., Rother M. (2009) Selenoproteins in Archaea and Gram-positive bacteria. Biochim. Biophys. Acta. 1790, 1520–1532 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Romero H., Salinas G., Gladyshev V. N. (2006) Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 7, R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Talarico T. L., Casas I. A., Chung T. C., Dobrogosz W. J. (1988) Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32, 1854–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Whitehead K., Versalovic J., Roos S., Britton R. A. (2008) Genomic and genetic characterization of the Bile Stress Response of Probiotic Lactobacillus reuteri ATCC 55730. Appl. Environ. Microbiol. 74, 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin Y. P., Thibodeaux C. H., Pena J. A., Ferry G. D., Versalovic J. (2008) Probiotic Lactobacillus reuteri Suppress Proinflammatory Cytokines via c-Jun. Inflamm. Bowel Dis. 14, 1068–1083 [DOI] [PubMed] [Google Scholar]
- 21. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 22. Bianga J., Szpunar J. (2013) ICP-MS-assisted identification of selenium-containing proteins in 2D gels using a new capillary HPLC–ICP MS interface and Orbitrap tandem mass spectrometry. J. Analyt. Atomic Spectrom. 28, 288–292 [Google Scholar]
- 23. Bianga J., Ballihaut G., Pécheyran C., Touat Z., Preud'Homme H., Mounicou S., Chavatte L., Lobinski R., Szpunar J. (2012) Detection of selenoproteins in human cell extracts by laser ablation-ICP MS after separation by polyacrylamide gel electrophoresis and blotting. J. Analyt. Atomic Spectrom. 27, 25–32 [Google Scholar]
- 24. Schaumlöffel D., Ruiz Encinar J., Łobiński R. (2003) Development of a sheathless interface between reversed-phase capillary HPLC and ICPMS via a microflow total consumption nebulizer for selenopeptide mapping. Analyt. Chem. 75, 6837–6842 [DOI] [PubMed] [Google Scholar]
- 25. Rayman M. P. (2004) The use of high-selenium yeast to raise selenium status: how does it measure up? Br. J. Nutr. 92, 557–573 [DOI] [PubMed] [Google Scholar]
- 26. Ma S., Caprioli R. M., Hill K. E., Burk R. F. (2003) Loss of selenium from selenoproteins: conversion of selenocysteine to dehydroalanine in vitro. J. Am. Soc. Mass Spectrom. 14, 593–600 [DOI] [PubMed] [Google Scholar]
- 27. Calomme M., Hu J., Van Den Branden K., Vanden Berghe D. A. (1995) Seleno-Lactobacillus an organic selenium source. Biol. Trace Element Res. 47, 379–383 [DOI] [PubMed] [Google Scholar]
- 28. Alzate A., Cañas B., Pérez-Munguía S., Hernández-Mendoza H., Pérez-Conde C., Gutiérrez A. M., Cámara C. (2007) Evaluation of the inorganic selenium biotransformation in selenium-enriched yogurt by HPLC-ICP-MS. J. Agric. Food Chem. 55, 9776–9783 [DOI] [PubMed] [Google Scholar]
- 29. Alzate A., Fernández-Fernández A., Pérez-Conde M. C., Gutiérrez A. M., Cámara C. (2008) Comparison of biotransformation of inorganic selenium by Lactobacillus and Saccharomyces in lactic fermentation process of yogurt and kefir. J. Agric. Food Chem. 56, 8728–8736 [DOI] [PubMed] [Google Scholar]
- 30. Nakajima H., Amano W., Fujita A., Fukuhara A., Azuma Y. T., Hata F., Inui T., Takeuchi T. (2007) The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J. Biol. Chem. 282, 26562–26574 [DOI] [PubMed] [Google Scholar]
- 31. Lu X., Galkin A., Herzberg O., Dunaway-Mariano D. (2004) Arginine deiminase uses an active-site cysteine in nucleophilic catalysis of L-arginine hydrolysis. J. Am. Chem. Soc. 126, 5374–5375 [DOI] [PubMed] [Google Scholar]
- 32. Wessjohann L. A., Schneider A., Abbas M., Brandt W. (2007) Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 388, 997–1006 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y., Gladyshev V. N. (2005) An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 21, 2580–2589 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.