Abstract
In order to map the extracellular or membrane proteome associated with the vasculature and the stroma in an embryonic organism in vivo, we developed a biotinylation technique for chicken embryo and combined it with mass spectrometry and bioinformatic analysis. We also applied this procedure to implanted tumors growing on the chorioallantoic membrane or after the induction of granulation tissue. Membrane and extracellular matrix proteins were the most abundant components identified. Relative quantitative analysis revealed differential protein expression patterns in several tissues. Through a bioinformatic approach, we determined endothelial cell protein expression signatures, which allowed us to identify several proteins not yet reported to be associated with endothelial cells or the vasculature. This is the first study reported so far that applies in vivo biotinylation, in combination with robust label-free quantitative proteomics approaches and bioinformatic analysis, to an embryonic organism. It also provides the first description of the vascular and matrix proteome of the embryo that might constitute the starting point for further developments.
The vasculature has an important role in embryonic development and tissue homeostasis (1). It is also involved in disease processes in which defective or excessive vascularization is observed, such as chronic inflammation and tumor growth (2). The extracellular matrix and stroma fibroblasts also participate in these processes and play an important role in tissue remodeling (3, 4). For therapeutic intervention, the molecular repertoire of the vasculature and stroma must be known.
The chicken embryo is a model organism that has been particularly well studied in different types of experiments. For example, studies on the chicken embryo have had significant effects on developmental biology. Techniques such as the creation of a quail-chick chimera were developed to study the migration and the fate of cell populations in intact embryos (5). This has led to the elucidation of the origins and fate of neural crest cells, the discovery of hemangioblasts, and a better understanding of neural tube patterning (6, 7). Furthermore, techniques for gain and loss-of-function studies, promoter analysis, and transgenesis have recently been developed, together with methods for the isolation of embryonic stem cells (8–10). In addition, the sequencing of the chicken genome has been completed.
The chicken embryo, in particular the chorioallantoic membrane (CAM), has also been used to gain insights into the different steps of vessel formation and is an experimental in vivo tumor model for cancer research (11, 12). It has been utilized for the evaluation of angiogenic and anti-angiogenic compounds within its vasculature and to study the growth, angiogenesis, and metastasis of human tumors (13–15). By implanting tumor cells onto the CAM 6 days after fertilization of the egg, a model that simulates key features of human tumor growth and vascularized tumors can be generated, allowing rapid research into human tumor progression (13–15).
Functional genomic strategies have recently been applied to characterize the vascular proteome. Neri and colleagues developed a method for the detection of antigens accessible in the blood stream, based on the terminal perfusion of tumor-bearing rodents with a reactive ester of biotin (16, 17). This in vivo labeling procedure is followed by the recovery of biotinylated proteins from normal organs and tumors, which are then proteolytically digested and submitted to comparative mass spectrometry analysis. This approach has been extended to the ex vivo perfusion of surgically resected human colon cancer, thereby directly revealing the overexpression of markers in the tumor matrix or vessels (18). Proteomic technologies have matured to a level enabling the accurate and reproducible quantitation of peptides and proteins. Today, with recent advances in mass spectrometry, label-free quantitative proteomics approaches are considered as reliable and efficient methods for studying protein expression level changes in complex mixtures (19).
In order to characterize the extracellular and membrane proteome associated with the vasculature and the stroma in an embryonic organism in vivo, we utilized an in vivo biotinylation technique in the chicken embryo and combined it with high-resolution mass spectrometry and bioinformatics analysis to determine expression patterns and endothelial cell expression signatures. We also performed an analysis on neovessels growing (i) after the implantation of tumor cells into the chicken CAM and (ii) within granulation tissue after tissue wounding. This is the first study reported so far that provides a description of the vascular and matrix proteome in an embryonic organism by using the in vivo biotinylation method. This might constitute the starting point for further developments.
EXPERIMENTAL PROCEDURES
Tissue Biotinylation
Brown Leghorn eggs were cultured at 38 °C for 3 days; then shells were cracked and the egg contents were transferred to 10-cm-diameter cell culture dishes. Embryos were cultured for another 7 days, and then 1-cm2 injuries were inflicted to the CAM in the form of superficial scalpel cuts and subsequent scraping-off of the epithelium. The wound area was then covered with a nylon grid. At the opposite location, the CAM epithelium was scratched with a scalpel and covered with a nylon grid, and 10 μl of glioblastoma slurry (300000 cells/μl) was poured onto the grid center. Petri dishes with chicken embryos were then returned to the incubator for another 6 days of incubation, and biotinylation was performed. Then the chest of the E16 embryo was opened, the right pulmonary artery was canulated, and the embryo was perfused with 15 ml heparinized Ringer's solution. After that, 15 ml of 1 mg/ml sulfo-NHS-LC-LC-biotin (1 mg/ml) was injected into the embryonic circulation and quenched by subsequent injection of Tris-glycine buffer (20). Finally, amine buffer was eluted with Ringer's solution, and embryonic tissues were cut off and analyzed. After protein extraction, lysates were mixed with streptavidin-linked beads pre-washed twice with lysis buffer. After incubation, the bead slurry was washed and transferred to Eppendorf tubes, and PNGase F and Sialidase were added for deglycosylation. The beads were then transferred back to the washing column, washed again and mixed with Ultra Pure™ water, and transferred to 1-ml syringes connected to needles clogged with cotton gauze (21). Biotinylated proteins were eluted with water heated to 70 °C. Collected eluates were condensated on a SpeedVac for electrophoresis or frozen and lyophilized for subsequent mass spectrometric analysis. A more detailed description is provided in the supplemental “Experimental Procedures” section.
Protein Sample Processing
Protein samples were resuspended in Laemmli sample buffer and loaded onto a one-dimensional SDS-PAGE gel (1.5 mm by 8 cm) for one-shot analysis of the entire mixture. No fractionation was performed, and the electrophoretic migration was stopped as soon as the protein sample (10 μg) entered the separating gel. The gel was briefly stained with Coomassie Blue, and a single band containing the whole sample was cut out.
The 18 gel slices, each containing one protein sample replicate (control CAM, intestine, kidney, liver, wounded CAM, or tumor CAM), were washed by two cycles of incubation in 100 mm ammonium bicarbonate (15 min, 37 °C) followed by 100 mm ammonium bicarbonate/acetonitrile (1:1) (15 min, 37 °C). Proteins were digested by 0.6 μg of modified sequencing grade trypsin (Promega, Madison, WI) in 50 mm ammonium bicarbonate overnight at 37 °C. The resulting peptides were extracted from the gel via incubation in 50 mm ammonium bicarbonate (15 min, 37 °C) and twice in 10% formic acid/acetonitrile (1:1) (15 min, 37 °C). The three collected extractions were pooled with the initial digestion supernatant, dried in a SpeedVac, and resuspended with 14 μl of 5% acetonitrile and 0.05% trifluoroacetic acid (TFA).
Nano-LC-MS/MS Analysis
The resulting peptides were analyzed via nano-LC-MS/MS using an Ultimate3000 system (Dionex, Amsterdam, The Netherlands) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Five microliters of each sample were loaded on a C-18 pre-column (300 μm inner diameter × 5 mm, Dionex) at 20 μl/min in 5% acetonitrile and 0.05% TFA. After 5 min of desalting, the pre-column was switched online with the analytical C-18 column (75 μm inner diameter × 15 cm, PepMap C18, Dionex) equilibrated in 95% solvent A (5% acetonitrile, 0.2% formic acid) and 5% solvent B (80% acetonitrile, 0.2% formic acid). Peptides were eluted using a 5% to 50% gradient of solvent B over 105 min at a 300 nl/min flow rate. The LTQ-Orbitrap Velos was operated in data-dependent acquisition mode with XCalibur software. Survey MS scans were acquired in the Orbitrap in the 300–2000 m/z range with the resolution set to a value of 60,000. The 20 most intense ions per survey scan were selected for collision-induced dissociation fragmentation, and the resulting fragments were analyzed in the linear trap (LTQ). Dynamic exclusion was employed within 60 s to prevent repetitive selection of the same peptide.
Database Search and Data Validation
Mascot Daemon software (version 2.3.2, Matrix Science, London) was used to perform database searches, and the Extract_msn.exe macro provided with Xcalibur (version 2.0 SR2, Thermo Fisher Scientific) was used to generate peaklists. The following parameters were set for creation of the peaklists: parent ions in the mass range of 400–4500, no grouping of MS/MS scans, and threshold at 1000. A peaklist was created for each analyzed sample, and individual Mascot (version 2.3.01) searches were performed. Data were searched against Gallus gallus (chicken) entries in the Uniprot protein database (released September 21, 2011; 145,173 sequences) or against Gallus gallus (chicken) and Homo sapiens (human) entries from the same protein database. Carbamidomethylation of cysteines was set as a fixed modification, and oxidation of methionine and modification of lysines by NHS-LC-biotin (+467.5525 Da) were set as variable modifications. The specificity of trypsin digestion was set for cleavage after K or R, and two missed trypsin cleavage sites were allowed. The mass tolerances in MS and MS/MS were set to 5 ppm and 0.6 Da, respectively, and the instrument setting was specified as “ESI-Trap.” In order to calculate the false discovery rate (FDR), the search was performed using the “decoy” option in Mascot.
Peptide identifications extracted from Mascot result files were validated with in-house software if their score was greater than the Mascot homology threshold (when available; otherwise the Mascot identity threshold was used) for a given Mascot p value which was adjusted to get a final peptide FDR of 5%. The FDR at the peptide level was calculated as described by Navarro and Vazquez (22). Using this method, the p value was automatically adjusted to obtain an FDR of 5% at the peptide level. Validated peptides were assembled into proteins groups following the principle of parsimony (Occam's razor), which involves the creation of a minimal list of protein groups explaining the list of peptide spectrum matches. Protein groups were then re-scored for the protein validation process. For each peptide match belonging to a protein group, the difference between its Mascot score and its homology threshold (or identity threshold) was computed for a given p value (automatically adjusted to increase the discrimination between target and decoy matches), and these “score offsets” were then summed to obtain the protein group score. Protein groups were validated based on this score to obtain an FDR of 1% at the protein level (FDR = number of validated decoy hits/(number of validated target hits + number of validated decoy hits) × 100).
Data Quantification
The quantification of proteins was performed using the label-free module implemented in the MFPaQ software (23). For each sample, the software uses the validated identification results and extracted ion chromatograms (XICs) of the identified peptide ions in the corresponding raw nano-LC-MS files, based on their experimentally measured retention time and monoisotopic m/z values. The time value used for this process was retrieved from Mascot result files, based on an MS2 event matching the peptide ion. If several MS2 events were matched to a given peptide ion, the software checked the intensity of each corresponding precursor peak in the previous MS survey scan. The time of the MS scan that exhibited the highest precursor ion intensity was attributed to the peptide ion and then used for XIC extraction, as well as for the alignment process. Peptide ions identified in all the samples to be compared were used to build a retention time matrix in order to align LC-MS runs. If some peptide ions were sequenced via MS/MS and validated only in some of the samples to be compared, their XIC signal was extracted in the nano-LC-MS raw file of the other samples using a predicted retention time value calculated from this alignment matrix via a linear interpolation method. The quantification of peptide ions was performed based on calculated XIC area values. To perform normalization of a group of comparable runs, the software computed XIC area ratios for all the extracted signals between a reference run and all the other runs of the group and used the median of the ratios as a normalization factor. In order to perform protein relative quantification in different samples, a protein abundance index (PAI) was calculated, defined as the average of XIC area values for at most three intense reference tryptic peptides identified for the protein (the three peptides exhibiting the highest intensities across the different samples were selected as reference peptides, and these same three peptides were used to compute the PAI of the protein in each sample; if only one or two peptides were identified and quantified in the case of low-abundant proteins, the PAI was calculated based on their XIC area values). In the case of SDS-PAGE fractionation, integration of quantitative data across the fractions was performed as indicated in the text by summing the PAI values for fractions adjacent to the fraction with the best PAI (the same three consecutive fractions for all the samples to be compared). For differential studies (here, between the CAM samples and tumor samples and between the CAM samples and the wound samples), a Student's t test on the PAI values was used for statistical evaluation of the significance of variations in expression level. For proteins with missing PAI values, an in-house script inferred these values by assigning a random value according to observed intensity variation. In CAM–tumor sample comparison and in CAM and wound sample comparison, a 2-fold change and p value of 0.05 were used as combined thresholds to define biologically regulated proteins. A volcano plot is used to represent these varying proteins.
The quantification values obtained (PAI) for all proteins in CAM, intestine, kidney, and liver samples were transformed into a matrix that was used in the R software to organize the proteins into cluster groups. For the three replicates, the median values of PAI were calculated and divided by the maximum value for the same protein between the four median values (CAM, intestine, kidney, and liver). All values were then centroided via this method to obtain values between 0 and 1.
This matrix from CAM, intestine, kidney, and liver experiments (three replicates per sample) was used to generate clusters using “Ward's hierarchical cluster analysis” in R Commander (V1.6-2). The clustering parameters were set to use the Ward method for hierarchical clustering; the distance measure was Euclidean, and the group number was set at 10.
The percentages of membrane and extracellular proteins in the different samples were evaluated using Protein Center software V3.8.2017 (Proxeon Bioinformatics, Seattle, WA).
Human-chick ortholog identification, endothelial cell expression signatures, angiogenic expression signatures, and the abstract scanning for relevance to angiogenesis were carried out using methods described elsewhere (24) (for more details, see the supplemental “Experimental Procedures” section).
All other procedures (histochemistry, immunohistochemistry, Western blotting, etc.) were done according to standard protocols. A detailed description of the materials and methods can be found in the supplemental “Experimental Procedures” section.
RESULTS
Implantation of Tumor Cells and Induction of Granulation Tissue
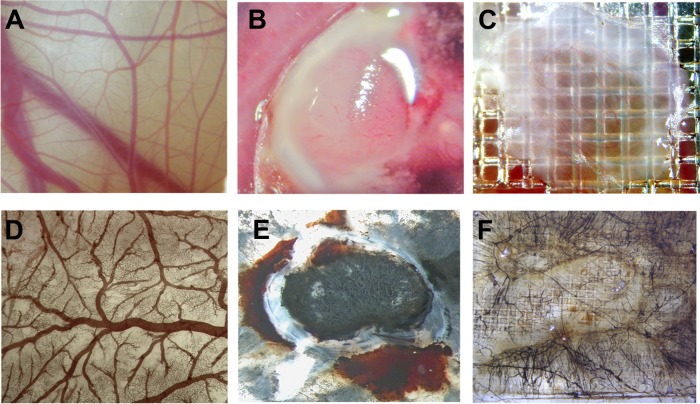
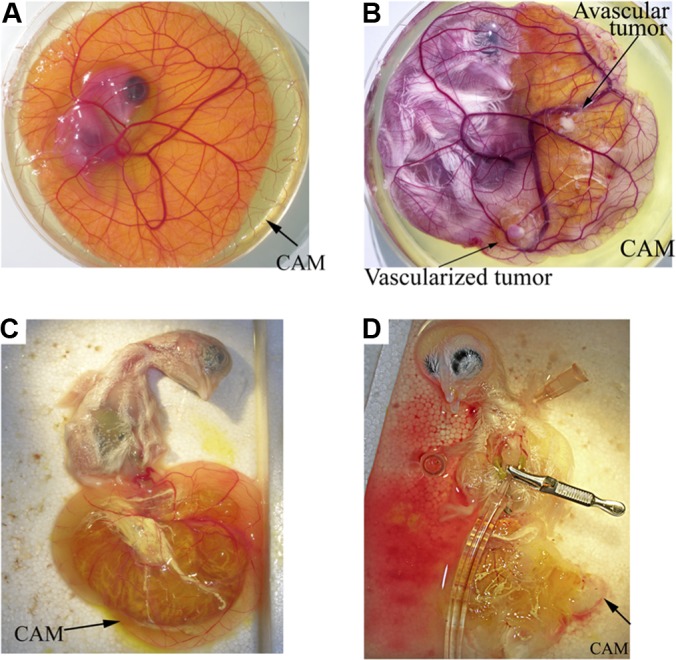
After 10 days of culture in shell-less conditions, embryos develop CAMs with dense vasculature, as can be seen in Fig. 1A. On day 10, the U87 glioma cells were implanted onto the CAM, which grew into a highly vascularized tumor 4 days later (Fig. 1B). A wound was inflicted on another area of the CAM and was covered with a square nylon grid. As shown in Fig. 1C, newly formed blood vessels grew through the grid.
Fig. 1.
Visualization of perfusion in the chicken CAM. Sixteen-day-old embryos were injected with colloidal carbon, which visualized the functional perfusion of vessels in implanted tumor and wound, and perfusion of the CAM vessels was visualized. A, normal live CAM vessels. Only macrovessels can be identified. D, fixed CAM tissue stained for erythrocyte hemoglobin with DAB reveled a high-density network of capillaries. B, E, CAM engrafted with U87 tumor cells (B, live; E, injected with colloidal carbon and co-stained for haamoglobin). C, F, wounded CAM (C, live; F, injected with colloidal carbon and co-stained for hemoglobin). All vessels were filled with carbon particle, which indicates that thy were functionally perfused.
To verify the perfusion of neovessels, colloidal carbon particles were injected into CAM vein 5 days after U87 glioma cell implantation or wound incision (Figs. 1D–1F). Carbon particles localized to all normal vessels of the CAM and neovessels of both granulation and U87 tumor tissues. This indicates that the biotinylation reagent, when injected into the chicken circulation, can be delivered into the entire vasculature of normal, granulation, or xenografted tumor tissues.
Embryo Perfusion and in Vivo Biotinylation
The right pulmonary artery of E16 embryos was cannulated, and the embryo yolk, CAM, and embryo circulation connected with the pulmonary artery via the ductus arteriosus were washed and perfused with a biotin solution as described under “Experimental Procedures” (Figs. 2A–2D). The flow rate was adjusted to 1 ml/min to allow optimal perfusion and preserve vessel integrity. The biotinylation reaction was stopped by injection of Tris-buffered glycine buffer. After biotinylation, different tissues (CAM, liver, small intestine, kidney), implanted tumors, or granulation tissue were removed and processed for immunostaining or protein extraction as indicated under “Experimental Procedures” (for perfusion in real time, see the video included in the supplementary materials).
Fig. 2.
Biotinylation in vivo in the live chick. The chick embryo was grown in a Petri dish until E16 was perfused with Ringer's solution to eliminate blood cells and plasma proteins, which allowed subsequent biotinylation. A, chicken embryo and extra-embryonic membranes at day 6 of development. B, implantated tumor (U87) on the CAM on day 16 of development. C, pulling the embryo inside-out through the CAM opening submerged in the buffer bath allowed direct access to chicken chest with minimal injury to the tissue. D, cannulation of the heart and perfusion.
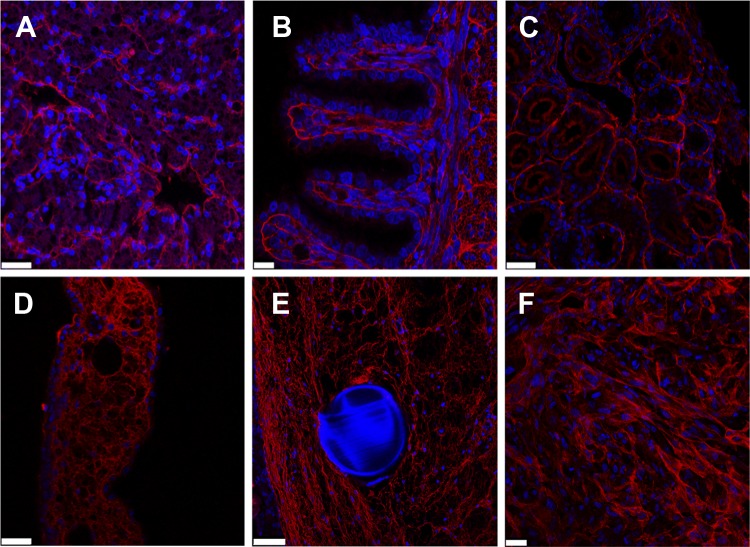
Significant biotinylation of blood vessels was seen in the different tissues (Figs. 3A–3F). Fast red staining of biotinylated proteins was found in all organs and tissues examined (liver, intestine, kidney, CAM, wound, and xenografted tumor). The staining of biotin was pronounced in all tissues and was localized to the vessels or the extracellular matrix (basal lamina, intercellular matrix). In the liver (Fig. 3A), staining was prominent around sinusoidal vessels. In the intestine (Fig. 3B), strong immunoreactivity was found in intestinal villosities and underlining tissues. In the kidney (Fig. 3C), immunoreactivity was detected around tubules and glomeruli. In the CAM, a trabecular pattern of staining was seen (Fig. 3D). Because of the leaky nature of neovessels, additional biotin reactivity was observed in and around the granulation tissue (Fig. 3E). Similarily, in xeonografted tumors, strong positive biotin staining was detected around tumor cells and in the matrix (Fig. 3F).
Fig. 3.
Immunostaining of in vivo biotinylated tissues. Various biotinylated tissues were sectioned for staining with streptavidin: A, liver; B, intestine; C, kidney; D, CAM; E, granulation tissue; F, xenografted tumor (U87). Biotinylated proteins (red) were detected in basement membranes of the different tissues. Images were taken with a Zeiss confocal microscope. Scale bars: B, 10 μm; A, C, D, 25 μm; E, 50 μm. A grid was used in the wound experiment (blue autofluorescence) (E). Nuclei were stained with DAPI (blue).
To visualize more precisely the staining pattern of biotin in relation to blood vessels and other tissue elements, double staining was performed using SNA isolectin (endothelial cells), anti-desmin antibodies (pericytes), or anti-α smooth muscle actin (smooth muscle cells and myofibroblasts) antibodies (14) (Figs. 4A–4F). In the CAM tissue, double staining for biotinylated proteins and pericytes/vascular smooth muscle cells indicated highly significant biotinylation in and around blood vessels, evidenced by the staining around pericytes (Figs. 4A and 4B). In tumors, double staining was detected around blood vessels (endothelial cells), which indicates biotinylation of proteins in the basement membrane (Figs. 4C and 4D). However, biotinylated proteins were also found at a distance from blood vessels and around tumor cells, which indicates penetration of biotinylating reagent into the tumor. In the wound, double staining of the biotinylated proteins and α-smooth muscle actin–positive myofibroblasts showed significant biotinylation in non-vascular structures (Fig. 4E). Furthermore, in tissue such as the kidney, biotinylation of interstitial tissue around α-smooth muscle actin–positive pericytes was seen (Fig. 4F). This indicates that in growing tissues like tumor and wound, in which vessels are characterized by the extensive extravasation of macromolecules, the in vivo biotinylation technique also labels distinct matrix proteins.
Fig. 4.
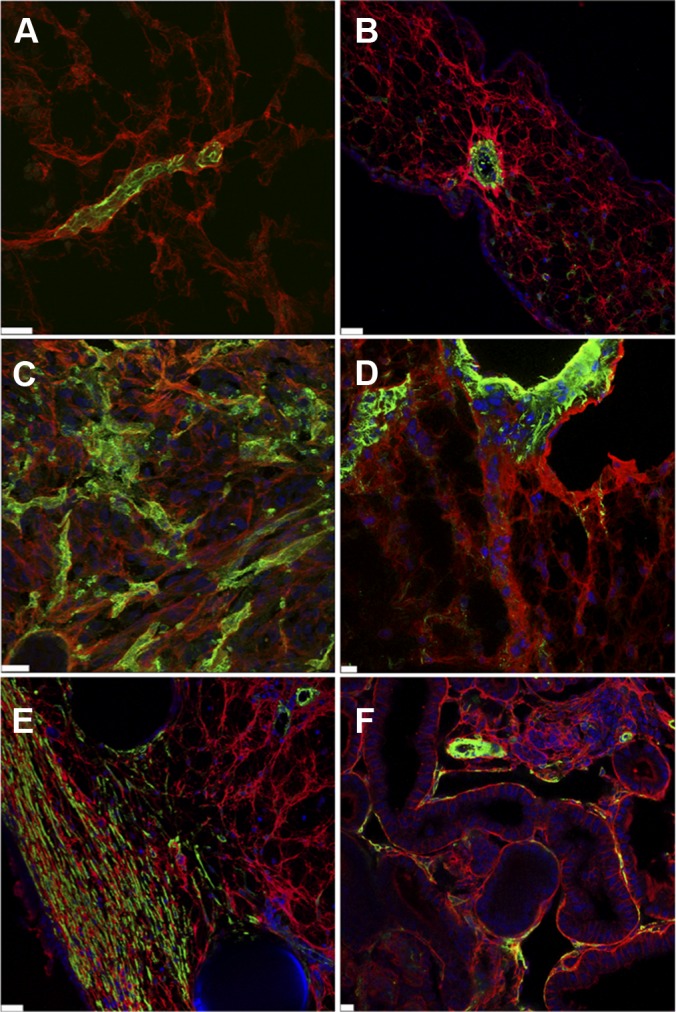
Co-labeling of biotinylated protein with vascular, stroma, or tumor antigens. Tissues from normal CAM (A, B), engrafted tumor (C, D), wound tissue (E), and kidney (F) were immunostained using neutravidin-rhodamin (red) and anti-desmin (green; A), anti-α smooth muscle actin (green; B, D, E, and F), or SNA-lectin (green; C). Images were taken with a Zeiss confocal microscope. Scale bars: B, D–F, 10 μm; A, 20 μm; C, 25 μm. Nuclei were stained with DAPI (blue).
Purification and Identification of Biotinylated Proteins
Proteins from total tissue lysate and those purified on the streptavidin beads were separated on SDS-PAGE gels and revealed as a continuous smear with no discrete banding (lanes 1, 5, and 6, supplemental Fig. S1). Evidence for purification quality was found; except for cellular carboxylases that covalently harbor endogenous biotin, no other proteins were purified from total tissue lysates obtained from organs of non-biotinylated embryos. As another quality indicator, β-actin, the most abundant intracellular protein, was not detected via Western blot in a purified biotinylated protein mixture. The elution of biotinylated proteins was aided by a novel and recently described technique in which strong biotin–streptavidin bonds are dissociated in pure water heated to 70 °C (21). The addition of biotin before elution prevented destabilization of the streptavidin tetramer, which prevented the overrepresentation of single proteins that would mask less well represented proteins in the subsequent mass spectrometry analysis.
For all the protein samples isolated, we performed deglycosylation and two SDS-PAGE gel separations. A “one-shot” mass spectrometry analysis was also performed by loading the samples on a one-dimensional gel without protein fractionation (the whole sample was collected in one slice of gel). After trypsin digestion, the samples were injected onto a nano-LC-MSMS/LTQ-Orbitrap Velos system (see “Experimental Procedures”). Information for single-peptide-based protein identifications (sequence identified, the precursor m/z and charge, score/E-value), appropriately labeled MS/MS spectra, masses detected, and fragment assignments is provided in the supplemental material.
We performed mass spectrometry on three independent pools of the different tissues including CAM (C), wound (W), tumor (T), kidney (K), intestine (I), and liver (L). Each pool comprised seven to nine samples from independently perfused embryos. A total of 1264 proteins were detected from all organs combined, and these are listed in supplemental Table S1 (sheet S1A).
High Number of Extracellular and Membrane Proteins
When samples were analyzed with ProteinCenter software (v3.5.2.1, Proxeon Bioinformatics, Seattle, WA), a significant number of membrane and extracellular proteins were detected. Among these proteins, between 59.7% (liver) and 69.2% (intestine) were extracellular or membrane. The number of extracellular/membrane proteins identified with Gene Ontology annotation ranged between 67.76% (tumor) and 80.38% (intestine) (Table I).
Table I. Quantification of membrane and extracellular matrix proteins.
| Total protein number | E and/or M | E and M | E only | M only | Others | Without Gene Ontology CC[1] terms | % E + M | % E + M/protein with Gene Ontology CC annotation | |
|---|---|---|---|---|---|---|---|---|---|
| CAM | 1044 | 626 | 75 | 242 | 459 | 418 | 167 | 59.96 | 71.38 |
| Intestine | 1031 | 713 | 96 | 261 | 548 | 318 | 144 | 69.16 | 80.38 |
| Kidney | 1172 | 799 | 85 | 217 | 667 | 373 | 170 | 68.17 | 79.74 |
| Liver | 1213 | 724 | 69 | 178 | 615 | 489 | 175 | 59.69 | 69.75 |
| Tumor | 621 | 372 | 47 | 183 | 236 | 249 | 72 | 59.90 | 67.76 |
| Wound | 659 | 395 | 57 | 193 | 259 | 264 | 94 | 59.94 | 69.91 |
Note: the numbers of extracellular (E) and membrane proteins (M) were analyzed using ProteinCenter software. CC, cellular content.
Quantification of Proteins
The quantification of proteins was performed using the label-free module implemented in the MFPaQ software (23). For the relative quantification analysis of protein expression, a label-free quantification method was chosen because it did not require the labeling of proteins (19). This method is based on integration of the MS signals of the three best peptides of each protein (PAI values) (see “Experimental Procedures”). The quantified proteomic PAI data were divided into two separate groups: (i) CAM, tumor, and wound group (CTW); and (ii) CAM, intestine, kidney, and liver group (CIKL). Data values can be found in supplemental Table S1 (sheets S1B and S1C, respectively).
Sample Reproducibility and Distribution
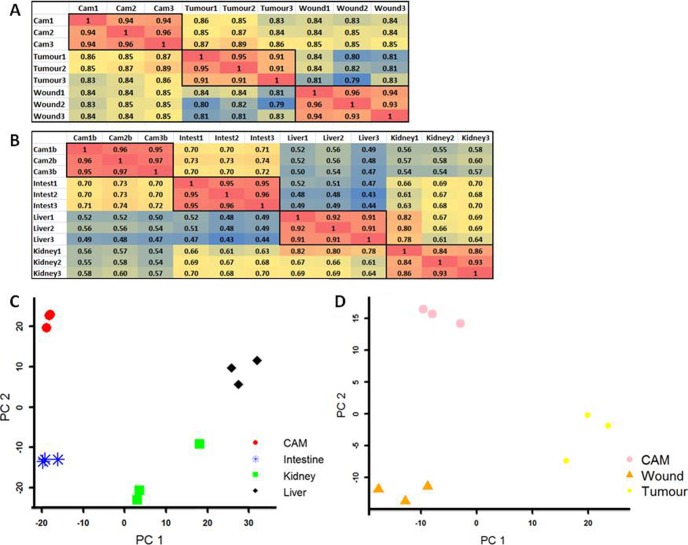
To test the reproducibility of biological replicates from within each tissue and between the different tissues, Pearson correlation coefficients and principal component analyses were carried out. The results of the different groups can be seen in Fig. 5. Pearson correlation results for all biological replicates indicated correlations greater than or equal to 0.84 (Figs. 5A (CTW) and 5B (CIKL)), which shows strong correlation and reproducibility of the biological replicates. Fig. 5C portrays an independent principal component plot of the CIKL pool and reveals a kidney replicate with more variation than seen in the other kidney samples (which is the minimum 0.84 Pearson correlation coefficient (Fig. 5B)). Fig. 5D shows an independent principal component analysis of the CAM, wound, and tumor samples. The largest variation in the first principal component shows that the wound and CAM samples had less global gene expression difference than the tumor samples, as expected.
Fig. 5.
Reproducible biological replicates. A and B show heat-maps of the Pearson correlation coefficient results of all biological replicates from both pools. For replicates from the same tissue type, a minimum correlation of 0.84 was seen. C portrays an independent principal component plot of the CIKL pool. The higher resolution reveals a kidney replicate with variation relative to the other kidney samples (the minimum 0.84 correlation from B). D shows an independent principal component analysis of the CAM, wound, and tumor samples. The largest variation in the first principal component shows that the wound and CAM samples showed less global gene expression difference than the tumor samples.
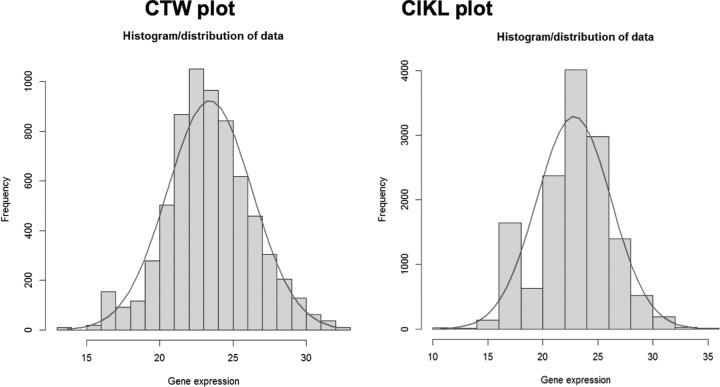
In order to test the proteomic PAI data for Gaussian/normal distributions, a histogram for each pool was produced (Fig. 6). The results show a bell-like shape for both pools, characteristic of a normal distribution, which enables the use of valid parametric test statistics for differential expression with these data (Student's t test).
Fig. 6.
Normal distribution of PAI proteomic quantified data. Logarithmic to the base 2 PAI values follow an approximate Gaussian distribution, enabling the use of Student's t test for differential expression experiments.
Human Ortholog Identification
As genes related by evolution, particularly orthologs, usually have conserved function between vertebrates, and because human is the most characterized model species, human orthologs of chicken proteins were sought in order to further investigate their biology. All except 26 proteins were assigned a human ortholog using the PAI data set, and these can be viewed in sheets S1B and S1C of supplemental Table S1. The proteins not assigned were either chicken- or egg-specific proteins. When only the non-redundant set of human orthologs was considered, 51% were either extracellular or membrane genes. This is in line with the percentage of cell surface and extracellular matrix proteins as found by Rybak et al., which ranged between 20% and 50% for all organs from an in vivo mouse biotinylation experiment (17). A total of 1157 non-redundant human orthologs was found for both CIKL and CTW combined. The CIKL and CTW pools contained 1052 and 670 non-redundant human orthologs, respectively.
Functional Enrichment for Protein Sets Detected through in Vivo Biotinylation
Gene Ontology analysis using DAVID on all proteins identified showed principally, for cellular components (p value < 0.001), enrichment in the extracellular matrix and extracellular region categories (>10% of all proteins) (Fig. 7). For biological processes (p value < 0.0001), the noted categories were metabolic process, primary metabolic process, cellular process, cell communication, signal transduction, protein metabolic process, developmental process, cell adhesion, immune system process, transport, system development, cell surface receptor linked, response to stimulus, and cell–cell adhesion (>10% of all proteins) (supplemental Fig. S2A). For molecular function (p value < 0.0001), catalytic activity, hydrolase activity, receptor activity, structural molecule activity, transferase activity, and oxidoreductase activity were the most represented (>10% of all proteins) (supplemental Fig. S2B). Most categories are related to components and processes of the membrane and extracellular space in agreement with the in vivo biotinylation.
Fig. 7.
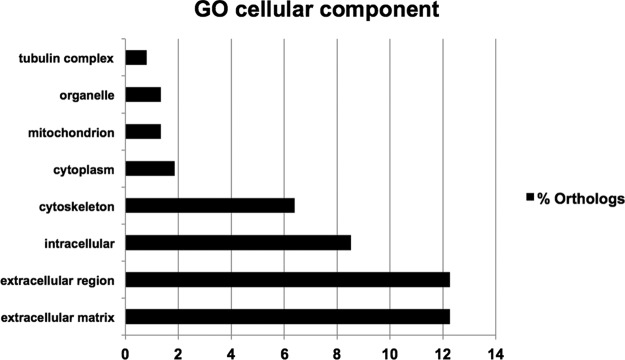
Gene Ontology categories of proteins identified in the proteomic analysis. Gene Ontology analyses were performed using DAVID to evidence cellular component categories. The figure depicts the main categories associated with the extracellular compartment (matrix, membrane).
Tumor and Wound Differential Expression
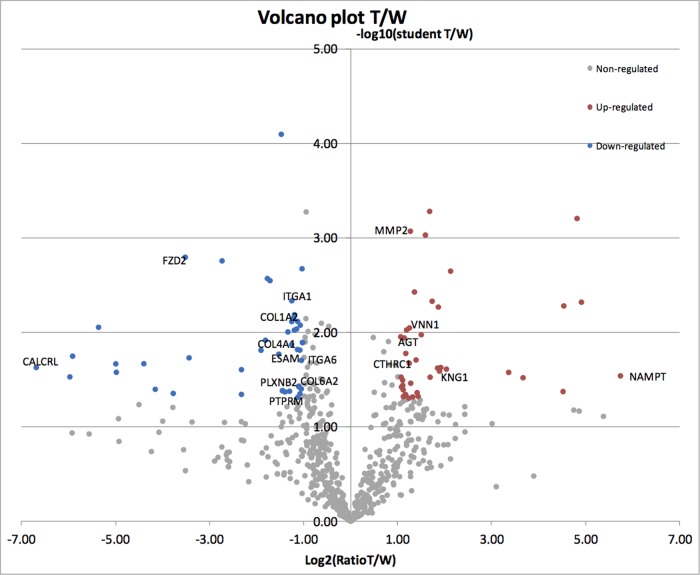
As indicated above, a Student's t test differential protein expression experiment was carried out on the CTW group. This method compares the number of peptides between two samples and allows a more precise quantitative analysis. We first compared tumor and wound in this analysis (Fig. 8). When tumor and wound were compared and two statistical tests were performed, 39 proteins were found to be overexpressed (>2-fold) and 44 proteins were less expressed (>2-fold) after tumor implantation (supplemental Table S2). Among the proteins overexpressed were several matrix molecules and receptors (ANXA5, ITGB5, collagen triple helix repeat containing 1, MSLN, and VNN2). Furthermore, complement proteins, coagulation factors, protease, and their inhibitors were detected (angiotensinogen, MMP2, F9, SERPINA10, SERPINA1, C3, F2, A2ML1,1 CFB, and KNG1). However, some carrier proteins, intracellular binding proteins, or enzymes were surprisingly highly overexpressed, such as NAMPT (53-fold), OAS3 (30-fold), POMGNT1 (28-fold), and PDIA3 (23-fold).
Fig. 8.
Representation of tumor versus wound quantification. The regulated proteins in blue and red get a fold > 2 and a t test < 0.05 Volcano plot.
Among the proteins overexpressed in wound relative to tumor, a number of extracellular matrix molecules, adhesion receptors, and signaling receptors were found (supplemental Table S2). These include FZD2 (11.5-fold), ESAM (2.9-fold), PTPRM (2.6-fold), PLXNB2 (2.5-fold), COL4A1 (2.4-fold), ITGA1 (2.4-fold), COL1A1/2 (2.3-fold), COL6A2 (2.215-fold), and ITGA6 (2.1-fold). The highest fold change was observed for CALCRL (104-fold).
Among proteins that were differentially expressed in the tumor sample relative to the CAM with the same stringency (supplemental Fig. S3A), we found among the most decreased proteins CD151 (107-fold), F13A1 (75-fold), TSEN54 (55-fold), carbonic anhydrase XIII (49-fold), PFN2 (31-fold), CALCRL (31-fold), carnitine palmitoyltransferase 1A (28-fold), and FREM2 (24-fold). Some vascular-associated proteins (CD93, MCAM, PCAM-1, and VWA1) were also found in this group. However, the decrease was much less for these molecules. Furthermore, several cell surface or adhesion receptors were also down-regulated (ciliary neurotrophic factor receptor, ELTD1, FLRT2, HAPLN3, MMRN2, and PTRRM).
In the wound samples, we found 33 proteins that were overexpressed relative to the CAM (supplemental Fig. S3B). The most up-regulated proteins (fold change (FC) > 4) included ATP-binding cassette, sub-family E (OABP), member 1; ANK1; bone morphogenetic protein 1 2,3-bisphosphoglycerate mutase; eukaryotic translation initiation factor 3, subunit B; eukaryotic translation initiation factor 5; EPB41; EPB42; H1F0; HNRNPM; LMNA; NAP1L1; OLA1; PABPC4; SLC4A1; SLC25A43; TSPO; and ZP1.32. The most down-regulated proteins (FC > 4) included capping protein (actin filament) muscle Z-line beta, complement factor I, F13A1, MYL6, NAMPT, POMGNT1, PLSCR1, PPIB, PSCA, RAC2, SGCD, TNFAIP6, and TNFRSF6B.
Finding Tissue Enriched Proteins
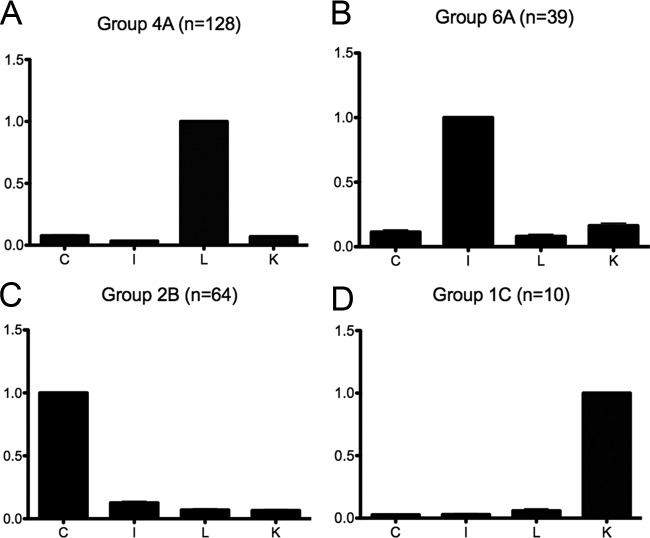
We furthermore conducted a statistical analysis of the results via Ward's hierarchical clustering method using R software (v2.12; R Commander V1.6-2) to generate clusters representing the expression profiles of the identified proteins over the different samples. For the CIKL group, 17 clusters were found. Among the 17 clusters, 4 were found to display overexpression in one tissue relative to the three others (Figs. 9A–9D). These four clusters depict liver, intestine, CAM, and kidney enriched proteins, respectively. Supplemental Table S3 depicts all 17 clusters. In the following, four clusters exhibiting enrichment in one organ relative to the others are discussed.
Fig. 9.
Four major clusters of the CIKL group. The figure depicts the four major clusters in which overexpression is found in one tissue but not in all the others. (A) Liver, (B) Intestine, (C) CAM and (D) Kidney.
In the first cluster (liver expression, Fig. 9A), 128 proteins were found. Among these were a number of extracellular matrix molecules, cell surface-associated molecules, and receptors such as CD36, cell adhesion molecule with homology to L1CAM, COL15A1, ESAM, FABP1, HAPLN1, LAMC3, LYVE1, NGFR, PCDH11X, and PTX3. We emphasize that LYVE-1 is highly expressed in liver sinusoids and was detected via our biotinylation method. Furthermore, several enzymes that may be found in the extracellular compartment were detected (e.g., ECE1, GGT1). It is of note that ECE1 has two activities, one intracellular and one extracellular at the liver plasma membrane. Also, BRP44 has been shown in gene expression analysis to be overexpressed in the liver (Gene Expression Atlas). Furthermore, many metabolic enzymes were detected. Among these, carnitine palmitoyltransferase 1A had specific annotation pertaining to the liver tissue.
In the second cluster (intestine, Fig. 9B), 39 proteins were found. Among these, a number of extracellular matrix molecules, cell surface-associated molecules, receptors, and secreted proteins were found, such as ADAMTS-like 1, ANGPTL2, caveolin 1, CDH17, chromogranin B, catenin beta 1 cathepsin D, desmonglein 2, EGF containing fibulin-like extracellular matrix protein 1, elastin microfibril interfacer 3, FAT3, FBLN5, FBN2, FGL2, FNDC1, GPA33, GRM7, LRIG1, MDGA1, MEGF6, MXRA5, NEGR1, neuroligin 3, NTNG1, THBS3, TNC, and TNR. Furthermore, some intracellular molecules and enzymes were also detected (contactin associated protein-like 1, CYP2E1, GDPD1, KIAA1731, LIPH, MOXD1, MYLK, PTGR1, and TLN1).
In the third cluster (CAM, Fig. 9C), 64 proteins were found. Among these were a number of extracellular matrix molecules, cell surface-associated molecules, receptors, and secreted proteins such as A2ML1, AMBP, ANXA1, ANXA2, CD99, COL17A1, calponin 2, desmonglein 1, FAT2, FSTL1, LCN15, LUM, METRNL, ORM2, PSCA, SPARC, SERPIN, SERPINB3, SDK2, SPINT1, and TNFRSF6B.
In the fourth cluster (kidney, Fig. 9D), only 10 proteins were found. It is of note that the kidney-specific angiotensin-converting enzyme was detected in the kidney in our analysis.
In addition, we looked for proteins in the CIKL group where peptides were found in one tissue but not in the others. For CAM, intestine, kidney, and liver, 15, 11, 8, and 9 proteins were identified, respectively (supplemental Table S4). Among the interesting proteins were PSCA, FAT2, A2ML1, NUCB2, LYG2, and TNFRSF6B (CAM); NTNG1, GPLD1, MEGF6, MDGA1, TNRLRIG1, and GPA33 (intestine); FLT4 and RIMS2 (kidney); and FABP2 and PYGL (liver). In addition, liver-specific metabolic enzymes were also detected.
Tumor-specific Peptides (Human Genes)
To identify proteins that were derived from the tumor, we screened for peptides that matched human proteins exactly but not chicken proteins (supplemental Table S5). Peptides with exact matches to only human proteins (49 proteins) were included in the list (Table II). Among these proteins were a number of extracellular/membrane proteins. These included proteins already associated with tumors, such as COL1A1, COL4A1, FN1, LOXL1, TGFBI, TNC, and VIM. Furthermore, annexins (ANXA1, ANXA2, and ANXA5), which are also membrane proteins, were detected. Additionally, several metabolic enzymes were detected, including glycolytic enzymes (aldolase A, fructose-bisphosphate; aldolase C, fructose-bisphosphate; enolase 2; GAPDH; LDHA; LDHAL68; PKM2; and TPI). It is of note that the tumor-specific PKM2 was detected in our analysis, which is an enzyme that contributes to the Warburg effect in tumor cells. Furthermore, heat-shock proteins (HSP90AB1, HSPA5, and HSPD1) and the antioxidant protein PRDX4 were also detected.
Table II. List of proteins from the tumor/CAM sample that were unambiguously identified as human.
| Gene | Number of peptides | Products |
|---|---|---|
| Extracellular, secreted, or membranar proteins | ||
| ANXA5 | 3 | Annexin A5 |
| FN1 | 3 | Fibronectin 1 |
| TGFBI | 3 | Transforming growth factor, beta-induced, 68 kDa |
| TNC | 3 | Tenascin C |
| ANXA1 | 2 | Annexin A1 |
| ANXA2 | 2 | Annexin A2 |
| COL1A1 | 2 | Collagen, type I, alpha 1 |
| ITGA5 | 2 | Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| COL4A1 | 1 | Collagen, type IV, alpha 1 |
| LOXL1 | 1 | Lysyl oxidase-like 1 |
| PDIA4 | 1 | Protein disulfide isomerase family A, member 4 |
| PDIA6 | 1 | Protein disulfide isomerase family A, member 6 |
| PRDX4 | 1 | Peroxiredoxin 4 |
| SBSN | 1 | Suprabasin |
| Glycolytic or fermentation enzyme | ||
| PCCA | 9 | Propionyl CoA carboxylase, alpha polypeptide |
| GAPDH | 7 | Glyceraldehyde-3-phosphate dehydrogenase |
| PKM2 | 7 | Pyruvate kinase, muscle |
| PC | 5 | Pyruvate carboxylase |
| ENO1 | 4 | Enolase 1 (alpha) |
| ALDOA | 3 | Aldolase A, fructose-bisphosphate |
| LDHA | 3 | Lactate dehydrogenase A |
| MCCC1 | 3 | Methylcrotonoyl-CoA carboxylase 1 (alpha) |
| TPI1 | 2 | Triosephosphate isomerase 1 |
| ALDOC | 1 | Aldolase C, fructose-bisphosphate |
| ENO2 | 1 | Enolase 2 (gamma, neuronal) |
| GPI | 1 | Glucose-6-phosphate isomerase |
| LDHAL6B | 1 | Lactate dehydrogenase A-like 6B |
| PGK1 | 1 | Phosphoglycerate kinase 1 |
| Cytoskeleton proteins | ||
| VIM | 7 | Vimentin |
| LMNA | 1 | Lamin A/C |
| MYH14 | 1 | Myosin, heavy chain 14, non-muscle |
| MYH9 | 1 | Myosin, heavy chain 9, non-muscle |
| TUBB8 | 1 | Tubulin, beta 8 |
| Stress proteins | ||
| AKR1B1 | 2 | Aldo-keto reductase family 1, member B1 (aldose reductase) |
| HSPD1 | 2 | Heat shock 60-kDa protein 1 (chaperonin) |
| HSP90AB1 | 1 | Heat shock protein 90-kDa alpha (cytosolic), class B member 1 |
| HSPA5 | 1 | Heat shock 70-kDa protein 5 (glucose-regulated protein, 78 kDa) |
| PRDX4 | 1 | Peroxiredoxin 4 |
| Others | ||
| PHEX | 1 | Phosphate regulating endopeptidase homolog, X-linked |
| AEBP1 | 1 | AE binding protein 1 |
| EEF2 | 1 | Eukaryotic translation elongation factor 2 |
Note: This table lists glioma genes identified by peptide matching. Genes had peptides specific to human (glioma cells) with eight genes matching at least seven different human specific peptides. Ten genes had promiscuous but human-specific peptides (data not shown).
We next performed a literature search for glioma involvement of human-specific peptide genes. Keyword searches using “glioma” and “glioblastoma” revealed that 17 of the 48 genes had published evidence of glioma involvement, and these are listed with PubMed ID numbers in Table III. These results indicate good correlation of the identified proteins to known genes involved in glioma pathobiology.
Table III. Literature search of glioma involvement of human-specific peptide genes.
| Gene | Total abstracts | Abstracts matching keywords | PubMed ID of glioma or glioblastoma matching publications |
|---|---|---|---|
| TNC | 149 | 11 | 18794852 20622113 7694284 11920587 18757408 16292494 11313993 16388320 8548761 19459858 19147558 |
| VIM | 238 | 9 | 3027087 8381971 12169273 16944923 11082283 18046501 21317457 12084347 18947333 |
| FN1 | 407 | 5 | 20819642 10783396 21980357 10470109 1409674 |
| ANXA2 | 221 | 5 | 20018898 21033036 21976520 10777578 7518469 |
| HSPA5 | 285 | 4 | 10085239 21112319 18708359 18403493 |
| ITGA5 | 227 | 4 | 16569642 20514406 18992284 18464290 |
| COL4A1 | 122 | 2 | 10382266 9506531 |
| COL1A1 | 459 | 1 | 1409674 |
| HSPD1 | 230 | 1 | 20978188 |
| GAPDH | 188 | 1 | 17597534 |
| TGFBI | 159 | 1 | 20419098 |
| ANXA1 | 146 | 1 | 19767728 |
| ENO1 | 108 | 1 | 19795461 |
| KRT1 | 72 | 1 | 16944923 |
| KRT10 | 63 | 1 | 16944923 |
| ALDOC | 24 | 1 | 2209624 |
| SBSN | 9 | 1 | 21574646 |
Note: This table lists the 17 genes and PubMed ID numbers for those having matched glioma-related keyword searches.
Endothelial Cell Expression of Orthologs (EndoFactor)
The human orthologs of the chicken proteins were then analyzed for their endothelial cell expression (we call this their EndoFactor) using methods similar to those of Herbert et al., who developed an analysis of endothelial gene expression signatures (25). In this work, we combined cDNA libraries with a Roche 454 RNA-seq hypoxia liver endothelium library to identify endothelial- expressed genes within the proteomic data. For the full set of orthologs, an analysis was performed that matched gene symbols between orthologs and endothelial enriched genes. A criterion was used to rank the genes: (i) a gene was log2(FC) ≥ 1 (2-fold) up-regulated in endothelial cells, or (ii) genes showed an endothelial specific profile by having zero counts in the non-endothelial pool. Two hundred and five genes with an enriched endothelial expression profile were found (supplemental Table S6, sheet S6A). Seventy-nine of these genes were membrane, 46 extracellular, and 80 intracellular. Membrane genes with the highest FC (log2(FC) > 2.5) were KDR, CDH5, HSD17B2, CALCRL, ELTD1, CD93, LOC100652938, TEK, SLC12A2, NRCAM, PVRL3, PECAM1, CUBN, FAT4, LYVE1, THBD, PODXL, PTPRB, CDH13, INSR, CD109, PLXNA4, PLXND1, EPHB1, PTPRG, ACP2, ENG, CD47, TMEM119, STT3A, PLXNA2, CXADR, CNTNAP1, FABP2, ADAM17, ADAM10, FREM2, IL31RA, NRP1, ITGA6, ENPP2, SLC44A1, CD9, and LRRN4. Extracellular proteins with the highest FC (log2(FC) > 2.5) were ANGPT2, MMRN1, VWF, SRPX, MMRN2, PROS1, ectonucleotide pyrophosphatase/phosphodiesterase 6, EGF containing fibulin-like extracellular matrix protein 1, capping protein (actin filament) muscle Z-line alpha 1, COL15A1, alpha-2-macroglobulin, C7, chromosome 8 ORF 84, GPC5, glycosylphosphatidylinositol, THBS1, TXN, asporin, MAMDC2, SPINK5, IL1RAP, SERPINB5, PON2, CTNNB1, and SERPINB9.
Angiogenic Screen and Possible Vascular Targets
The proteomic technique used here was preferentially employed to acquire membrane and extracellular proteins from a developing chicken embryo. Because angiogenesis is a process pivotal to both embryonic development and tumor growth, an angiogenic cDNA library screen was performed to compare tumor/fetal and normal bulk public tissue libraries to rank human orthologs as potential vascular targets. This was also combined with the endothelial cDNA/454 screen, and 136 genes had a positive FC in both endothelial and angiogenic screens (supplemental Table S6, sheet S6B). Several genes among these are ANGPT2, ANGPTL2, ANXA5, COL15A1, EPHB1, EPHB2, IGFBP7, IL31RA, JAM3, DKK3, MMP15, PLXNA2, PLXNA4, PTX3, SULT1E1, SPARC, PXDN, COL4A1, NID1, and VIM. Some of these genes are already published as tumor endothelial markers/vascular targets. For instance, the genes COL4A1, SPARC, VIM, and IGFBP7 were reported by van Beijnum et al. as tumor angiogenesis genes (26), and the genes PXDN, PLXNA2, NID1, and DKK3 were reported as endothelial markers of brain tumors (27) or colon tumors (28). Therefore, in this list there could be other potential tumor endothelial genes.
Literature Scanning to Determine Angiogenesis Involvement
To provide further evidence regarding whether known angiogenesis-related genes were in fact being selected with the proteomic technique, angiogenesis and tumor angiogenesis keywords were searched against total publications for a gene to derive an “AngioScore” (the percentage of publications matching a word to the total number of publications) (supplemental Table S6, sheet S6C). Forty-three of these had an AngioScore ≥ 50. The best AngioScores were found for KDR (94), CDH5 (90), FLT4 (86), endothelin receptor type A (81), PROCR (80), TEK (77), ECE1 (74), carbonic anhydrase IX (74), amine oxidase copper containing 3 (70), COL18A1 (70), VCAM1 (70), PECAM1 (70), NRP2 (69), SERPINB5 (68), TUSC5 (67), NRP1 (66), ANGPT2 (66), MCAM (65), cell adhesion molecule 1 (65), IGFBP7 (63), ADAM metallopeptidase domain 17 (63), ANG (62), and TIE1 (62). It is of note that many of the classic, well-known endothelial genes or angiogenesis factors/receptors were present in both lists (EndoFactor and AngioScore). Furthermore, we also detected recently identified angiogenesis regulators such as LOXL2.
We also compared the percentages of proteins with positive EndoFactors or AngioScores for the different tissues tested (Table IV, supplemental Table S7). All proteins with a positive EndoFactor (EndoFactor log2(F) ≥ 1) were included, and for the AngioScores, a cut-off of 50 was chosen. It is interesting that the percentages of proteins with endothelial cell signatures were relatively constant at a total of 20%. This indicates that one-fifth of proteins identified exhibit an endothelial cell expression signature.
Table IV. Number of proteins with endothelial gene signature in the different tissues.
| Total number of proteins | EndoFactor positive | AngioScore > 50 | |
|---|---|---|---|
| CAM | 1295 | 670 | 49 |
| Kidney | 1090 | 191 | 43 |
| Liver | 1097 | 193 | 42 |
| Intestine | 1101 | 191 | 43 |
| Tumor | 664 | 106 | 30 |
| Wound | 670 | 114 | 31 |
Experimental Validation of Genes
We could not perform immunohistology on chicken tissue because of the lack of specific chicken antibodies for these proteins. Therefore, we analyzed the expression of some of the human proteins in tumors grown on the chicken CAM derived from human U87 glioma cells (Fig. 10).
Fig. 10.
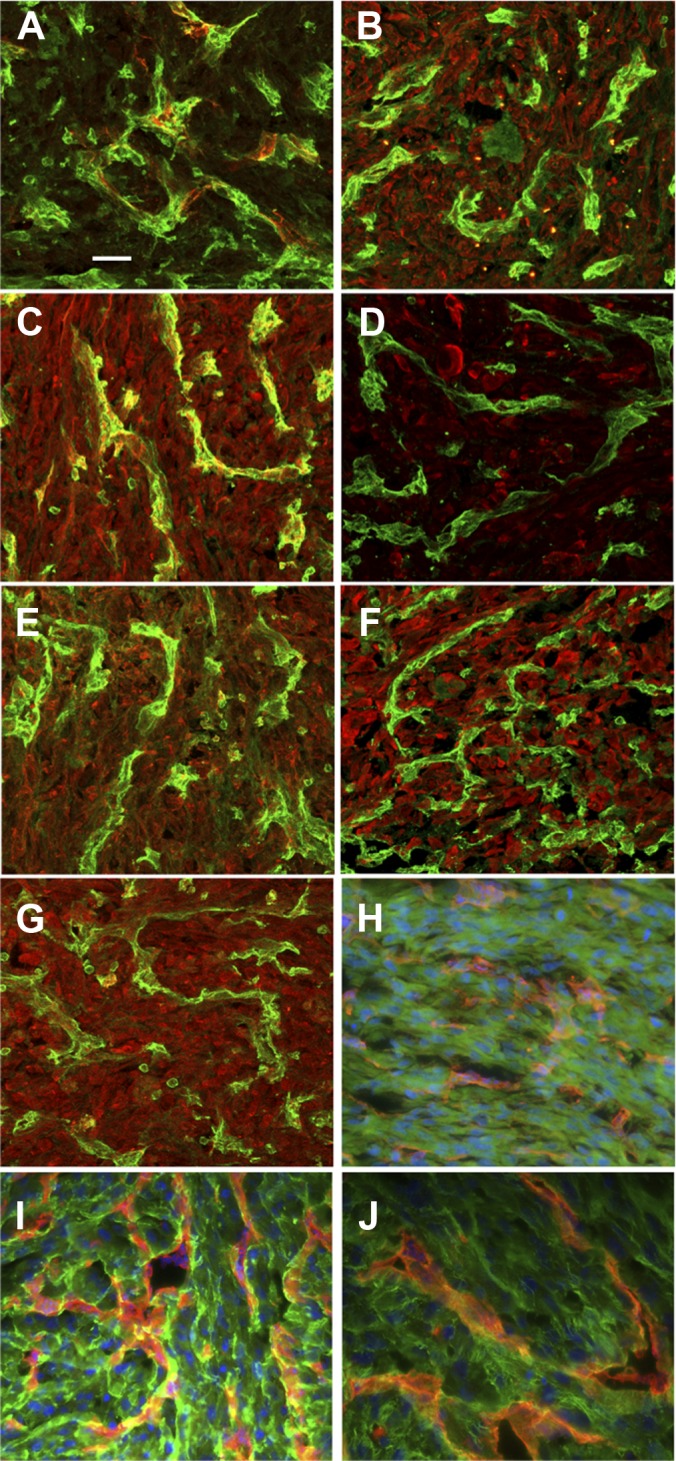
Expression of proteins from tumor xenografts derived from human glioma cells. Immunostaining was performed for selected proteins associated to vessels or the extracellular matrix. Immunostaining for (A) Col IV, (B) FSTL1, (C) HRNR, (D) LOXL2, (E) Plexin D1, (F) SIRPA, and (G) SRPX. Vessels are stained with a FITC conjugated-lectin (SNA) in panels A–G. Tenacin C, TGFβi, and fibronectin are respectively, in panels H, I, and J. Vessels are stained with a biotin-conjugated lectin (SNA) in panels H–J and were detected with rhodamin-conjugated streptavidin. The magnification is 60 ×, and the scale bar is 25 μm. As a nonspecific control, non-immune IGG was used; this did not reveal significant staining (data not shown).
The selection of tumor enriched proteins for validation was based on four techniques: (i) Peptides that matched human proteins exactly but not chicken were priority candidates, as they were of human origin. (ii) As tumor peptides can match both human and chicken genes, differential gene expression of proteins between tumor and CAM identified further candidate genes. (iii) Protein expression profiles of tumor, wound, and CAM samples were clustered using the Cluster Affinity Search Tool to derive tumor enriched candidate genes. (iv) Finally, public Unigene cDNA libraries, literature scans, and cancer databases were surveyed to rank the candidate genes. Following this strategy, 10 genes were chosen for validation, and these are listed in Table V.
Table V. Tumour enriched candidate genes.
| Gene | Product | Gene Ontology cell component | Analysis | Human peptides | % Unigene brain ESTs | Rembrandt database | Total abstracts | Glioma keyword abstracts |
|---|---|---|---|---|---|---|---|---|
| TNC | Tenascin C | Extracellular | Human peptides | 3 | 12 | Up in glioma | 156 | 15 |
| FN1 | Fibronectin 1 | Extracellular | Human peptides | 3 | 5 | Up in glioma | 433 | 7 |
| TGFBI | Transforming growth factor, beta-induced, 68 kDa | Membrane | Human peptides | 3 | 19 | Up in glioma | 172 | 5 |
| HRNR | Hornerin | Extracellular | Human peptides | 2 | 0 | Not found | 14 | 0 |
| COL4A1 | Collagen, type IV, alpha 1 | Extracellular | Human peptides | 1 | 4 | Up in glioma | 132 | 2 |
| SIRPA | Signal-regulatory protein alpha | Membrane | Cast cluster | 0 | 39 | Not differential | 81 | 4 |
| SRPX | Sushi-repeat containing protein, X-linked 2 | Extracellular | Cast cluster | 0 | 17 | Not differential | 19 | 1 |
| PLXND1 | Plexin D1 | Membrane | Cast cluster | 0 | 15 | Not differential | 20 | 0 |
| FSTL1 | Follistatin-like 1 | Extracellular | Cast cluster | 0 | 9 | Up in glioma | 25 | 0 |
| LOXL2 | Lysyl oxidase-like 2 | Extracellular | Cast cluster | 0 | 7 | Up in glioma | 37 | 0 |
Note: This table lists the 10 candidate tumor enriched genes chosen for immunohistochemistry validation. Attributes reported are the number of human-specific peptides matching a gene, the percentage of total Unigene cluster expressed sequence tags (ESTs) expressed in brain for a gene, a Rembrandt database glioma enriched profile, total PubMed article abstracts, and the number of abstracts that contain one or more of the following keywords: glioma, glioblastoma, astrocytoma, and neuroblastoma.
Immunostaining was performed by immunofluorescence staining for selected proteins associated to vessels or the extracellular matrix. We performed immunostaining for Col IV (a), FSTL1 (b), HRNR (c), LOXL2 (d), Plexin D1 (e), SIRPA (f), SRPX (g), Tenacin C (h), TGF-βi (i), and fibronectin (j). These were selected for several reasons. FSTL1, HRNR, LOXL2, PLXND1, and SRPX are not known to be expressed in glioma, and SIRPA has low expression. TGF-βi, human tenascin, and fibronectin had excellent peptide coverage and were well detected via immunolabeling. Blood vessels were stained using SNA-1 isolectin as previously described (14). Human proteins were detected in tumor cells, in the tumor matrix, or around blood vessels, but not in blood vessels. In addition, HRNR exhibited a vessel-associated staining pattern. These results are in agreement with the mass spectrometry data from the U87 xenografts obtained after in vivo biotinylation.
We then performed additional staining on human tumor samples from oligo-astrocytoma (supplemental Fig. S4). Tumor samples were stained with anti-FSTL1, anti-SIRPA, anti-HRNR, and anti-SRPX antibodies. We used sheep anti-CD31 for co-staining with mouse anti-COL IV. FSTL1, SIRPA, and SRPX were tumor and stroma associated without vessel staining. In contrast, HRNR was found associated to the vasculature, and for Col IV a vascular staining was observed.
DISCUSSION
Novel Approach
In this research, we applied in vivo biotinylation combined with high-resolution mass spectrometry and bioinformatic analyses to study the vascular and matrix proteome in the chicken embryo. This is the first time such an approach has been used in an embryonic organism. We provide, in addition, the first description of the vascular and matrix proteome in an embryonic organism. We performed our analysis not only on several embryonic organs, but also on tumor cells xenografted into the chicken CAM and granulation tissue induced by wounding. In contrast to previous techniques, we purified the biotynylated proteins by means of mild warm-water elution, which significantly reduced the background and prevented the overrepresentation of single proteins that would mask less well-represented proteins in the mass spectroscopy analysis. Also, proteins were deglycosylated, as described elsewhere (16), to increase the pool of peptides available for spectroscopy and proteomic fingerprinting.
There have been several publications reporting proteomic studies on the chicken embryo. These include, for example, studies on the chicken at stage 29 of development (29), embryo chicken vitelline membrane (30), the facial development of E3-E5 embryos (31), subcutaneous gel in avian hatchlings (32), the embryonic chicken retina (33, 34), the chicken egg yolk plasma and granules (35), the chicken egg white (36, 37), cerebrospinal fluid (38), embryonic chicken gonadal primordial germ cells (39), matrix vesicles from chicken femur (40), the calcified eggshell layer (41), and the chicken cardiovascular system (42). These studies mainly used standard proteomic techniques such as one-dimensional or two-dimensional gel electrophoresis of total proteins, often coupled with MALDI-TOF-MS. These techniques provide information on a global scale of proteins found in a total tissue extract, but not in tissue-specific compartments. Furthermore, these techniques cannot distinguish specific components of plasma membrane and extracellular matrix from intracellular proteins.
In contrast to these studies, our procedure involved the in vivo biotinylation of membrane or extracellular proteins associated with the vasculature and stroma. It is impossible to avoid also detecting intracellular or yolk proteins because of vascular leakage or cell and tissue injury after perfusion. Our technique is based on the method published by Neri and collaborators, who pioneered the detection and analysis of extracellular or cell membrane proteins via in vivo biotinylation in adult rodents (16, 17). They have applied their technique to identify novel, potential therapeutic targets in tumors grown in mice. More recently, they have also applied their technique to human colon cancer via ex vivo perfusion (18). However, our procedure differs from theirs in several respects. Firstly, we removed biotinylated proteins from the column after binding to streptavidin not by means of trypsin digestion, but through the elution of water at 70 °C, as it has been previously shown that water at 70 °C is able to reversibly break the interaction between biotin and streptavidin (21). Secondly, by using this method and all the required controls, we showed that our proteins were indeed biotinylated and that this protein fraction was largely extracellular and membrane associated. This is different from trypsin-clipping on columns where only digested peptides, and not entire biotinylated proteins, are eluted from columns, thus making it impossible to determine whether proteins are biotinylated or not after elution. In addition, we also deglycosylated our biotinylated proteins in a manner similar to that reported by the Neri laboratory (43).
Enrichment of Extracellular and Membrane Proteins
A great number of proteins identified were found to belong to cellular components or biological processes of the extracellular space when a DAVID Gene Ontology analysis was performed. This is in agreement with the immunohistology, which evidenced significant in vivo biotinylation in the extracellular space around vessels and in the surrounding tissue. However, in the case of tumor vasculature, we detected significant quantities of intracellular proteins. This is most likely due to a high necrosis rate and, as a consequence, a release of intracellular components into the interstitium, which occurs in a fast-growing tumor. The intracellular proteins detected reflect the active metabolic demand and the Warburg effect that occurs in tumor cells. In particular, we found that PKM2, which had one of the best peptide coverages, was specifically expressed in tumor cells and not in transformed cells (44–46). Furthermore, LDH was also detected, which reflects active lactate production in tumor cells.
Differential Expression
Differential protein expression analysis was performed via a label-free quantitative proteomic approach as described under “Experimental Procedures.” We emphasize that our results led to identifications of proteins that are very characteristic for a given tissue. When protein clusters were analyzed for overexpression in a specific organ, the greatest number of proteins was found in the liver. This was expected, as the liver is an organ that has many biosynthetic and metabolic functions.
Liver Enriched Proteins
Most important, liver-enriched proteins found in this study have also been reported to be enriched in the liver in other transcriptomic studies and databases. For example, LYVE-1, detected in this analysis, is known to be highly overexpressed in liver sinusoids (47). FABP1 is found, as expected, to be enriched in liver. Fatty acid binding proteins are a family of small, highly conserved carrier proteins that bind long-chain fatty acids and other hydrophobic ligands. This protein can also be found outside the cell and is able to bind bile acids (48). It is thought that the roles of fatty acid binding proteins include fatty acid uptake, transport, and metabolism and are regulated by liver-enriched transcription factors HNF3β and C/EBPα (49). The fatty acid translocase CD36 was found, as might be expected, in the liver. Fatty acid translocase CD36 (FAT/CD36) mediates the uptake and intracellular transport of long-chain fatty acids in diverse cell types. It has a role in liver pathology, including hepatic steatosis and non-alcoholic fatty liver disease (50). The endothelial cell adhesion molecule (ESAM) is a 55-kDa type I transmembrane glycoprotein of the JAM family, which is a member of the immunoglobulin superfamily, that is found in endothelial cells. ESAM has been detected in the liver sinusoids and is responsible for leukocyte migration in the liver (51). ESAM also might be present on hematopoietic stem cells in the liver; evidence in support of this has been reported in murine fetal liver (52). Endothelin converting enzyme (ECE1) has been described in the liver and may be up-regulated after injury, inflammation, or allograft rejection (53, 54). It has two activities, one intracellular and another extracellular at the liver plasma membrane. Brain protein 44 has been shown in gene expression analysis to be overexpressed in the liver (Gene Expression Atlas). LAMC3 is a component of basement membranes. It is also found in other tissues, such as skin, heart, lung, and the reproductive tracts, and is a prominent element of the apical surfaces found on ciliated epithelial cells. It has been detected in the retinal vasculature and is thought to play an important role in vessel development (55). LAMC3 has not yet been described in the liver but could be present in liver basement membranes or in the liver vasculature.
Intestine Enriched Proteins
The intestinal enriched cluster exhibited a number of proteins that have previously been described in the digestive system. For example, GPA33 is a transmembrane protein, found overexpressed in the digestive system and colon (Gene Expression Atlas Database).
The leucine-rich repeats and Ig-like domains 1 (LRIG1) gene is a transmembrane protein that interacts with ErbBs and promotes its degradation. It is a key player in the control of intestinal homeostasis. LRIG1 has recently been found to be enriched at the crypt base and in the progenitor compartment of the small intestine and colon. It limits the size of the intestinal progenitor compartment by dampening EGF/ErbB-triggered stem cell expansion (56).
Chromogranin B encodes a tyrosine-sulfated secretory protein abundant in peptidergic endocrine cells and neurons. This protein might serve as a precursor for regulatory peptides. Angiogenic properties of this molecule have also been described (57). This gene has been described as present in the small intestine of the chicken (58). The MAM domain containing glycosylphosphatidylinositol anchor 1 (MDGA1) is a transmembrane glycosylphosphatidylinositol anchored protein that has a role in the nervous system at the plasma membrane. It has also been found in the instestine and is down-regulated in inflammatory bowel disease. A protein classically found in the central nervous system that we identified in the intestine cluster is neuroligin 3. This protein encodes a member of a family of neuronal cell surface proteins. Members of this family may act as splice site-specific ligands for beta-neurexins and might be involved in the formation and remodeling of central nervous system synapses (59). It is interesting that this protein is also detected in the intestine. It might play a role in innervation of the gut.
CAM Enriched Proteins
The CAM cluster exhibited a number of proteins that are involved in adhesion or other extracellular functions. Annexins (ANXA2, ANXA1) were detected. These proteins have multiple functions such as Ca2+ binding, interaction with phospholipids, and recognition of soluble factors such as CXCL12 (60). Among the matrix molecules found in this study, lumican and collagen 17 were overexpressed. SIDEKICK2 encodes a protein that is a member of the immunoglobulin superfamily. The protein contains two immunoglobulin domains and 13 fibronectin type III domains and might serve as an adhesion receptor (61). Another adhesion molecule is FAT2, which is an integral membrane protein characterized by the presence of cadherin-type repeats and plays a role in the control of planar cell polarity (62). We also detected CD99, which has been described as expressed in immune cells and is a key mediator of the transendothelial migration of neutrophils and dendritic cells (63, 64). We found orosomucoid (alpha1-acid glycoprotein), which is needed to maintain the high capillary permselectivity required for normal homeostasis. It was detected in endothelial cells as well as in the liver. Follistatin-like 1, an activin-binding protein, was also found in our analysis. Finally, proteases and their inhibitors were also overexpressed in the CAM cluster (AMBP, SPARC, SPINT1, and SERPINB3).
Kidney Enriched Proteins
In the kidney cluster, only a limited amount of overexpressed proteins was detected. It is of note that angiotensin-converting enzyme was found in this group. Angiotensin-converting enzyme is synthesized in the kidney and is a major regulator of vessel tonus (65).
t Test of Tumor versus Wound
Several interesting observations were made when wound and tumor expression were compared using Volcano plots. Among the proteins overexpressed were several matrix molecules and receptors (ANAXA5, ITGB5, collagen triple helix repeat containing 1, MSLN, and VNN2). Furthermore, complement and coagulation factors, protease, and their inhibitors were detected (angiotensinogen, MMP2, F9, SERPINA10, SERPINA1, C3, F2, A2ML1, CFB, and KNG1). However, some carrier proteins, intracellular binding proteins, or enzymes were surprisingly highly overexpressed, such as NAMPT, OAS3, POMGNT1, and PDIA3. This most likely was due to a high necrosis rate, which leads to the release of intracellular components into the interstitium, as occurs in a fast-growing tumor.
NAMPT was the most tumor-enriched protein. NAMPT is the gene symbol of the enzyme nicotinamide phosphoribosyltransferase, which is important in the biosynthesis of nicotinamide-adenine-dinucleotide. It catalyzes the condensation of nicotinamide and 5-phosphoribosyl-pyrophosphate to produce nicotinamide mononucleotide and inorganic pyrophosphate, and it is the rate-limiting enzyme in the salvage pathway of nicotinamide-adenine-dinucleotide synthesis. In the literature, NAMPT has been shown to be up-regulated in colon cancer at both mRNA and protein levels (66, 67). In addition, it has been shown to be up-regulated in glioma and has been proposed as a serum prognostic marker of glioblastoma (68). It has been shown that adipokine, the extracellular isoform of NAMPT, promotes angiogenesis and is a potent activator of in vivo neovascularization of the chick CAM (68). Furthermore, Notch1-dependent production of FGF2 is induced in NAMPT-primed endothelial angiogenesis. Several groups have shown that NAMPT/Visfatin can influence angiogenesis (69–72). NAMPT has also been shown to induce proliferation and tube formation in human umbilical vein endothelial cells (73). Lastly, the compound FK866 has been reported to inhibit NAMPT and starve tumor cells that rely on nicotinamide-adenine-dinucleotide metabolism for energy. This is a potential therapy for cancer (74, 75).
CALCRL, H1F0, EPB41, ANK1, and NMT were most overexpressed in wound versus tumor. CALCRL belongs to the adrenomedullin receptor complex that is involved in vasodilation and has been shown to be up-regulated in hypoxia, which, in conjunction with adrenomedullin, can enhance endothelial cell survival and migration. CALCRL has also an effect of increasing vessel permeability and can facilitate the migration of white blood cells (76), which suggests a feasible role for CALCRL in wound healing, as white blood cells are delivered to a wound to substantiate an immune response and provide macrophages that perform phagocytosis of necrotic tissue.
Retinoic acid (vitamin A) is also implicated in normal wound healing (77–79). H1F0 is up-regulated in mice in response to retinoic acid (80) and might constitute a possible link to wound healing and granulation tissue formation.
EPB41 has also been shown to have a pivotal role in wound healing in mice. It was demonstrated that EPB41 knockout mice show a phenotype of wound healing impairment (81). In the same study, it was also shown that keratinocytes with silenced EPB41 exhibit reduced adhesion, cell spreading, migration, and motility. In addition, DLG1 has a role in wound healing, and it is proposed that the transglutaminase EPB41 binds to DLG1 (82). Ankyrin 1 could play a role in wound healing by initiating endothelial cell proliferation and/or adhesion via its interaction with CD44v10 and IP3 receptors on endothelial cell lipid rafts (83).
It is interesting that the glycolysis pathway gene BPGM is up-regulated in wound tissue (84). NMT1 is an enzyme known to catalyze the attachment of special lipids onto proteins, which is a pivotal process in cellular growth. This process is also involved in the development of cells from the leukocytic lineage known to be essential for rapid cell proliferation.
Another gene significantly overexpressed in wound relative to tumor is FZD2. This gene is a member of a family that encodes seven-transmembrane domain proteins that are receptors for the wingless type MMTV integration site family of signaling proteins and encodes a protein that is coupled to the β-catenin canonical signaling pathway. This pathway plays a role in tissue regeneration and is regulated during these processes (85).
Forty-nine proteins detected in tumor samples were unambiguously of human origin and thus were derived from tumor cells. Many of these proteins were extracellular, matrix, or matrix-associated proteins. The highest peptide coverage (>2) was found for ANXA5, FN1, TGFBI, TNC, ANAXA1, A2, type I collagen (COL1A1), and integrin α5. This indicates that our biotinylation procedure in the chicken embryo allows the identification of tumor-derived matrix or matrix-associated proteins when human tumor cells are xenografted into the CAM. Besides matrix proteins, the high percentage of glycolytic enzymes reflects active tumor metabolism and the Warburg effect (PCCA; GAPDH; PKM2; aldolase A, fructose-bisphosphate; and LDH). Some cytoskeletal proteins and heat shock proteins were also detected. Our in vivo biotinylation method should detect exclusively extracellular and membrane proteins, but this was not the case. It is likely that cell death or injury led to extracellular deposits of intracellular proteins that were then detected via our in vivo biotinylation method.
Endothelial Cell Signatures
Endothelial cell data filtering of our proteomic data revealed that many well-known angiogenesis regulators or vascular associated proteins were detected in our proteomic analysis. It is interesting that the percentages of proteins with endothelial cell signatures were relatively constant at a total of 20%. This indicates that one-fifth of proteins identified exhibit an endothelial cell signature. Endothelial cell enrichment for proteins with the highest FC were SLC12A2, PECAM1, CDH5, CALCRL, ELTD1, CD93, PODXL, TEK, INSR, PTPRB, CD47, KDR, TT3A, endoglin, coxsackie virus and adenovirus receptor, ADAM metallopeptidase domain 17, and NRP1. We also used another bioinformatic data mining approach to identify genes implicated in angiogenesis (AngioScore). The highest AngioScores were observed for many angiogenesis regulators and receptors such as VEGFR2 (KDR), CDH5, TEK, PECAM1, VCAM, NRP1, ANGPT2, TIE-1 ANG, endoglin, ANGPTL1, and THBS1. It is interesting that recently validated angiogenesis regulators were identified here, such as LOXL2 (86). Through the combination of in vivo proteomics and a cDNA library angiogenic screen, potential novel tumor angiogenesis genes were identified, as evidenced by the presence of previously published tumor endothelial markers. In summary, we found that in this high-throughput proteomic study, both EndoFactor and data mining strategies adequately identified many proteins involved in angiogenesis. We emphasize that we detected many of the classical angiogenesis factors and receptors. These molecules are often expressed at lower abundance levels than matrix molecules and are subsequently more difficult to detect. This indicates that the biotinylation procedure we used in this study is sensitive enough to detect these proteins. Eighty percent of proteins, however, did not exhibit an endothelial cell signature. This indicates that the in vivo biotinylation will evidence a majority of proteins that are not strictly endothelial cell associated and are derived from other stroma or tissue compartments.
We performed immunostainings of 10 proteins detected in our study in glioblastoma-implanted CAM and human astrocytoma. For five of these proteins, there was no published evidence of an association with glioma. In particular, HRNR, which is found in regenerating, psoriatic, and healthy human skin and has a role in keratinocyte cornification (87), was detected in glioma cells and in association with tumor vessels. Thus, it might constitute a new glioma and vascular marker.
All in all, we describe in this article for the first time the use of the in vivo biotinylation method combined with high-resolution mass spectrometry and bioinformatic analysis in an embryo to study the vascular and matrix proteome. This technique allows the identification of a significant number of proteins associated with the vasculature, the stroma, and the extracellular matrix. The results also show that a number of classical angiogenesis regulators were identified; these are generally proteins that are found in lower abundance than extracellular matrix molecules. Furthermore, there is evidence of organ/tissue-specific distributions of a number of proteins. Several proteins in this list are known to be classically found in a given tissue, a finding that validates our proteomic analysis. This study represents also the first description of the proteome associated with the vasculature and the stroma in an embryonic organism. The study and the methodology described in our article might constitute a fertile ground for further developments. Up-scaling of the protein amounts will certainly allow an increase in the number of proteins detected, especially of those of lower abundance, which might still be of functional importance.
Supplementary Material
Acknowledgments
We thank Karima Chaoui for help in the first MS analysis (Institut de Pharmacologie et Biologie Structurale, CNRS, Université de Toulouse, France). We acknowledge the Computational Biology Research Group, Oxford, for use of their hardware and services: Steve Taylor, Simon McGowan, and Zong-Pei Han.
Footnotes
* This work was supported by grants from the European Commission (STROMA, FP6, LSHC-CT-2003-5032, STROMA Consortium) and the Agence National de la Recherche (ANR, “projet blanc”) to A.B. (Endothelial Growth Cones, EGC). This work was also supported by grants to B.M. from the CNRS, the Agence Nationale de la Recherche (ANR/INCa Programme Plates-formes technologiques du vivant), the Fondation pour la Recherche Médicale (Programme Grands Equipements), the Cancéropole Grand Sud-Ouest, and the Région Midi-Pyrénées. W.K. was a recipient of a post-doctoral fellowship from the Lefoulon-Delalande Foundation. F.S. was a recipient of a post-doctoral fellowship from the ANR.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- A2ML1
- alpha-2-macroglobulin-like 1
- ANG
- ankyrin 1, angiogenin, ribonuclease, RNase A family, 5
- ANGPT2
- angiopoietin 2
- ANGPTL
- angiopoietin-related protein
- ANX
- annexin
- CALCRL
- calcitonin receptor-like
- CDH
- cadherin
- COL
- collagen
- CYP2E1
- cytochrome P450, family 2, subfamily E, polypeptide 1
- ECE1
- endothelin converting enzyme 1
- ELTD1
- EGF, latrophilin, and seven transmembrane domain containing 1
- EPB
- erythrocyte membrane protein band
- ESAM
- endothelial cell adhesion molecule
- F13A1
- coagulation factor XIII, A1 polypeptide
- FABP
- fatty acid binding protein
- FAT2
- FAT tumor suppressor homolog 2
- FAT3
- FAT tumor suppressor homolog
- FBLN5
- fibulin 5
- FBN2
- fibrillin 2
- FGL2
- fibrinogen-like 2
- FLRT2
- fibronectin leucine rich transmembrane protein 2
- FLT4
- fms-related tyrosine kinase 4
- FN1
- fibronectin 1
- FNDC1
- fibronectin type III domain containing 1
- FRAS1
- Fraser syndrome 1
- FREM2
- FRAS1 related extracellular matrix protein 2
- FRM2
- fatty acid repression mutant 2
- FSTL1
- follistatin-like 1
- FZD2
- frizzled family receptor 2
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GDPD1
- glycerophosphodiester phosphodiesterase domain containing 1
- GGT1
- gamma-glutamyltransferase 1
- GPA33
- glycoprotein A33
- GPC5
- glypican 5
- GRM7
- glutamate receptor, metabotropic 7
- H1F0
- H1 histone family, member 0
- HAPLN1
- hyaluronan and proteoglycan link protein 1
- HAPLN3
- hyaluronan and proteoglycan link protein 3
- HNRNPM
- heterogeneous nuclear ribonucleoprotein M
- HRNR
- hornerin
- HSD17B2
- hydroxysteroid (17-beta) dehydrogenase 2
- HSP90AB1
- heat shock protein 90-kDa alpha (cytosolic), class B member 1
- HSPA5
- heat shock 70-kDa protein 5
- HSPD1
- heat shock 60kDa protein 1
- IGFBP7
- insulin-like growth factor binding protein 7
- IL1RAP
- interleukin 1 receptor accessory protein
- IL31RA
- interleukin 31 receptor A
- INSR
- insulin receptor
- ITGA1
- integrin, alpha 1
- ITGA6
- integrin, alpha 6
- JAM3
- junctional adhesion molecule 3
- KDR
- kinase insert domain receptor
- KIAA1731
- centrosomal protein KIAA1731
- KNG1
- kininogen 1
- LAMC3
- laminin, gamma 3
- LRIG1
- leucine-rich repeats and immunoglobulin-like domains protein 1
- LRRN4
- leucine rich repeat neuronal 4
- LCN15
- lipocalin 15
- LDHA
- lactate dehydrogenase A
- LIPH
- lipase, member H
- LMNA
- lamin A
- LOXL1
- lysyl oxidase-like 1
- LOXL2
- lysyl oxidase-like 2
- LUM
- lumican
- LYVE1
- lymphatic vessel endothelial hyaluronan receptor 1
- MAMDC2
- MAM domain containing 2
- MCAM
- melanoma cell adhesion molecule
- MDGA1
- MAM domain containing glycosylphosphatidylinositol anchor 1
- MEGF6
- multiple EGF-like-domains 6
- METRNL
- meteorin, glial cell differentiation regulator-like
- MMP2
- matrix metallopeptidase 2
- MMP15
- matrix metallopeptidase 15
- MMRN1
- multimerin 1
- MMRN2
- multimerin 2
- MOXD1
- monooxygenase, DBH-like 1
- MXRA5
- matrix-remodelling associated 5
- MYL6
- myosin, light chain 6, alkali, smooth muscle and non-muscle
- MYLK
- myosin light chain kinase
- NAMPT
- nicotinamide phosphoribosyltransferase
- NAP1L1
- nucleosome assembly protein 1-like 1
- NEGR1
- neuronal growth regulator
- NGFR
- nerve growth factor receptor
- NRCAM
- neuronal cell adhesion molecule
- NRP1
- neuropilin 1
- NTNG1
- netrin G1
- OAS3
- 2′-5′-oligoadenylate synthetase 3
- OLA1
- Obg-like ATPase 1
- ORM2
- orosomucoid 2
- PABPC4
- poly(A) binding protein, cytoplasmic 4
- PCDH11X
- protocadherin 11 X-linked
- PDIA3
- protein disulfide isomerase family A, member 3
- PECAM-1
- platelet endothelial cell adhesion molecule 1
- PFN2
- profilin 2
- PKM2
- pyruvate kinase isozymes M2
- PLSCR1
- phospholipid scramblase 1
- PLXNA2
- plexin A2
- PLXNA4
- plexin A4
- PLXNB2
- plexin B2
- PLXND1
- plexin D1
- PODXL
- podocalyxin-like
- POMGNT1
- protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase
- PON2
- paraoxonase 2
- PRDX4
- peroxiredoxin 4
- PROCR
- endothelial protein C receptor
- PROS1
- protein S (alpha)
- PSCA
- prostate stem cell antigen
- PTGR1
- prostaglandin reductase 1
- PTPRB
- protein tyrosine phosphatase, receptor type B
- PTPRG
- protein tyrosine phosphatase, receptor type G
- PTPRM
- protein tyrosine phosphatase, receptor type M
- PTX3
- pentraxin 3
- PVRL3
- poliovirus receptor-related 3
- RAC2
- ras-related C3 botulinum toxin substrate 2
- SDK2
- sidekick cell adhesion molecule 2
- SERPINB3
- serpin peptidase inhibitor, clade B
- SERPINB5
- serpin peptidase inhibitor, clade B (ovalbumin), member 5
- SERPINB9
- serpin peptidase inhibitor, clade B (ovalbumin), member 9
- SGCD
- sarcoglycan, delta
- SIRPA
- signal-regulatory protein alpha
- SLC4A1
- solute carrier family 4, anion exchanger, member 1
- SLC12A2
- solute carrier family 12
- SLC25A43
- solute carrier family 25, member 43
- SPARC
- secreted protein, acidic, cysteine-rich
- SPINK5
- serine peptidase inhibitor, Kazal type 5
- SPINT1
- serine peptidase inhibitor, Kunitz type 1
- SRPX
- sushi-repeat containing protein, X-linked
- STT3A
- STT3, subunit of the oligosaccharyltransferase complex, homolog A
- SULT1E1
- sulfotransferase family 1E, estrogen-preferring, member 1
- TEK
- tyrosine kinase, endothelial
- TGFBI
- transforming growth factor-β induced protein
- THBD
- thrombomodulin
- THBS1
- thrombospondin 1
- THBS3
- thrombospondin 3
- TIE1
- tyrosine kinase with immunoglobulin-like and EGF-like domains 1
- TLN1
- talin 1
- TMEM119
- transmembrane protein 119
- TNC
- tenascin C
- TNFAIP6
- tumor necrosis factor, alpha-induced protein 6
- TNR
- tenascin R
- TNFRSF6B
- tumor necrosis factor receptor superfamily, member 6b, decoy
- TPI
- Triose phosphate isomerase
- TRAP1
- TNF receptor-associated protein 1
- TSEN54
- tRNA splicing endonuclease 54 homolog
- TSPO
- translocator protein
- TUSC5
- tumor suppressor candidate 5
- TXN
- thioredoxin
- VCAM1
- vascular cell adhesion molecule 1
- VMN
- vimentin
- VNN1
- vanin 1
- VWF
- von Willebrand factor
- VWA1
- von Willebrand factor A domain containing 1
- ZP1.32
- zona pellucida glycoprotein 1.
REFERENCES
- 1. Carmeliet P. (2005) Angiogenesis in life, disease and medicine. Nature 438, 932–936 [DOI] [PubMed] [Google Scholar]
- 2. Folkman J. (1995) Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1, 27–31 [DOI] [PubMed] [Google Scholar]
- 3. Glaros T., Larsen M., Li L. (2009) Macrophages and fibroblasts during inflammation, tissue damage and organ injury. Front. Biosci. 14, 3988–3993 [DOI] [PubMed] [Google Scholar]
- 4. Kalluri R., Zeisberg M. (2006) Fibroblasts in cancer. Nat. Rev. 6, 392–401 [DOI] [PubMed] [Google Scholar]
- 5. Le Douarin N. M. (2008) Developmental patterning deciphered in avian chimeras. Dev. Growth Differ. 50 Suppl 1, S11–S28 [DOI] [PubMed] [Google Scholar]
- 6. Le Douarin N. M., Creuzet S., Couly G., Dupin E. (2004) Neural crest cell plasticity and its limits. Development 131, 4637–4650 [DOI] [PubMed] [Google Scholar]
- 7. Dieterlen-Lievre F., Jaffredo T. (2009) Decoding the hemogenic endothelium in mammals. Cell Stem Cell 4, 189–190 [DOI] [PubMed] [Google Scholar]
- 8. Stern C. D. (2005) The chick; a great model system becomes even greater. Dev. Cell 8, 9–17 [DOI] [PubMed] [Google Scholar]
- 9. Sauka-Spengler T., Barembaum M. (2008) Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 87, 237–256 [DOI] [PubMed] [Google Scholar]
- 10. Kondoh H., Uchikawa M. (2008) Dissection of chick genomic regulatory regions. Methods Cell Biol. 87, 313–336 [DOI] [PubMed] [Google Scholar]
- 11. Ribatti D. (2008) The chick embryo chorioallantoic membrane in the study of tumor angiogenesis. Rom. J. Morphol. Embryol. 49, 131–135 [PubMed] [Google Scholar]
- 12. Kilarski W. W., Bikfalvi A. (2007) Recent developments in tumor angiogenesis. Curr. Pharm. Biotechnol. 8, 3–9 [DOI] [PubMed] [Google Scholar]
- 13. Saidi A., Javerzat S., Bellahcene A., De Vos J., Bello L., Castronovo V., Deprez M., Loiseau H., Bikfalvi A., Hagedorn M. (2008) Experimental anti-angiogenesis causes upregulation of genes associated with poor survival in glioblastoma. Int. J. Cancer 122, 2187–2198 [DOI] [PubMed] [Google Scholar]
- 14. Ossowski L. (1988) In vivo invasion of modified chorioallantoic membrane by tumor cells: the role of cell surface-bound urokinase. J. Cell Biol. 107, 2437–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagedorn M., Javerzat S., Gilges D., Meyre A., de Lafarge B., Eichmann A., Bikfalvi A. (2005) Accessing key steps of human tumor progression in vivo by using an avian embryo model. Proc. Natl. Acad. Sci. U.S.A. 102, 1643–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosli C., Rybak J. N., Neri D., Elia G. (2008) Quantitative recovery of biotinylated proteins from streptavidin-based affinity chromatography resins. Methods in molecular biology (Clifton, N.J.) 418, 89–100 [DOI] [PubMed] [Google Scholar]
- 17. Rybak J. N., Ettorre A., Kaissling B., Giavazzi R., Neri D., Elia G. (2005) In vivo protein biotinylation for identification of organ-specific antigens accessible from the vasculature. Nat. Methods 2, 291–298 [DOI] [PubMed] [Google Scholar]
- 18. Conrotto P., Roesli C., Rybak J., Kischel P., Waltregny D., Neri D., Castronovo V. (2008) Identification of new accessible tumor antigens in human colon cancer by ex vivo protein biotinylation and comparative mass spectrometry analysis. Int. J. Cancer 123, 2856–2864 [DOI] [PubMed] [Google Scholar]
- 19. Gautier V., Mouton-Barbosa E., Bouyssie D., Delcourt N., Beau M., Girard J. P., Cayrol C., Burlet-Schiltz O., Monsarrat B., Gonzalez de Peredo A. (2012) Label-free quantification and shotgun analysis of complex proteomes by one-dimensional SDS-PAGE/nanoLC-MS: evaluation for the large scale analysis of inflammatory human endothelial cells. Mol. Cell. Proteomics 11, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beckstead J. H. (1994) A simple technique for preservation of fixation-sensitive antigens in paraffin-embedded tissues. J. Histochem. Cytochem. 42, 1127–1134 [DOI] [PubMed] [Google Scholar]
- 21. Holmberg A., Blomstergren A., Nord O., Lukacs M., Lundeberg J., Uhlen M. (2005) The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis 26, 501–510 [DOI] [PubMed] [Google Scholar]
- 22. Navarro P., Vazquez J. (2009) A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome Res. 8, 1792–1796 [DOI] [PubMed] [Google Scholar]
- 23. Mouton-Barbosa E., Roux-Dalvai F., Bouyssie D., Berger F., Schmidt E., Righetti P. G., Guerrier L., Boschetti E., Burlet-Schiltz O., Monsarrat B., Gonzalez de Peredo A. (2010) In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol. Cell. Proteomics 9, 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herbert J. M., Buffa F. M., Vorschmitt H., Egginton S., Bicknell R. (2009) A new procedure for determining the genetic basis of a physiological process in a non-model species, illustrated by cold induced angiogenesis in the carp. BMC Genomics 10, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herbert J. M., Stekel D., Sanderson S., Heath V. L., Bicknell R. (2008) A novel method of differential gene expression analysis using multiple cDNA libraries applied to the identification of tumour endothelial genes. BMC Genomics 9, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Beijnum J. R., Dings R. P., van der Linden E., Zwaans B. M., Ramaekers F. C., Mayo K. H., Griffioen A. W. (2006) Gene expression of tumor angiogenesis dissected: specific targeting of colon cancer angiogenic vasculature. Blood 108, 2339–2348 [DOI] [PubMed] [Google Scholar]
- 27. Madden S. L., Cook B. P., Nacht M., Weber W. D., Callahan M. R., Jiang Y., Dufault M. R., Zhang X., Zhang W., Walter-Yohrling J., Rouleau C., Akmaev V. R., Wang C. J., Cao X., St Martin T. B., Roberts B. L., Teicher B. A., Klinger K. W., Stan R. V., Lucey B., Carson-Walter E. B., Laterra J., Walter K. A. (2004) Vascular gene expression in nonneoplastic and malignant brain. Am. J. Pathol. 165, 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. St Croix B., Rago C., Velculescu V., Traverso G., Romans K. E., Montgomery E., Lal A., Riggins G. J., Lengauer C., Vogelstein B., Kinzler K. W. (2000) Genes expressed in human tumor endothelium. Science 289, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 29. Agudo D., Gomez-Esquer F., Diaz-Gil G., Martinez-Arribas F., Delcan J., Schneider J., Palomar M. A., Linares R. (2005) Proteomic analysis of the Gallus gallus embryo at stage-29 of development. Proteomics 5, 4946–4957 [DOI] [PubMed] [Google Scholar]
- 30. Mann K. (2008) Proteomic analysis of the chicken egg vitelline membrane. Proteomics 8, 2322–2332 [DOI] [PubMed] [Google Scholar]
- 31. Mangum J. E., Farlie P. G., Hubbard M. J. (2005) Proteomic profiling of facial development in chick embryos. Proteomics 5, 2542–2550 [DOI] [PubMed] [Google Scholar]
- 32. McLean L., Doherty M. K., Deeming D. C., Beynon R. J. (2004) A proteome analysis of the subcutaneous gel in avian hatchlings. Mol. Cell. Proteomics 3, 250–256 [DOI] [PubMed] [Google Scholar]
- 33. Mizukami M., Kanamoto T., Souchelnytskyi N., Kiuchi Y. (2008) Proteome profiling of embryo chick retina. Proteome Sci. 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lam T. C., Li K. K., Lo S. C., Guggenheim J. A., To C. H. (2006) A chick retinal proteome database and differential retinal protein expressions during early ocular development. J. Proteome Res. 5, 771–784 [DOI] [PubMed] [Google Scholar]
- 35. Farinazzo A., Restuccia U., Bachi A., Guerrier L., Fortis F., Boschetti E., Fasoli E., Citterio A., Righetti P. G. (2009) Chicken egg yolk cytoplasmic proteome, mined via combinatorial peptide ligand libraries. J. Chromatogr. A 1216, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 36. D'Ambrosio C., Arena S., Scaloni A., Guerrier L., Boschetti E., Mendieta M. E., Citterio A., Righetti P. G. (2008) Exploring the chicken egg white proteome with combinatorial peptide ligand libraries. J. Proteome Res. 7, 3461–3474 [DOI] [PubMed] [Google Scholar]
- 37. Mann K. (2007) The chicken egg white proteome. Proteomics 7, 3558–3568 [DOI] [PubMed] [Google Scholar]
- 38. Parada C., Gato A., Aparicio M., Bueno D. (2006) Proteome analysis of chick embryonic cerebrospinal fluid. Proteomics 6, 312–320 [DOI] [PubMed] [Google Scholar]
- 39. Han B. K., Kim J. N., Shin J. H., Kim J. K., Jo D. H., Kim H., Han J. Y. (2005) Proteome analysis of chicken embryonic gonads: identification of major proteins from cultured gonadal primordial germ cells. Mol. Reprod. Dev. 72, 521–529 [DOI] [PubMed] [Google Scholar]
- 40. Balcerzak M., Malinowska A., Thouverey C., Sekrecka A., Dadlez M., Buchet R., Pikula S. (2008) Proteome analysis of matrix vesicles isolated from femurs of chicken embryo. Proteomics 8, 192–205 [DOI] [PubMed] [Google Scholar]
- 41. Mann K., Macek B., Olsen J. V. (2006) Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics 6, 3801–3810 [DOI] [PubMed] [Google Scholar]
- 42. Bon E., Steegers R., Steegers E. A., Ursem N., Charif H., Burgers P. C., Luider T. M., Dekker L. J. (2010) Proteomic analyses of the developing chicken cardiovascular system. J. Proteome Res. 9, 268–274 [DOI] [PubMed] [Google Scholar]
- 43. Kamada H., Fugmann T., Neri D., Roesli C. (2009) Improved protein sequence coverage by on resin deglycosylation and cysteine modification for biomarker discovery. Proteomics 9, 783–787 [DOI] [PubMed] [Google Scholar]
- 44. Reinacher M., Eigenbrodt E. (1981) Immunohistological demonstration of the same type of pyruvate kinase isoenzyme (M2-Pk) in tumors of chicken and rat. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 37, 79–88 [DOI] [PubMed] [Google Scholar]
- 45. Brinck U., Eigenbrodt E., Oehmke M., Mazurek S., Fischer G. (1994) L- and M2-pyruvate kinase expression in renal cell carcinomas and their metastases. Virchows Arch. 424, 177–185 [DOI] [PubMed] [Google Scholar]
- 46. Steinberg P., Klingelhoffer A., Schafer A., Wust G., Weisse G., Oesch F., Eigenbrodt E. (1999) Expression of pyruvate kinase M2 in preneoplastic hepatic foci of N-nitrosomorpholine-treated rats. Virchows Arch. 434, 213–220 [DOI] [PubMed] [Google Scholar]
- 47. Mouta Carreira C., Nasser S. M., di Tomaso E., Padera T. P., Boucher Y., Tomarev S. I., Jain R. K. (2001) LYVE-1 is not restricted to the lymph vessels: expression in normal liver blood sinusoids and down-regulation in human liver cancer and cirrhosis. Cancer Res. 61, 8079–8084 [PubMed] [Google Scholar]
- 48. Oyama Y., Takeda T., Hama H., Tanuma A., Iino N., Sato K., Kaseda R., Ma M., Yamamoto T., Fujii H., Kazama J. J., Odani S., Terada Y., Mizuta K., Gejyo F., Saito A. (2005) Evidence for megalin-mediated proximal tubular uptake of L-FABP, a carrier of potentially nephrotoxic molecules. Lab. Invest. 85, 522–531 [DOI] [PubMed] [Google Scholar]
- 49. Wu Y. L., Peng X. E., Wang D., Chen W. N., Lin X. (2012) Human liver fatty acid binding protein (hFABP1) gene is regulated by liver-enriched transcription factors HNF3beta and C/EBPalpha. Biochimie (Paris) 94, 384–392 [DOI] [PubMed] [Google Scholar]
- 50. Miquilena-Colina M. E., Lima-Cabello E., Sanchez-Campos S., Garcia-Mediavilla M. V., Fernandez-Bermejo M., Lozano-Rodriguez T., Vargas-Castrillon J., Buque X., Ochoa B., Aspichueta P., Gonzalez-Gallego J., Garcia-Monzon C. (2011) Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut 60, 1394–1402 [DOI] [PubMed] [Google Scholar]
- 51. Khandoga A., Huettinger S., Khandoga A. G., Li H., Butz S., Jauch K. W., Vestweber D., Krombach F. (2009) Leukocyte transmigration in inflamed liver: a role for endothelial cell-selective adhesion molecule. J. Hepatol. 50, 755–765 [DOI] [PubMed] [Google Scholar]
- 52. Yokota T., Oritani K., Butz S., Kokame K., Kincade P. W., Miyata T., Vestweber D., Kanakura Y. (2009) The endothelial antigen ESAM marks primitive hematopoietic progenitors throughout life in mice. Blood 113, 2914–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khimji A. K., Shao R., Rockey D. C. (2008) Divergent transforming growth factor-beta signaling in hepatic stellate cells after liver injury: functional effects on ECE-1 regulation. Am. J. Pathol. 173, 716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lattmann T., Ortmann J., Horber S., Shaw S. G., Hein M., Barton M. (2006) Upregulation of endothelin converting enzyme-1 in host liver during chronic cardiac allograft rejection. Exp. Biol. Med. (Maywood) 231, 899–901 [PubMed] [Google Scholar]
- 55. Li Y. N., Radner S., French M. M., Pinzon-Duarte G., Daly G. H., Burgeson R. E., Koch M., Brunken W. J. (2012) The gamma3 chain of laminin is widely but differentially expressed in murine basement membranes: expression and functional studies. Matrix Biol. 31, 120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wong V. W., Stange D. E., Page M. E., Buczacki S., Wabik A., Itami S., van de Wetering M., Poulsom R., Wright N. A., Trotter M. W., Watt F. M., Winton D. J., Clevers H., Jensen K. B. (2012) Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 14, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fournier I., Gaucher D., Chich J. F., Bach C., Shooshtarizadeh P., Picaud S., Bourcier T., Speeg-Schatz C., Strub J. M., Van Dorsselaer A., Corti A., Aunis D., Metz-Boutigue M. H. (2011) Processing of chromogranins/secretogranin in patients with diabetic retinopathy. Regul. Pept. 167, 118–124 [DOI] [PubMed] [Google Scholar]
- 58. Salvi E., Buffa R., Renda T. G. (1996) Ontogeny, distribution and amine/peptide content of chromogranin A- and B-immunoreactive endocrine cells in the small and large intestine of the chicken. Anat. Embryol. (Berl.) 194, 89–98 [DOI] [PubMed] [Google Scholar]
- 59. Sudhof T. C. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jung Y., Shiozawa Y., Wang J., Patel L. R., Havens A. M., Song J., Krebsbach P. H., Roodman G. D., Taichman R. S. (2011) Annexin-2 is a regulator of stromal cell-derived factor-1/CXCL12 function in the hematopoietic stem cell endosteal niche. Exp. Hematol. 39, 151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hayashi K., Kaufman L., Ross M. D., Klotman P. E. (2005) Definition of the critical domains required for homophilic targeting of mouse sidekick molecules. FASEB J. 19, 614–616 [DOI] [PubMed] [Google Scholar]
- 62. Viktorinova I., Konig T., Schlichting K., Dahmann C. (2009) The cadherin Fat2 is required for planar cell polarity in the Drosophila ovary. Development 136, 4123–4132 [DOI] [PubMed] [Google Scholar]
- 63. Torzicky M., Viznerova P., Richter S., Strobl H., Scheinecker C., Foedinger D., Riedl E. (2012) Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31) and CD99 are critical in lymphatic transmigration of human dendritic cells. J. Invest. Dermatol. 132, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 64. Bixel M. G., Petri B., Khandoga A. G., Khandoga A., Wolburg-Buchholz K., Wolburg H., Marz S., Krombach F., Vestweber D. (2007) A CD99-related antigen on endothelial cells mediates neutrophil but not lymphocyte extravasation in vivo. Blood 109, 5327–5336 [DOI] [PubMed] [Google Scholar]
- 65. Crowley M. J., Powers B. J., Myers E. R., McBroom A. J., Sanders G. D. (2012) Angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers for treatment of ischemic heart disease: future research needs prioritization. Am. Heart J. 163, 777–782 [DOI] [PubMed] [Google Scholar]
- 66. Hufton S. E., Moerkerk P. T., Brandwijk R., de Bruine A. P., Arends J. W., Hoogenboom H. R. (1999) A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett. 463, 77–82 [DOI] [PubMed] [Google Scholar]
- 67. Van Beijnum J. R., Moerkerk P. T., Gerbers A. J., De Bruine A. P., Arends J. W., Hoogenboom H. R., Hufton S. E. (2002) Target validation for genomics using peptide-specific phage antibodies: a study of five gene products overexpressed in colorectal cancer. Int. J. Cancer 101, 118–127 [DOI] [PubMed] [Google Scholar]
- 68. Reddy P. S., Umesh S., Thota B., Tandon A., Pandey P., Hegde A. S., Balasubramaniam A., Chandramouli B. A., Santosh V., Rao M. R., Kondaiah P., Somasundaram K. (2008) PBEF1/NAmPRTase/Visfatin: a potential malignant astrocytoma/glioblastoma serum marker with prognostic value. Cancer Biol. Ther. 7, 663–668 [DOI] [PubMed] [Google Scholar]
- 69. Kim S. R., Bae Y. H., Bae S. K., Choi K. S., Yoon K. H., Koo T. H., Jang H. O., Yun I., Kim K. W., Kwon Y. G., Yoo M. A., Bae M. K. (2008) Visfatin enhances ICAM-1 and VCAM-1 expression through ROS-dependent NF-kappaB activation in endothelial cells. Biochim. Biophys. Acta 1783, 886–895 [DOI] [PubMed] [Google Scholar]
- 70. Kim S. R., Bae S. K., Choi K. S., Park S. Y., Jun H. O., Lee J. Y., Jang H. O., Yun I., Yoon K. H., Kim Y. J., Yoo M. A., Kim K. W., Bae M. K. (2007) Visfatin promotes angiogenesis by activation of extracellular signal-regulated kinase 1/2. Biochem. Biophys. Res. Commun. 357, 150–156 [DOI] [PubMed] [Google Scholar]
- 71. Bae Y. H., Bae M. K., Kim S. R., Lee J. H., Wee H. J., Bae S. K. (2009) Upregulation of fibroblast growth factor-2 by visfatin that promotes endothelial angiogenesis. Biochem. Biophys. Res. Commun. 379, 206–211 [DOI] [PubMed] [Google Scholar]
- 72. Bae Y. H., Park H. J., Kim S. R., Kim J. Y., Kang Y., Kim J. A., Wee H. J., Kageyama R., Jung J. S., Bae M. K., Bae S. K. (2011) Notch1 mediates visfatin-induced FGF-2 up-regulation and endothelial angiogenesis. Cardiovasc. Res. 89, 436–445 [DOI] [PubMed] [Google Scholar]
- 73. Adya R., Tan B. K., Punn A., Chen J., Randeva H. S. (2008) Visfatin induces human endothelial VEGF and MMP-2/9 production via MAPK and PI3K/Akt signalling pathways: novel insights into visfatin-induced angiogenesis. Cardiovasc. Res. 78, 356–365 [DOI] [PubMed] [Google Scholar]
- 74. Hasmann M., Schemainda I. (2003) FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63, 7436–7442 [PubMed] [Google Scholar]
- 75. Garten A., Petzold S., Korner A., Imai S., Kiess W. (2009) Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol. Metab. 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fonseca C., Abraham D., Ponticos M. (2009) Neuronal regulators and vascular dysfunction in Raynaud's phenomenon and systemic sclerosis. Curr. Vasc. Pharmacol. 7, 34–39 [DOI] [PubMed] [Google Scholar]
- 77. Freiman M., Seifter E., Connerton C., Levenson S. M. (1970) Vitamin A deficiency and surgical stress. Surg. Forum 21, 81–82 [PubMed] [Google Scholar]
- 78. Shapiro S. S., Mott D. J. (1981) Modulation of glycosaminoglycan biosynthesis by retinoids. Ann. N. Y. Acad. Sci. 359, 306–321 [DOI] [PubMed] [Google Scholar]
- 79. Cohen B. E., Gill G., Cullen P. R., Morris P. J. (1979) Reversal of postoperative immunosuppression in man by vitamin A. Surg. Gynecol. Obstet. 149, 658–662 [PubMed] [Google Scholar]
- 80. Balmer J. E., Blomhoff R. (2002) Gene expression regulation by retinoic acid. J. Lipid Res. 43, 1773–1808 [DOI] [PubMed] [Google Scholar]
- 81. Chen L., Hughes R. A., Baines A. J., Conboy J., Mohandas N., An X. (2011) Protein 4.1R regulates cell adhesion, spreading, migration and motility of mouse keratinocytes by modulating surface expression of beta1 integrin. J. Cell Sci. 124, 2478–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. O'Neill A. K., Gallegos L. L., Justilien V., Garcia E. L., Leitges M., Fields A. P., Hall R. A., Newton A. C. (2011) Protein kinase C alpha promotes cell migration through a PDZ-dependent interaction with its novel substrate discs large homolog 1 (DLG1). J. Biol. Chem. 286, 43559–43568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Singleton P. A., Bourguignon L. Y. (2004) CD44 interaction with ankyrin and IP3 receptor in lipid rafts promotes hyaluronan-mediated Ca2+ signaling leading to nitric oxide production and endothelial cell adhesion and proliferation. Exp. Cell Res. 295, 102–118 [DOI] [PubMed] [Google Scholar]
- 84. Xu R. H., Pelicano H., Zhou Y., Carew J. S., Feng L., Bhalla K. N., Keating M. J., Huang P. (2005) Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 65, 613–621 [PubMed] [Google Scholar]
- 85. Zhang Y., Goss A. M., Cohen E. D., Kadzik R., Lepore J. J., Muthukumaraswamy K., Yang J., DeMayo F. J., Whitsett J. A., Parmacek M. S., Morrisey E. E. (2008) A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat. Genet. 40, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bignon M., Pichol-Thievend C., Hardouin J., Malbouyres M., Brechot N., Nasciutti L., Barret A., Teillon J., Guillon E., Etienne E., Caron M., Joubert-Caron R., Monnot C., Ruggiero F., Muller L., Germain S. (2011) Lysyl oxidase-like protein-2 regulates sprouting angiogenesis and type IV collagen assembly in the endothelial basement membrane. Blood 118, 3979–3989 [DOI] [PubMed] [Google Scholar]
- 87. Henry J., Hsu C. Y., Haftek M., Nachat R., de Koning H. D., Gardinal-Galera I., Hitomi K., Balica S., Jean-Decoster C., Schmitt A. M., Paul C., Serre G., Simon M. (2011) Hornerin is a component of the epidermal cornified cell envelopes. FASEB J. 25, 1567–1576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.