Abstract
Comprehensive and quantitative information of the thermophile proteome is an important source for understanding of the survival mechanism under high growth temperature. Thermoanaerobacter tengcongensis (T. tengcongensis), a typical anaerobic thermophilic eubacterium, was selected to quantitatively evaluate its protein abundance changes in response to four different temperatures. With optimized procedures of isobaric tags for relative and absolute quantitation quantitative proteomics (iTRAQ), such as peptide fractionation with high-pH reverse phase (RP) high performance liquid chromatography (HPLC), tandem MS acquisition mode in LTQ Orbitrap Velos MS, and evaluation of the quantification algorithms, high quality of the quantitative information of the peptides identified were acquired. In total, 1589 unique proteins were identified and defined 251 as the temperature-dependent proteins. Analysis of genomic locations toward the correspondent genes of these temperature-dependent proteins revealed that more than 30% were contiguous units with relevant biological functions, which are likely to form the operon structures in T. tengcongensis. The RNA sequencing (RNA-seq) data further demonstrated that these cluster genes were cotranscribed, and their mRNA abundance changes responding to temperature exhibited the similar trends as the proteomic results, suggesting that the temperature-dependent proteins are highly associated with the correspondent transcription status. Hence, the operon regulation is likely an energy-efficient mode for T. tengcongensis survival. In addition, evaluation to the functions of differential proteomes indicated that the abundance of the proteins participating in sulfur-respiration on the plasma membrane was decreased as the temperature increased, whereas the glycolysis-related protein abundance was increased. The energy supply in T. tengcongensis at high temperature is, therefore, speculated not mainly through the respiration chain reactions.
Thermophiles are organisms that live at relatively high temperatures approximately over 50 °C or more. Studies on the survival mechanisms of these organisms has drawn great attention, because the relevant knowledge helps us to understand how life can thrive under extreme temperatures, what their potential is in biotechnology, and whether anaerobic thermophiles contain information regarding the early evolutionary life forms on earth. Traditionally, most investigations have focused on the stability of protein structures or enzyme activity. Based on many crystal structures of thermophilic enzymes reported, several factors responsible for their thermo stability have been suggested, such as selected amino acid substitutions (1, 2), hydrophobic cores (3, 4), buried polar contacts and ion pairs (5, 6), as well as interactions among subunits (7, 8). On the other hand, it is becoming a realization that even though the thermal-tolerant mechanism for a single protein is completely elucidated, its biological significance is still partially understood, because the life activity of a bacterium relies heavily on the coordination of the molecular networks within cells. Overall, large-scale analyses, such as genomics, transcriptomics and proteomics, are emerging in the field of thermophile biology to investigate the importantly thermophilic factors.
Over 80 genomes of thermophilic prokaryotes have been completely sequenced and decoded in the NCBI database. On the basis of such genomics data, investigators are able to acquire much more information of thermophily, which is unavailable in the traditional databases. Using principal component analysis to deal with the genomic data from thermophilic and mesophilic bacilli, Takami et al. claimed that the G+C content in rRNA and asymmetric amino acid substitutions were possibly linked with the thermo-adaptation of the organisms (9). By integrating omic techniques such as, gene array and mass spectrometry, Trauger et al. monitored global molecular changes of Pyrococcus furiosus in response to temperature changes, and found 11 genes were significantly up-regulated and 107 genes were down-regulated when the culture temperature increased from 72 to 95 °C (10). It merits noting that with the proteomic techniques being rapidly developed, quantitatively and sensitively analyzing the temperature-dependent proteins is well recognized for exploring the thermophilic mechanism.
Thermoanaerobacter tengcongensis (T. tengcongensis) is a thermophilic eubacterium isolated from Tengchong, China. It is an anaerobic, Gram-negative, rod-shaped bacterium, and is able to survive in temperatures ranging from 50 to 80 °C (11). The genome sequence of T. tengcongensis was decoded in 2001, comprising 2.69 Mb in length and 2588 predicted proteins (12), and the protein profile of this thermophile at optimal temperature were carefully characterized (13). To evaluate the proteomic response to the culture temperatures, our laboratory measured the differential proteins at three temperatures, 55, 75, and 80 °C, using two-dimensional gel electrophoresis (2DE)1 and MALDI-TOF/TOF MS. A total of 23 unique proteins were found in abundance changes within the temperature range (14), including four proteins closely related with redox regulation and chaperon. Because of limitations of the technique, however, only a few of proteins with semiquantitative estimation for the differential 2DE spots responsive to temperature changes were obtained, which makes it difficult to draw a convincing conclusion regarding the molecular mechanism of thermo-survival for T. tengcongensis. This prompts us to employ a comprehensively quantitative proteomics strategy for seeking overall temperature-dependent proteins.
Isobaric tags for relative and absolute quantitation (iTRAQ) is a widely accepted quantitative proteomics that allows a single MS analysis with multiple samples, leading to significant reduction of the experimental errors generated from individual experiments (15). This method lowers chemical noise and improves quantification accuracy, benefits statistical analysis, and augments the data confidence (16, 17). On the other hand, we have to fully acknowledge that some technique challenges are critical to achieve the satisfactory data based on the current iTRAQ-labeling quantification protocol. First of all, because iTRAQ quantification is relied on to measure the labeled peptides containing reporter tags and peptides, a MS/MS spectrum ideally contains only a single peptide with minimum interference by other peptides (18). Hence, efficient separation for peptides and setting proper parameters in the mass spectrometer with high resolution are prerequisites to improve the iTRAQ data quality. Despite several quantitative algorithms available to analyze iTRAQ data, there is lack of a program generally accepted in the iTRAQ data analysis. Hence, on the basis of the high quality iTRAQ data achieved, systematic evaluation of their quantitative results elicited from different algorithms is helpful to correctly interpret the quantitative information.
In this study, we first optimized the experimental procedures for iTRAQ-based quantitative proteomics, by comparison of two different HPLC methods for peptide fractionation, examination of the performance of two different ion fragmentation modes in the LTQ Orbitrap Velos MS for peptide identification and quantification, and evaluation of the quantitative data estimated from different algorithms. We then cultured T. tengcongensis at four temperatures, 55, 65, 75, and 80 °C, and identified 1589 unique proteins in total, including 251 proteins significantly changed in their abundance responding to temperature rising. From analyzing the correspondent genes to these temperature-dependent proteins, we discovered that approximately one-third of the gene loci were clustered in the T. tengcongensis genome. We further employed the RNA-seq approach to confirm the findings, and deduced that some operon structures might play important roles in the adaptation of T. tengcongensis to a high temperature environment.
EXPERIMENTAL PROCEDURES
Culturing T. tengcongensis at Different Temperatures
T. tengcongensis strain MB4T was cultured in media as previously described (14). The bacteria were first inoculated at 75 °C to the log phase, then distributed to the media (at a ratio of 1:50) prewarmed at four different temperatures, 55, 65, 75, and 80 °C, respectively. The bacterial cells were harvested at log phase by centrifugation at 4000 × g at 4 °C, and the pellets were washed with the buffer of 50 mm Tris-HCl, pH 7.8. The washed bacteria were stored at −80 °C until use.
Protein Extraction and Peptide Labeling by iTRAQ
The harvested T. tengcongensis were lysed in the buffer, containing 8 m urea, 4% CHAPS, 10 mm dithiotreitol, and 40 mm Tris-HCl, pH 8.0, with sonication in ice. After centrifugation at 12,000 × g at 4 °C, the supernatants were reduced and alkylated by 10 mm dithiotreitol and 55 mm iodoacetamide. The treated proteins were precipitated in 80% acetone at −20 °C overnight, and the precipitants were resuspended in 0.8 m urea and 500 mm tetraethylammonium bicarbonate (TEAB), pH 8.5. The protein concentrations were determined using the Bradford method followed by a 16 h trypsin digestion at 37 °C. The tryptic peptides were labeled by the 8-plex iTRAQ reagents (AB Sciex, Foster City, CA) following the manufacturer's protocol. After 2 h of labeling reactions, the reaction solvents were removed by Speed-vacuum, and the labeled peptides were dissolved in 20 mm NH4FA, pH 10, for the following experiments.
Peptide Fractionation by the Two Different HPLC Approaches
To evaluate the fractionation efficiency, two different HPLC approaches were adopted, high-pH RP (19) and strong cation exchange (SCX), to separate the bacterial peptides. In the high-pH RP HPLC procedure, the bacterial peptides were loaded onto a RP column (Luna C18, 4.6 mm inner diameter × 250 mm length, Phenomenex, CA), and eluted by a step linear elution program, 0–10 min equilibrated in 100% solution A (2% acetonitrile (can) and 20 mm NH4FA, pH 10), 10–15 min fast elution from 0–12% of solution B (80% ACN and 20 mm NH4FA, pH 10), 15–50 min linear elution from 12–56% of solution B; and 50–55 min washing elution from 56–80% of solution B. In the SCX HPLC procedure, the bacterial peptides were loaded onto a SCX column (Luna SCX, 4.6 mm × 250 mm, Phenomenex), and eluted by step linear elution program, 0–10 min equilibrated in buffer A (25% ACN, 20 mm KCl and 10 mm KH2PO4, pH 3.0), 10–15 min fast elution from 0–5% of buffer B (25% ACN, 1 m KCl and 10 mm KH2PO4, pH 3.0), 15–50 min linear elution from 5–30% of solution B; and 50–55 min washing elution from 30–80% of solution B. All the HPLC procedures were manipulated in a Prominence HPLC system (Shimadzu, Nakagyo-ku, Kyoto, Japan) with the flow rate at 1.0 ml/min and the peptides were monitored at 214 nm. The fractioned peptides were collected at one tube/min during the linear elution period.
Peptide Identification by Nano RP HPLC and Mass Spectrometry
The peptide contents in the collected fractions were first evaluated by MALDI-TOF/TOF MS (Bruker Daltonics, Billerica, MA), and the fractions were further pooled to average the peptide content. The eluted fractions were delivered onto a nano RP column (5-μm Hypersil C18, 75 μm × 150 mm, Thermo Fisher Scientific, Waltham, MA) mounted in a Prominence Nano HPLC system (Shimadzu, Nakagyo-ku, Kyoto, Japan), and were eluted with ACN gradient from 5–40% containing 0.1% formic acid, for 95 min at 400 nL/min. The eluates were directly entered LTQ Orbitrap Velos MS (Thermo Fisher Scientific), setting in positive ion mode and data-dependent manner with full MS scan from 350–1800 m/z, resolution at 60,000, MS/MS scan with minimum signal threshold 5000, isolation width at 2 Da. To evaluate the performance of this mass spectrometry on the iTRAQ labeled samples, two MS/MS acquisition modes, higher collision energy dissociation (HCD) and collision induce dissociation (CID), were employed. And to optimize the MS/MS acquisition efficiency of HCD, normalized collision energy (NCE) was systemically examined from 25–70%.
Database Searches for Peptide and Protein Identification
The raw MS/MS data were converted into MGF format by Proteome Discoverer 1.2 (Thermo Fisher Scientific, Waltham, MA). And the exported MGF files were searched by Mascot 2.3 (Matrix Science, Boston, MA) against the database with all 2588 predicted proteins in T. tengcongensis downloaded from NCBI (NCBI reference sequence: NC_003869.1). An automatic decoy database search was performed. Several parameters in Mascot were set for peptide searching, including iTRAQ 8-plex for quantification, tolerance of one missed cleavage of trypsin, carbamidomethylation for cysteine as fixed modification, oxidation for methionine as variable modification. The precursor mass tolerance was 10 ppm, and the product ion tolerance was either 0.5 Da for CID in LTQ or 0.02 Da for HCD in Orbitrap, respectively.
Quantitative Data Analysis for the iTRAQ Labeling Peptides
A unique protein with at least two unique peptides, with a false discovery rate (FDR) < 0.01, was qualified for further quantification data analysis. The fold changes in protein abundance were defined as the median ratio of all significantly matched spectra with tag signals. For the sake of high quality in quantitative analysis, especially for the T. tengcongensis peptides labeled with iTRAQ, three software programs, Isobar (20), IsobariQ (21), and ProteinPilot (AB Sciex, Foster City, CA), were carefully evaluated against the same original MS/MS data. Based on the software analysis, the CVs distribution of all the quantified proteins and the quantitative results derived from duplicated injections were compared in parallel. The software with the best performance of the quantitative analysis was chosen for whole data analysis in this study.
RNA-seq to the T. tengcongensis mRNA
The total RNAs were prepared with the Trizol reagent according to the manufacturer's instructions (Invitrogen, Grand Island, NY), and the integrity of total RNA was checked using Agilent Technologies 2100 Bioanalyzer (Agilent, Santa Clara, CA). After removing rRNA using mRNA-only prokaryotic mRNA isolation kit (Epicenter Biotech, Madison, WI), the remaining RNAs were incubated with the fragmentation buffer to interrupt RNAs to short fragments. Random hexamer-primers were used to synthesize the first-strand cDNA, and the second-strand cDNA was synthesized with DNA polymerase I. The short fragments were purified with QIAquick PCR extraction kit (Qiagen, Valencia, CA), and were ligated to Illumina sequencing adapters (Illumina, San Diego, CA), followed by amplification with PCR. Using Illumina HiSeqTM 2000 (Illumina), all the RNAs with adapters were sequenced (expected library size: 200 bp; read length: 90 nt; sequencing strategy: paired-end sequencing), and the qualified sequences were then mapped to the T. tengcongensis genome sequences using SOAP2 (22) with no more than two mismatched bases. The mRNA abundance was estimated by the number of uniquely mapped reads per kilobase per million reads (RPKM) method (23).
The quantitative proteomics and transcriptomics data from T. tengcongensis that we described here are available from the GigaScience database (24), the NCBI database with accession number SRP022748, and the ProteomeXchange with identifier PXD000264.
RESULTS
Optimizing the Approaches for Quantifying the Temperature-dependent Proteins Labeled with iTRAQ
At the initial phase of this study, we systematically optimized the experimental approaches to achieve high quality of quantitative proteomes. With regard to peptide separation, as shown in supplemental Fig. S1, compared with SCX/low-pH RP MS, high-pH RP/low-pH RP MS exhibited three technical advantages, 1) significantly higher number of the bacterial proteins identified, approximately over one fold, 2) almost covering the proteins identified by SCX/low-pH RP MS, ∼95%, and 3) relatively lower overlapped rates in the neighbor fractions. Therefore, the liquid chromatography of high-pH RP/low-pH RP was conducted for peptide separation in this study.
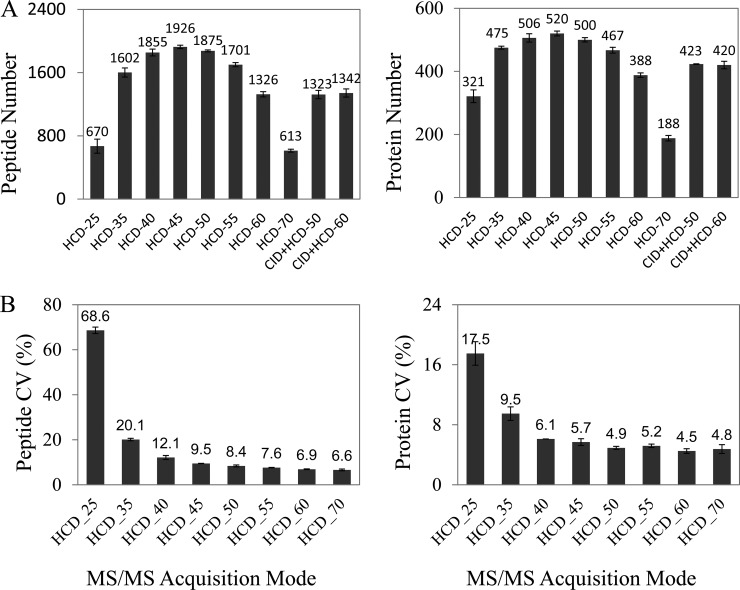
Dayon et al. suggested that combining CID and HCD activation modes in LTQ Orbitrap XL MS was applicable in iTRAQ quantification, because the CID spectra were beneficial for protein identification, whereas the HCD spectra were sensitive for the mass signal of reporter tags (25). However, because of the technique being improved in LTQ Orbitrap Velos MS (26, 27), we further inquired as to whether the performance in peptide detection of the CID plus HCD mode was different from the mode of HCD alone. As presented in Fig. 1A, the identification rates, either peptides or proteins, in HCD mode alone were significantly higher than that of CID plus HCD mode, indicating that the improved HCD mode is not only favorable in reporter tag quantification, but is also beneficial for peptide identification. We evaluated the identification results of the iTRAQ-labeled peptides in HCD mode with different NCE from 25% to 70%. As shown in Fig. 1A and 1B, 45% NCE resulted in the most identified peptides and proteins in all the NCEs, and at such NCE, the CVs were ∼10% for peptides and 5% for proteins, which are well accepted for the accurate iTRAQ quantification.
Fig. 1.
Optimizing the MS/MS settings in LTQ Orbitrap Velos MS for identification and quantification of the iTRAQ-labeled peptides. A, Comparison of the identified unique peptides (left panel) or proteins (right panel) between CID and HCD acquisition mode. B, Comparison of the CVs of the quantitative signals for the identified unique peptides (left panel) or proteins (right panel) using the different NCEs upon HCD acquisition mode.
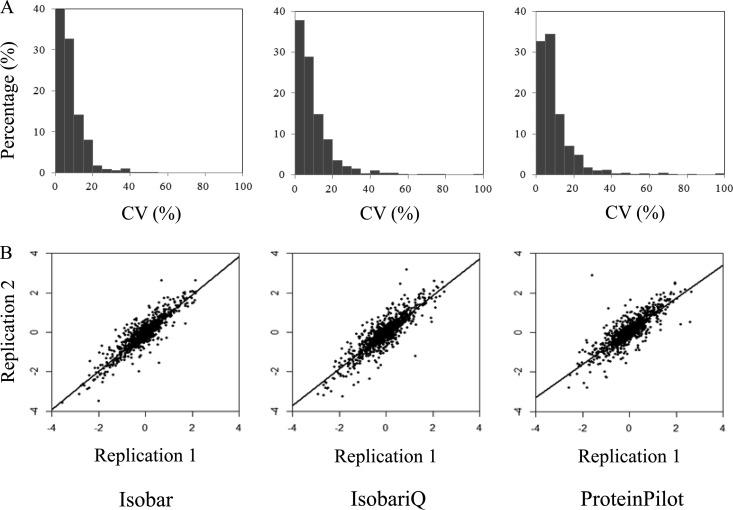
Considering no algorithm for analyzing iTRAQ data is generally accepted yet, we examined the same set of data for the iTRAQ labeled peptides of T. tengcongensis in parallel with three programs that are reported in iTRAQ data analysis, Isobar, IsobariQ, and ProteinPilot. Fig. 2 summarizes the typical evaluation results based on relative quantification between the tag signals of 116 and 114. Clearly, the CVs of the quantified proteins calculated by Isobar were lower than the values obtained from the other two programs. When the threshold of the CV was set at 20%, 96% of the quantified proteins estimated by Isobar analysis had fallen into range, whereas 90 and 89% of the quantified proteins analyzed with IsobariQ and ProteinPilot were within the range, respectively. As illustrated in Fig. 2B, the Pearson correlation was used to compare the duplicated quantification results. Although the slopes of the linear regressions were quite close, the correlation coefficients estimated by Isobar, IsobariQ, and ProteinPilot were still different from each other, 0.943, 0.919, and 0.893, respectively. We thus took Isobar for the next iTRAQ data quantitative analysis in this study.
Fig. 2.
Evaluation of three software programs, Isobar, IsobariQ and ProteinPilot, for quantitative analysis to the iTRAQ-labeled peptides. A data set of raw MS/MS was made for quantitative comparison of the differential proteins in T. tengcongensis cultured at 55 and 75 °C. The three software programs were employed to analyze the same dataset. A, The CVs distribution for all the proteins quantitatively determined. B, The correlation of the quantitative values for all the identified proteins between replicated injections.
Quantitative Estimation of the Temperature-dependent Proteins With iTRAQ Labeling
The digested peptide pool, consisting of equal amounts of iTRAQ labeled peptides collected from the four temperatures, was loaded to high-pH RP HPLC and separated to 36 eluted fractions. The eluted fractions were loaded on the nano RP HPLC coupled with LTQ Orbitrap Velos MS. In total, 732,234 MS/MS spectra were achieved from the two sample preparations and multiple loadings. Of the MS/MS spectra, ∼23% were identified as peptides (FDR<0.01), whereas less than 0.5% of the identified peptides were absent with the reporter tags. All the information regarding peptide or protein identification and quantification derived from Mascot and Isobar are shown in supplemental Tables S1, S2, S3A, and S3B. The reproducibility of identified peptides from multiple experiments (four technique replicates for the first sample preparation and three times for the second one) is summarized in supplemental Table S4, demonstrating that the overlapping rates for peptides and proteins were approximately 70 and 90%, respectively. A total of 1589 proteins, elicited from 13,223 unique peptides, were identified with quantitative information, which covered 61.4% of the 2588 predicted proteins of the T. tengcongensis genome.
In addition, we set four prerequisites to acquire the significant temperature-dependent proteins of T. tengcongensis. First, to avoid bias of iTRAQ labeling, 113 and 114 were alternatively labeled to the samples obtained from the 55 °C sample that served as the reference for estimation of the relative quantitative proteome. Second, a qualified protein for quantitative analysis should have at least two unique peptides labeled with iTRAQ. Third, correction of isotope impurities and normalization of intensity median were considered for judging the differential proteome. Finally, of all the seven replication experiments, a temperature-dependent protein should appear in the same change mode at least five times. According to the results statistically evaluated by Isobar, the threshold of fold changes in the relative abundance should be first defined to ensure which protein was significantly alternated in response to temperature change. Using volcano plot, a scatter-plot that allows evaluation of quantitative data from two parameters, such as fold change and the correspondent p values (28), we recognized that the threshold at fold change >1.5 and p value < 0.05 were representative to these T. tengcongensis proteins whose abundance significantly responded to the temperature change (supplemental Fig. S2). Next, it is reasoned that a temperature-dependent protein should represent its abundance change within a relatively large range of temperatures. We thus set the protein abundance at 55 °C as a reference, and defined a temperature-dependent protein that presents a significant abundance change against the reference regardless of which temperature was selected.
Through all the analysis above, as shown in supplemental Table S5, a total of 251 proteins were found to be temperature-dependent in T. tengcongensis, indicating that almost 10% of the bacterial proteins changed their abundance significantly in response to the increase in temperature. These proteins are generally divided into four classes according to the tendency modes of abundance change within the temperature range, 70 proteins were consecutively up-regulated, 117 were consecutively down-regulated, 44 were bell-regulated and 20 were in concave-regulated mode.
Functional Analysis Toward the Temperature-dependent Proteins
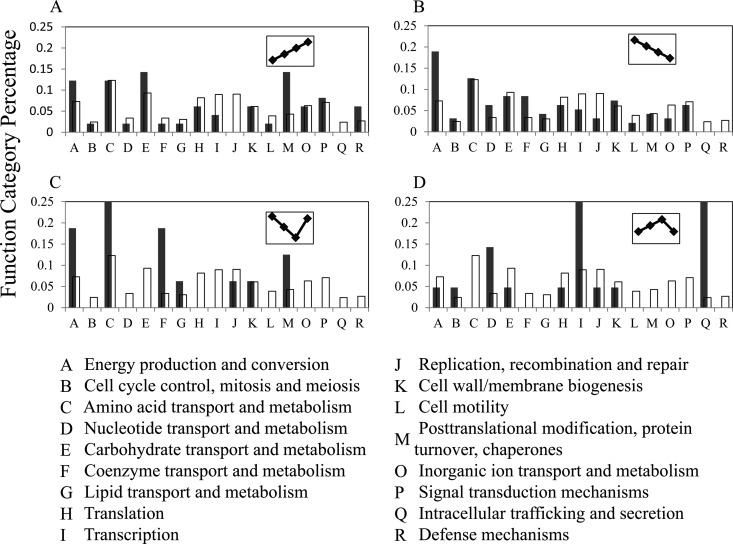
To understand the biological significance of the temperature-dependent proteins, a primary question is whether these proteins are relatively enriched in some functional groups. According to the analysis of Cluster of Orthologous Group (COG) (29), we could categorize the temperature-dependent proteins into 20 groups. Because three groups were not clearly defined in biological functions, the functional categories for the temperature-dependent proteins were simplified to 17 groups. And with the similar COG analysis, we analyzed the function distributions of the 2588 predicted proteins derived from the T. tengcongensis genome based on these 17 function groups. To find out if the proportion distributions of the functional genes in the temperature-dependent proteins were similar to that on the genomics scale in T. tengcongensis, we estimated the proportional distributions for the two data sets of the COG analysis, and further sought the statistical significance between the two distributions using the chi-square test with one degree of freedom. As shown in supplemental Table S6, the proportional distributions of the temperature-dependent proteins in four of the 17 functional groups were found significantly different from that at the genomic scale. This implies that some functional proteins rather than the whole genome gene expression are sensitive to temperature increases in T. tengcongensis. Specifically, in the consecutively up-regulated mode, the chaperon proteins occupy the largest portions as shown in Fig. 3A, including seven different chaperon proteins with the large changes at ∼5–50-fold (supplemental Table S5). This result is in good agreement with our early observation based on 2DE proteomics (14). The proteins participating in carbohydrate transport and metabolism (seven proteins) and energy production and conversion (six proteins) exhibit the higher portions as well. It is worth noting that all the proteins with higher COG distribution proportions mentioned above were identified in relatively larger numbers of MS/MS spectra, indicating that some T. tengcongensis proteins with higher absolute abundances are sensitive to temperature changes as well. In the consecutively down-regulated mode, similar to the up-regulated mode, some proteins involved in energy production and conversion have higher portion (18 proteins) as shown in Fig. 3B, whereas in contrast to the up-regulated mode, the proteins for nucleotide transport and metabolism (six proteins) and coenzyme transport and metabolism (eight proteins) display a higher percentile in COG distribution proportion. This leads to the assumption that the metabolisms related with energy production and conversion in T. tengcongensis may have multiple adaptive regulation, whereas more metabolic processes related to nucleotides and coenzymes may be required at relative lower temperature. In the concave-regulated mode shown in Fig. 3C, there is only one functional group, amino acid transport and metabolism, having the relative large number of the differential proteins (four proteins). Intriguingly, the maximum changes in protein abundance for most proteins in this mode present in the temperature range between 55 to 75 °C, whereas the differences of protein abundance between 75 to 80 °C group remain almost consistent in most cases. This means that the protein abundance at 55 °C in this mode is generally higher than that at higher temperatures. And in the bell-regulated mode, Fig. 3D, two functional groups contain relatively rich proteins response to temperature, transcription (six proteins), and intracellular trafficking and secretion (six proteins). All the proteins in this mode exhibit the highest abundance at 75 °C, and their abundance changes are relatively larger, ∼6–12-fold among the different temperatures. The analysis above toward the accurate quantitative proteomics thus provides a clue how to trace the functional molecules involved in the T. tengcongensis survival under high temperature.
Fig. 3.
Analysis of COGs to the temperature-dependent proteins of T. tengcongensis. The blank boxes represent distribution of the functional categories for the genes annotated from the T. tengcongensis genome, and the black boxes indicate distribution of the functional categories for the temperature-dependent proteins in this bacterium. A, The consecutively up-regulated proteins, B, the consecutively down-regulated proteins, C, the concave-regulated proteins, and D, the bell-regulated proteins.
Analysis of Genomic Locations Toward the Correspondent Genes of the Temperature-dependent Proteins
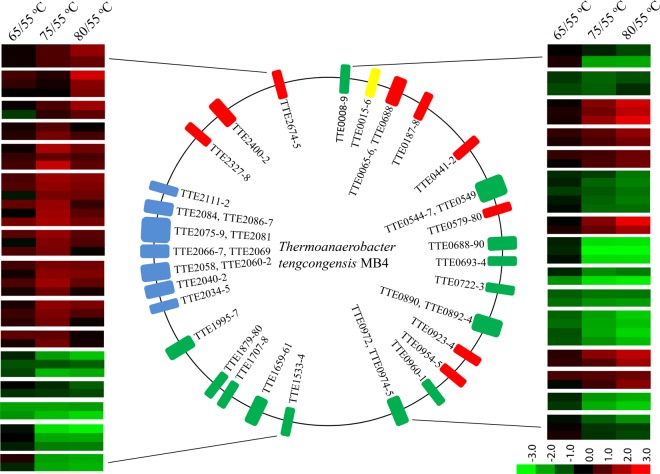
From localizing the genes derived from the temperature-dependent proteins onto the T. tengcongensis genome, we found a large portion of these genes clustered to certain regions, for instance, the genes in the consecutively down-regulated mode, TTE0544, TTE0545, TTE0546, TTE0547, and TTE0549 were located within a 6 kilobase pairs region. To classify the gene distribution of the temperature-dependent proteins acquired from the iTRAQ quantification experiment, we adopted the following filtration analysis. First of all, if a gene cluster is defined as at least two neighboring genes and tolerant to only one gene missed among at least three genes in a cluster, a total of 40 gene clusters covering 110 genes are delineated. Then these clusters are filtrated by two additional conditions, 1) the genes must be located on the same DNA sequence strand, either positive or negative, and 2) the distance between the neighboring members of a gene cluster must be within −50 and +100 base pairs. Thus, 30 gene clusters covering 80 genes are finally achieved as illustrated in Fig. 4 and supplemental Table S7. For instance, a cluster group with a higher density of temperature-dependent genes is located within length of 56 Kb, TTE2034 to TTE2112 (blue bars) including the seven gene clusters defined with 23 genes. Intriguingly, all the correspondent proteins exhibited the same abundance change tendency with the highest abundance at 75 °C. According to the COG analysis to these 23 genes, 11 of them have unknown functions. And for the rest, four proteins are transcription proteins, and five are from intracellular trafficking and secretion proteins.
Fig. 4.
The genomic localization for the genes correspondent to the temperature-dependent proteins that were filtered by cluster analysis. Central panel represents the genomics localization for the temperature-dependent proteins, red (the consecutively up-regulated proteins), blue (the bell-regulated proteins), yellow (the concave-regulated proteins), and green (the consecutively down-regulated proteins). Right and left panels represent the fold changes of abundance of each temperature-dependent protein. The right panel covers the cluster genes from TTE0008 to TTE0975, and the left panel contains the cluster genes from TTE1533 to TTE2675. The lower panel with color gradient represents the changes of protein abundance from down-regulated (green) to up-regulated (red).
Further analysis of the 30 gene clusters could bring some clues as to understanding how and why the temperature-dependent proteomes are generated. First, according to the iTRAQ quantitative data, the members in each gene cluster share similar abundance change modes in response to the culture temperature increase. Second, most of the gene clusters' members are expected to perform similar functions, such as in the up-regulated clusters, TTE0579 and TTE0580 as chaperon proteins, and TTE0065, TTE0066, and TTE0068 as cell wall and membrane biogenesis proteins, whereas in the down-regulated clusters, TTE0688, TTE0689, and TTE0690 as energy production and conversion proteins, and TTE1533 and TTE1534 as nucleotide transport and metabolism proteins. Third, with distance analysis of the neighboring genes to these clusters, most of the distances are within −10 to +50 base pairs as shown in supplemental Table S7, which are regular distances of operon structure genes in bacteria. Moreover, as compared with the cluster analysis on the operon data, Database of prOkaryotic OpeRons 2.0 (DOOR) (30, 31), more than 90% of the gene clusters defined by our filtration overlap with the theoretical prediction. All of these features lead to a hypothesis that gene transcription in some clusters is quite sensitive to temperature and regulation of the correspondent operons' activity and may exert the thermo-tolerant functions in T. tengcongensis.
Correlation Analysis Toward the Temperature-dependent mRNAs and Proteins
Generally it is accepted that mRNA formation is not always correlated with protein synthesis, particularly for middle and low abundance mRNAs and proteins (32). As a large amount of genes corresponding with the temperature-dependent proteins are likely to be elicited form the gene clusters, we suspect that transcription and translation of these genes may follow a synchronized mode in response to environmental temperature. If the hypothesis is correct, the correspondent mRNAs may display the similar modes in their abundance changes as the temperature-dependent proteins. To test it, we conducted the quantitative transcriptomics of T. tengcongensis by RNA-seq. With Illumina HiSeqTM 2000 sequencing and the raw data analysis by SOAP2, the RNA-seq data covered ∼97% (2501/2588) of the predicted genes, with more than two reads for each gene. Because the RNA-seq detection is much more sensitive than proteomic determination, the RNA-seq data is likely to delineate the entire figure of the cluster structures. In the 30 gene clusters defined, all of them were found in consecutive sequences of the mRNAs without any gap between two neighboring genes. Two typical operon structures derived from the RNA-seq data are illustrated in Fig. 5A, including the gene members and their abundance changes in response to temperature in the two clusters. We mapped the temperature-dependent mRNAs to the T. tengcongensis genome, and compared the modes of abundance changes due to temperature alteration for both mRNAs and proteins in the 30 clusters. As depicted in Fig. 5B which was generated by heat map analysis, of 80 temperature-dependent proteins in the 30 clusters, ∼90% (72/80) of them are well matched with the abundance change tendencies of mRNAs, whose abundance change folds are listed in supplemental Table S7. The results firmly support our hypothesis that the significant abundance changes of the temperature-dependent proteins in T. tengcongensis is tightly correlated with the transcriptive activity of some temperature-sensitive operons.
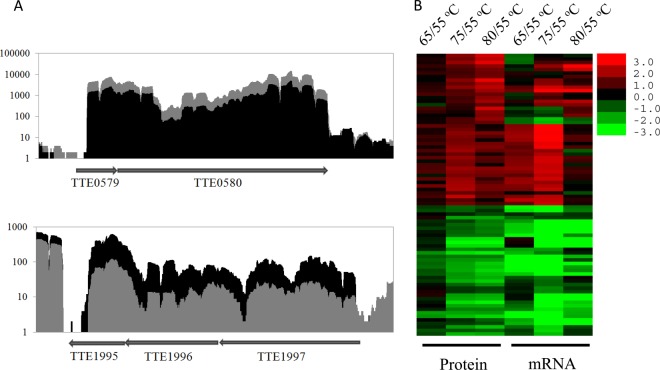
Fig. 5.
Correlation analysis of the quantitative mRNAs and proteins to the temperature-dependent proteins of T. tengcongensis. A, Sequence coverage plots based on the RNA-seq raw data to the two typical gene clusters elicited from the quantitative proteomics for the temperature-dependent proteins of T. tengcongensis. Upper panel shows the gene cluster of TTE0579-TTE0580 containing the consecutively up-regulated proteins; and lower panel shows the gene cluster of TTE1995-TTE1997 containing the consecutively down-regulated proteins. The gray shade represents the quantitative mRNAs at 75 °C, and the black shade represents the quantitative mRNAs at 55 °C. B, Heatmap analysis to the temperature-dependent proteins of T. tengcongensis on the basis of the relative abundance (log2 (ratios)) of mRNAs and proteins, i.e. the mRNA or protein abundance at certain temperature against the correspondent abundance at 55 °C. The small panel with color gradient represents the changes of protein abundance or mRNA abundance from down-regulated (green) to up-regulated (red).
Analysis of the Common Motifs in the Temperature-sensitive Operons
As a result of our hypothesis, we inquired as to whether there are common motifs shared by the temperature-sensitive operons. To find the motifs, we set more stringent criteria to select the temperature-sensitive operons. We selected only the clusters that contain the proteins with consistently increased or decreased abundance following temperature change. Also, the boundaries of the selected operon should well match the theoretical prediction of operons as well as the RNA-seq results. Thus, 16 operons were identfied after such filtration from the 30 clusters, seven in the up-regulated and nine in the down-regulated mode. We then chose the DNA sequences of −100 bp to the translation initiation site and conducted motif analysis using Multiple Em for Motif Elicitation (MEME) (33). The seven operons with the up-regulated mode, a sequence of GGGAGGGTAAAAGTA located at −70 to initiation sites was predicted in a common motif, while the nine operons with the down-regulated mode, two motifs of GAAGGAGA and TTTTGCAGAAAT located at −80 to −10 and −95 to −30 sites were predicted. The details of these three motifs are shown in supplemental Fig. S3. Using the algorithm of TOMTOM 4.9.0 (34), we further searched which DNA binding proteins could interact with these consensus sequences against the literature-based prokaryote regulatory interactions database, RegTransBase (35). Even though the search result was negative, no defined regulator was predicted to bind to these sequences. These consensus sequences thus lead to a new direction to explore the DNA interaction proteins with novel temperature-dependent motifs. Therefore, the quantitative proteomics to the T. tengcongensis cultured at different temperatures does not only provide the functional information to explore temperature-dependent proteins, but also offers the mechanical clues to understanding the expression regulation of these proteins.
DISCUSSION
As in the previous report, we employed 2DE and MALDI-TOF/TOF MS to explore the temperature-dependent proteins in T. tengcongensis cultured at three different temperatures, 55, 75, and 85 °C (14). In total, 23 unique proteins were selected as the temperature-dependent proteins. In the limited unique proteins, only four are related with energy production and two with chaperons. In contrast to the limited information, the iTRAQ approach described above indeed helps us to obtain much information that represents large numbers of the T. tengcongensis proteins with abundance changes during a wide range of culture temperatures. 1) With the strict cutoff settings, a total of 251 proteins were quantified as the temperature-dependent proteins, which included all the temperature-dependent proteins acquired in our earlier results. 2) Although the temperature-dependent proteins are broadly categorized to several functional groups, a large number of proteins with abundance changes mainly belong to limited functional groups, such as seven chaperon proteins with 5–50-fold abundance changes, 28 proteins involved in energy production and conversion, and 16 proteins participating in carbohydrate transport and metabolism. We reasoned that the improved quantification of proteomic analysis can build a solid base in further studies of thermophilic mechanisms.
Of the temperature-dependent proteins listed in supplemental Table S5, we found that more than 30% of the correspondent genes form the cluster structures on the T. tengcongensis genome. This feature led us to find out whether these temperature-dependent genes were structured as operon genes, which would be cotranscribed and coregulated as a single polycistronic mRNA. Therefore, we analyzed the correlation of the temperature-dependent genes at the transcription and translation level. The abundance correlation between mRNA and protein was reported to be poor after being systematically investigated in the yeast Saccharomyces cerevisiae by Ruedi Aebersold's group (32), and in the 23 human cell lines by Peter Nilsson group (36), however, some investigators claimed that the close correlations could be found between transcription and translation under some circumstances (37). For instance, with stringent data quality filtering, Lyris et al. found that in yeast the mRNA and protein abundance changes correlated very well for the abundance altered genes for some pathway components, whereas the overall correlation was quite poor (37). Sonya et al. observed that in Thalassiosira pseudonana, the stress-responsive up-regulated mRNA and protein were tightly correlated (38). In this study, we also found that 90% of the 80 cluster-distributed temperature-dependent proteins in T. tengcongensis displayed a similar abundance change mode as their corresponding mRNAs during temperature changes, which proved that the temperature-dependent responses of transcription and translation happened synchronically. Based on analyzing the solid data of quantitative mRNAs and proteins, we postulate that transcriptional regulation is likely an efficient way in which T. tengcongensis makes adaptive responses to environmental temperature changes.
It is a common phenomenon of cotranscription as well coregulation for the function-related genes in bacteria. As reported, E. coli possesses a set of specific defense genes against peroxides, mediated by the transcriptional regulator OxyR and SoxRS. Under oxidative stress, OxyR controls the gene expression such as HPI catalase, glutaredoxin, glutathione reductase, and NADPH-dependent alkyl hydroperoxide reductase, whereas SoxRS regulates at least ten genes' transcription abundance, including MnSOD, endonuclease IV, glucose-6-P DH, fumarase, aconitase, and ferredoxin reductase. Such global changes of gene expression lead to a coordinated and effective response that maintains the stability of the cellular equilibrium in E. coli under oxidative stress (39, 40). In our results, many of the genes belonging to different clusters, which have the same changing mode when temperature is increased, may perform similar biological functions. For example, six chaperon proteins, which located in three different clusters, TTE0579/TTE0580, TTE0954/TTE0955, and TTE2674/TTE2675, have high expression corresponding with rising temperature. These proteins have been well documented to perform protective function for proteins against thermal and other stress denaturation trends (41–44). We then come to the hypothesis that the temperature-dependent gene clusters with the similar regulation mode are regulated globally. And it reasoned that the upstream DNA sequences of these temperature-dependent clusters may contain some commonly conserved motifs that are potential binding sites for transcriptional regulators. As shown in supplemental Fig. S3, three conserved sequences are found in upstream of the temperature-dependent clusters. Discovery of the putative motifs supports our hypothesis, and also guides us to a new avenue to explore the regulatory proteins that should have high affinity to these DNA fragments.
The evidence revealing regulation of some T. tengcongensis operons responding to temperature changes deepens our knowledge of the structure and function of thermophile genomes. Analysis of the T. tengcongensis genome indicates that this bacterium has two thiamine pyrophosphate-dependent dehydrogenase complexes, TTE0186/TTE0187/TTE0188 and TTE0688/TTE0689/TTE0690, which are all composed of three proteins, two thiamine pyrophosphate (TPP) containing 2-oxoacid decarboxylase subunits (E1 alpha subunit, E1 beta subunit), and a lipoic acid-containing dihydrolipoamide acyltransferase (E2). These two complexes may function as pyruvate dehydrogenase complex (PDC) (45) or branched-chain alpha-keto acid dehydrogenase (BCKDH) complex (46), which could not be distinguished by the gene sequence homology. With the quantitative information of the temperature-dependent proteins, we are able to further annotate the correspondent functions of each dehydrogenase. As shown in the supplemental Table S5, all the proteins encoded by these two cluster genes exhibited as temperature-dependent, but their abundance changes showed opposite modes. To further identify their functions, we did a comparison of the temperature-dependent proteins functionally categorized as listed in Fig. 3. Similar to the enzymes involved in glycolysis, such as glyceraldehyde-3-phosphate dehydrogenase, oxaloacetate decarboxylase, dihydrolipoamide dehydrogenase, and acylphosphatase, the TTE0186/TTE0187/TTE0188 complex remained the mode of the up-regulated abundance responding to the increased temperature. This complex is likely to be a PDC because it catalyzes pyruvate converting to acetyl-CoA at the last step of glycolysis (47). As regards the TTE0688/TTE0689/TTE0690 complex, their abundance changes with temperature were similar to the enzymes participating in amino acid catabolism, such as aminotransferase, aminomutase, aminopeptidase, and oligoendopeptidase. BCKDH complex catalyzes the catabolic reaction of the branched-chain amino acids, isoleucine, leucine, and valine (46). The complex is thus deduced as the BCKDH complex in T. tengcongensis.
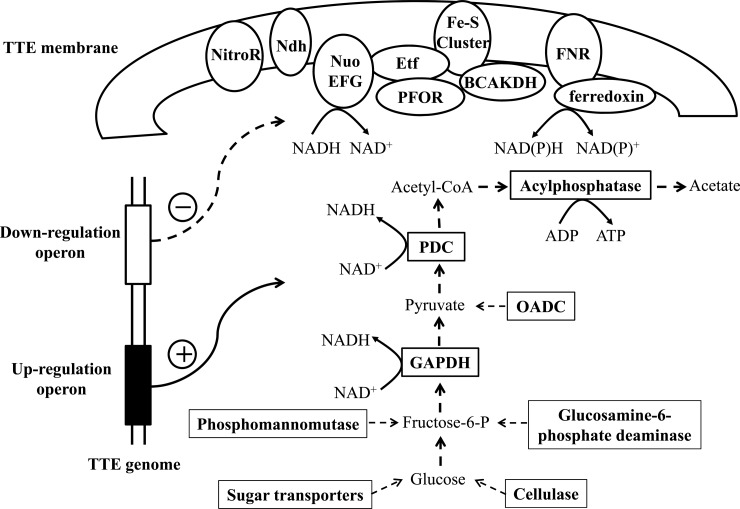
The genome of T. tengcongensis reveals a genetic potential for producing energy through glycolysis, incomplete tricarboxylic acid cycle, and a flexible respiratory pathway to use Fe(III), sulfur, and sulfate (12). Of the proteins that are consecutively down-regulated in response to increasing temperature with the functions of energy generation, ∼90% (16/18) of them are membrane or membrane-associated proteins, such as electron transfer flavoproteins, NADH:ubiquinone oxidoreductases, oxoglutarate ferredoxin oxidoreductases, nitroreductase, BCKDH complex, and Fe-S-cluster-containing hydrogenase (supplemental Table S5), which are essential in an anaerobic respiration system. More importantly, most these genes are located within five clusters on the bacterial genome. The RNA-seq measurement also offers solid data that suggests the correspondent mRNA abundance of these genes is lowered during a rise in temperature. The observation leads to that some genes encoding the key elements of the respiration system on the T. tengcongensis membrane are controlled by the temperature-sensitive regulators, resulting in reduction of the respiration capacity at higher temperature. In contrast to the lower abundance of the respiration relevant proteins, several proteins partaking in glycolysis and the related energy production, as mentioned above, were found to have significantly up-regulated their abundance. This is logically understandable that augmentation of the glycolytic pathway, by enhancing the catalytic capacity from either the key enzymes of glycolysis or conversion of other sources to glucose, is an efficient solution of energy supply for a thermophile's survival under environmental stress. Intriguingly, in an early observation, the growth rate of T. tengcongensis decreased dramatically from 0.26/h at 75 °C to 0.07/h at 80 °C (11), suggesting that all the synthesis processes required energy were attenuated above the optimal temperature. The lower abundance of the sulfur-respiration proteins at higher temperature may partially explain such phenotypic changes. Gathering the evidences above, we propose a model illustrated in Fig. 6 as regards to the survival mechanism of T. tengcongensis. During an environmental rise in temperature, the systems of transcriptional regulators to the gene clusters containing the sulfur-respiration and glycolysis are activated, and the coordinated expression network is formed, leading to attenuation of the respiration capacity on the membrane and augmentation of the energy generation through carbohydrate metabolism. The data presented here represents a global and quantitative survey to the gene abundance changes, mRNA or protein, in response to the altered temperatures, and delivers a solid base for a mechanical model of thermophile metabolism.
Fig. 6.
Schematic diagram to illustrate the hypothetical model derived from the temperature-dependent proteins in T. tengcongensis. In rectangle box, the proteins, all up-regulated during the increase in growth temperature, are located in the cytoplasm and involved in the glycolysis related pathways. In ellipse box, the proteins, all down-regulated responding the temperature rising, are located on the membrane or associated with membrane, and involved in the sulfur-respiration related system. The protein abbreviations: NitroR, nitroreductase; Ndh, NADH dehydrogenase FAD-containing subunit; Nuo EFG, NADH dehydrogenase/NADH: ubquinone oxidoreductase subunit E/F/G; Etf, electron transfer flavoprotein; PFOR, pyruvate: ferredoxin oxidoreductase and related 2-oxoacid: ferredoxin oxidoreductase; Fe-S Cluster, Fe-S-cluster-containing hydrogenase; BCKDHC, branched-chain alpha-keto acid dehydrogenase complex; FNR, ferredoxin-NADP(+) reductase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; OADC, oxaloacetate decarboxylase; PDC, pyruvate dehydrogenase complex.
Supplementary Material
Footnotes
* This work was supported by the National Basic Research Programs of China (2010CB912703 and 2009CB522204), and Nature Science Foundation of China (91131009).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S7.
1 The abbreviations used are:
- 2DE
- two-dimensional gel electrophoresis
- iTRAQ
- isobaric tags for relative and absolute quantitation
- RNA-seq
- RNA sequencing
- RP
- reverse phase
- SCX
- strong cation exchange
- HCD
- higher collision energy dissociation
- CID
- collision induce dissociation
- NCE
- normalized collision energy
- CV
- coefficient of variation
- RPKM
- reads per kilobase per million reads
- FDR
- false discovery rate
- COG
- cluster of orthologous group
- PDC
- pyruvate dehydrogenase complex
- BCKDH
- branched-chain alpha-keto acid dehydrogenase.
REFERENCES
- 1. Cannio R., Rossi M., Bartolucci S. (1994) A few amino acid substitutions are responsible for the higher thermostability of a novel NAD(+)-dependent bacillar alcohol dehydrogenase. Eur. J. Biochem. 222, 345–352 [DOI] [PubMed] [Google Scholar]
- 2. Arnórsdóttir J., Sigtryggsdottir A. R., Thorbjarnardóttir S. H., Kristjánsson M. M. (2009) Effect of proline substitutions on stability and kinetic properties of a cold adapted subtilase. J. Biochem. 145, 325–329 [DOI] [PubMed] [Google Scholar]
- 3. Chen J., Stites W. E. (2004) Replacement of staphylococcal nuclease hydrophobic core residues with those from thermophilic homologues indicates packing is improved in some thermostable proteins. J. Mol. Biol. 344, 271–280 [DOI] [PubMed] [Google Scholar]
- 4. Bezsudnova E. Y., Boyko K. M., Polyakov K. M., Dorovatovskiy P. V., Stekhanova T. N., Gumerov V. M., Ravin N. V., Skryabin K. G., Kovalchuk M. V., Popov V. O. (2012) Structural insight into the molecular basis of polyextremophilicity of short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Thermococcus sibiricus. Biochimie, 94, 2628–2638 [DOI] [PubMed] [Google Scholar]
- 5. Hakulinen N., Turunen O., Jänis J., Leisola M., Rouvinen J. (2003) Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability. Eur. J. Biochem. 270, 1399–1412 [DOI] [PubMed] [Google Scholar]
- 6. Taylor T. J., Vaisman II. (2010) Discrimination of thermophilic and mesophilic proteins. BMC Structural Biol. 10, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakka M., Iyer R. B., Bachas L. G. (2006) Intersubunit disulfide interactions play a critical role in maintaining the thermostability of glucose-6-phosphate dehydrogenase from the hyperthermophilic bacterium Aquifex aeolicus. Protein J. 25, 17–21 [DOI] [PubMed] [Google Scholar]
- 8. Pang J., Allemann R. K. (2007) Molecular dynamics simulation of thermal unfolding of Thermatoga maritima DHFR. Phys. Chem. Chem. Phys. 9, 711–718 [DOI] [PubMed] [Google Scholar]
- 9. Takami H., Takaki Y., Chee G. J., Nishi S., Shimamura S., Suzuki H., Matsui S., Uchiyama I. (2004) Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 32, 6292–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trauger S. A., Kalisak E., Kalisiak J., Morita H., Weinberg M. V., Menon A. L., Poole F. L., 2nd, Adams M. W., Siuzdak G. (2008) Correlating the transcriptome, proteome, and metabolome in the environmental adaptation of a hyperthermophile. J. Proteome Res. 7, 1027–1035 [DOI] [PubMed] [Google Scholar]
- 11. Xue Y., Xu Y., Liu Y., Ma Y., Zhou P. (2001) Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 51, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 12. Bao Q., Tian Y., Li W., Xu Z., Xuan Z., Hu S., Dong W., Yang J., Chen Y., Xue Y., Xu Y., Lai X., Huang L., Dong X., Ma Y., Ling L., Tan H., Chen R., Wang J., Yu J., Yang H. (2002) A complete sequence of the T. tengcongensis genome. Genome Res. 12, 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Xue Y., Feng X., Li X., Wang H., Li W., Zhao C., Cheng X., Ma Y., Zhou P., Yin J., Bhatnagar A., Wang R., Liu S. (2004) An analysis of the proteomic profile for Thermoanaerobacter tengcongensis under optimal culture conditions. Proteomics 4, 136–150 [DOI] [PubMed] [Google Scholar]
- 14. Wang J., Zhao C., Meng B., Xie J., Zhou C., Chen X., Zhao K., Shao J., Xue Y., Xu N., Ma Y., Liu S. (2007) The proteomic alterations of Thermoanaerobacter tengcongensis cultured at different temperatures. Proteomics 7, 1409–1419 [DOI] [PubMed] [Google Scholar]
- 15. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 16. Zieske L. R. (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 17. Gan C. S., Chong P. K., Pham T. K., Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 18. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”. J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 19. Gilar M., Olivova P., Daly A. E., Gebler J. C. (2005) Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J. Separation Sci. 28, 1694–1703 [DOI] [PubMed] [Google Scholar]
- 20. Breitwieser F. P., Müller A., Dayon L., Kocher T., Hainard A., Pichler P., Schmidt-Erfurth U., Superti-Furga G., Sanchez J. C., Mechtler K., Bennett K. L., Colinge J. (2011) General statistical modeling of data from protein relative expression isobaric tags. J. Proteome Res. 10, 2758–2766 [DOI] [PubMed] [Google Scholar]
- 21. Arntzen M. Ø., Koehler C. J., Barsnes H., Berven F. S., Treumann A., Thiede B. (2011) IsobariQ: software for isobaric quantitative proteomics using IPTL, iTRAQ, and TMT. J. Proteome Res. 10, 913–920 [DOI] [PubMed] [Google Scholar]
- 22. Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., Wang J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 [DOI] [PubMed] [Google Scholar]
- 23. Bullard J. H., Purdom E., Hansen K. D., Dudoit S. (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Z., Wen B., Wang Q., Tong W., Guo J., Bai X., Zhao J., Sun Y., Tang Q., Lin Z., Lin L., Liu S. (2013) Quantitative proteomics and transcriptomics data from the anaerobic thermophilic eubacterium Thermoanaerobacter tengcongensis. GigaScience Database, http://dx.doi.org/10.5524/100048
- 25. Dayon L., Pasquarello C., Hoogland C., Sanchez J. C., Scherl A. (2010) Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J. Proteomics 73, 769–777 [DOI] [PubMed] [Google Scholar]
- 26. Second T. P., Blethrow J. D., Schwartz J. C., Merrihew G. E., MacCoss M. J., Swaney D. L., Russell J. D., Coon J. J., Zabrouskov V. (2009) Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 81, 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pichler P., Köcher T., Holzmann J., Möhring T., Ammerer G., Mechtler K. (2011) Improved precision of iTRAQ and TMT quantification by an axial extraction field in an Orbitrap HCD cell. Anal. Chem. 83, 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Z., Wang Q., Lin L., Tang Q., Edwards J. L., Li S., Liu S. (2012) Comparative evaluation of two isobaric labeling tags, DiART and iTRAQ. Anal. Chem. 84, 2908–2915 [DOI] [PubMed] [Google Scholar]
- 29. Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., Krylov D. M., Mazumder R., Mekhedov S. L., Nikolskaya A. N., Rao B. S., Smirnov S., Sverdlov A. V., Vasudevan S., Wolf Y. I., Yin J. J., Natale D. A. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dam P., Olman V., Harris K., Su Z. C., Xu Y. (2007) Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 35, 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mao F., Dam P., Chou J., Olman V., Xu Y. (2009) DOOR: a database for prokaryotic operons. Nucleic Acids Res. 37, D459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey T. L., Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 34. Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S. (2007) Quantifying similarity between motifs. Genome Biol. 8, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kazakov A. E., Cipriano M. J., Novichkov P. S., Minovitsky S., Vinogradov D. V., Arkin A., Mironov A. A., Gelfand M. S., Dubchak I. (2007) RegTransBase–a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35, D407–D412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gry M., Rimini R., Strömberg S., Asplund A., Ponten F., Uhlen M., Nilsson P. (2009) Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics 10, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 38. Dyhrman S. T., Jenkins B. D., Rynearson T. A., Saito M. A., Mercier M. L., Alexander H., Whitney L. P., Drzewianowski A., Bulygin V. V., Bertrand E. M., Wu Z., Benitez-Nelson C., Heithoff A. (2012) The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One 7, e33768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farr S. B., Kogoma T. (1991) Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 55, 561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee H. S., Lee Y. S., Kim H. S., Choi J. Y., Hassan H. M., Chung M. H. (1998) Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: roles of FNR, ArcA, and Fur. Free Radical Biol. Med. 24, 1193–1201 [DOI] [PubMed] [Google Scholar]
- 41. Prasad J., McJarrow P., Gopal P. (2003) Heat and osmotic stress responses of probiotic Lactobacillus rhamnosus HN001 (DR20) in relation to viability after drying. Appl. Environ. Microbiol. 69, 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jain S., Graham C., Graham R. L., McMullan G., Ternan N. G. (2011) Quantitative proteomic analysis of the heat stress response in Clostridium difficile strain 630. J. Proteome Res. 10, 3880–3890 [DOI] [PubMed] [Google Scholar]
- 43. Li J. S., Bi Y. T., Dong C., Yang J. F., Liang W. D. (2011) Transcriptome analysis of adaptive heat shock response of. Streptococcus thermophilus. PLoS One 6, e25777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Carolis E., Posteraro B., Florio A. R., Colonna B., Prosseda G., Bugli F., Lorenzetti S. R., Fiscarelli E., Inzitari R., Iavarone F., Castagnola M., Fadda G., Sanguinetti M. (2011) Analysis of heat-induced changes in protein expression of Stenotrophomonas maltophilia K279a reveals a role for GroEL in the host-temperature adaptation. Int. J. Med. Microbiol. 301, 273–281 [DOI] [PubMed] [Google Scholar]
- 45. Mattevi A., Obmolova G., Schulze E., Kalk K. H., Westphal A. H., de Kok A., Hol W. G. (1992) Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science 255, 1544–1550 [DOI] [PubMed] [Google Scholar]
- 46. Singh V. K., Hattangady D. S., Giotis E. S., Singh A. K., Chamberlain N. R., Stuart M. K., Wilkinson B. J. (2008) Insertional inactivation of branched-chain alpha-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl. Environ. Microbiol. 74, 5882–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harada T., Satoh S., Yoshioka T., Ishizawa K. (2007) Anoxia-enhanced expression of genes isolated by suppression subtractive hybridization from pondweed (Potamogeton distinctus A. Benn.) turions. Planta 226, 1041–1052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.