Abstract
The mammalian F-box protein Fbw7 and its Caenorhabditis elegans counterpart Sel-10 have been implicated in the ubiquitin-mediated turnover of cyclin E as well as the Notch/Lin-12 family of transcriptional activators. Both unregulated Notch and cyclin E promote tumorigenesis, and inactivating mutations in human Fbw7 suggest that it may be a tumor suppressor. To generate an in vivo system to assess the consequences of such unregulated signaling, we generated mice deficient for Fbw7. Fbw7-null mice die around 10.5 days post coitus because of a combination of deficiencies in hematopoietic and vascular development and heart chamber maturation. The absence of Fbw7 results in elevated levels of cyclin E, concurrent with inappropriate DNA replication in placental giant trophoblast cells. Moreover, the levels of both Notch 1 and Notch 4 intracellular domains were elevated, leading to stimulation of downstream transcriptional pathways involving Hes1, Herp1, and Herp2. These data suggest essential functions for Fbw7 in controlling cyclin E and Notch signaling pathways in the mouse.
Successful organismal development requires precise control of gene expression. The posttranscriptional control of protein abundance has recently emerged as an important aspect of developmental control. The capacity to rapidly stabilize or destabilize key regulatory proteins through the ubiquitin-mediated proteolysis pathway is central to development. Ubiquitin is a 76-aa polypeptide added to cellular proteins in tandem chains as a means of targeting them for recognition and destruction by the 26S proteasome (1–3). Polyubiquitination is the rate-limiting step in regulating protein stability and involves a three-step cascade of ubiquitin-transfer reactions. First, in an energy-dependent reaction, a ubiquitin-activating enzyme (E1) activates the ubiquitin and transfers it to a ubiquitin-conjugating enzyme (E2). The E2 acts in concert with a specificity-determining component, E3, to transfer ubiquitin to the target protein. E3s are adapters that link the modular machinery of the E2 to specific substrates (3). The largest and most versatile class of E3s is the Skp1 Cul1 F-box protein E3 ubiquitin ligases (SCFs). SCFs are themselves modular complexes that consist in mammalian cells of Skp1, Cul1, Rbx1, and one of a family of dual domain F-box proteins. Unique regions of the F-box protein confer its ability to bind to substrates, whereas the F-box motif docks with Skp1, which in turn binds to the Cdc53 (Cul) scaffold. By means of interaction with Rbx1, Cdc53 connects to the E1/E2 portions of the pathway (4–10).
Fbw7 (10, 11) is an F-box protein that facilitates the ubiquitination of both cyclin E and Notch in vitro (12–16). In combination with cyclin-dependent kinase 2, cyclin E is a key regulator of the G1-to-S transition in mammalian cells. The deregulation of its expression is thought to be a critical step in the transition from a normal to a malignant cell (17–21). HeLa cells depleted of Fbw7 by small interfering RNA show stabilized cyclin E (14). Consistent with this observation, a mutation was found in the Fbw7 gene in a cell line derived from breast cancer in which high levels of cyclin E were observed (15). Loss of the fly Fbw7 homologue Archipelago resulted in overproliferation and increased cyclin E levels (13). Finally, the SCFFbw7 mediates the in vitro ubiquitination of cyclin E (14, 15).
The Notch protein provides an evolutionarily conserved pathway for intercellular communication and cell fate decisions during metazoan development (22). Notch is a large transmembrane receptor that undergoes three critical proteolytic processing events during its maturation and the elicitation of its downstream signals. A furin-like protease initially cleaves Notch into two fragments that heterodimerize to form the functional Notch receptor at the cell membrane. This form of Notch interacts with its DSL ligand (Delta, Serrate, Lag2) (23, 24). Ligand–receptor interaction induces a series of modifications whereby a metalloprotease cleaves and releases the extracellular domain (25, 26), whereas a γ-secretase subsequently cleaves and releases the Notch intracellular domain (NICD) (27–29). The liberation of the NICD enables its translocation to the nucleus, where it functions as a transcriptional regulator of C-promoter binding factor 1/recombination signal binding protein Jκ. Together, this complex activates the transcription of downstream target genes, among them the Hes and Herp families of basic helix–loop–helix transcriptional repressors (for review see refs. 30–33). After recombination signal binding protein Jκ transcriptional activation, the NICD is destroyed by ubiquitin-mediated proteolysis. Genetic and biochemical studies in Caenorhabditis elegans demonstrate that SCFFbw7 homologues facilitate the ubiquitination of the NICD (12, 16, 34).
There are four mouse Notch genes, and they are expressed in partially overlapping regions of the developing and adult mouse (35–46). Mice defective in Notch signaling generally die in utero with a variety of embryonic defects, including vascular abnormalities, disorganized somitogenesis, defective hematopoiesis, and heart anomalies (47–52). Disregulated expression of the NICD has been shown to lead to tumors in mice and humans (42, 53–55).
Here, we report the functional ablation of the mouse Fbw7 gene. Loss of Fbw7 results in embryonic lethality around 10.5 days postcoitus (ED10.5). Fbw7-/- embryos exhibit retarded development, likely associated with an array of defects in vascular and hematopoietic development. They further exhibit defects in heart development, marked by the failure of complete atrial and ventricular chamber formation and maturation. We observe increases in the amounts of embryonic Notch 1 and Notch 4 ICD proteins as well as cyclin E proteins in Fbw7 mutant embryos and placentas consistent with the stabilization of these proteins in vivo. In contrast to the hypothesized role of Fbw7 in human cancer, Fbw7 heterozygote mice are phenotypically normal and have not shown spontaneous tumor formation up to 1 year of age.
Materials and Methods
Construction of the Fbw7 Targeting Vector and Generation of Fbw7+/- Embryonic Stem (ES) Cells. We used the Recombination Cloning System to generate our construct to target the Fbw7 locus in ES cells (56). AB2.2 129Sv ES cells were passaged and manipulated on feeder cells as described (57). The targeting construct was linearized by PmeI restriction digestion and electroporated into ES cells. These cells were selected for 10 days in G418 (Invitrogen) and gancyclovir (kindly prescribed for research by Eric Tiblier, Smithville Regional Hospital, Austin, TX). One hundred ninety-two G418-, gancyclovir-resistant ES cell clones were screened by Southern blot after a HindIII digest of their genomic DNA. Twenty-two of these were found to contain the homologously recombined targeting construct with the distal loxP site intact. Two of these were further electroporated with either pCMV-CRE or pCMV-FLP and plated at low density (500–1,000 cells) on 10-cm feeder dishes. One hundred ninety-two clones were picked from each electroporation and screened by PCR. No clones in which Flp-mediated recombination could be detected were recovered, whereas 22 of 192 clones that were discovered to contain Cre-mediated excision were recovered, and four were injected into C57BL/6 blastocysts, giving rise to 16 chimeric males. These were backcrossed to C57BL/6 females to generate F1 Fbw7+/- animals, and one transmitted the desired allele to his offspring. All genotypes were verified by Expand Long Template PCR (Roche Molecular Biochemicals) from tail DNA prepared according to standard protocols (57). PCR primers were as follows: Fbw7 no. 1, 5′-GGACATAGTTCAGTGCTAACCCGAG-3′; Fbw7 no. 2, 5′-GCTAGGACTACTGCATTTCAGTGGC-3′; Fbw7 no. 3, 5′-GCACAGCCCAGTATCTGGGCCAAGC-3′; Fbw7 no. 4, 5′-CCCTGAGAGTGGCATCTCGAGAACC; Fbw7 no. 5, 5′-CCCTGGCTTAGCATATCAGCTATGG; Fbw no. 6, 5′-GCTGCAAACTCATAGTGAGAATCATGG-3′.
Histology and Immunocytohistochemistry. Embryos in the maternal decidua were fixed in 10% buffered formalin overnight at 4°C. Fixation was followed by dehydration in an ethanol series before paraffin embedding and sectioning. Antibodies used were cyclin E (M20, Santa Cruz Biotechnology), platelet endothelial cell adhesion marker 1 (Pharmingen), and Ki67 (Oncogene).
Whole Mount in Situ Hybridization. Embryos at ED9.5 were fixed in MEMFA (0.1M Mops, pH 7.4/200 mM EGTA, pH 8/100 mM MgSO4/3.7% formaldehyde) for 2 h at 24°C, then washed in ice-cold PBS, dehydrated in methanol series, and stored in 90% methanol at -20°C. Probes against cardiac actin and atrial natriuretic factor (58) and Nkx2.5 (59) were used as described.
RNA Isolation, Reverse Transcription, Real-Time PCR, and Analysis. RNA was isolated from embryos at ED10.5 and ED9.5 by using the RNAqueous micro kit (Ambion, Austin, TX). Reverse transcription (RT) was performed using the Message Sensor kit using 400–800 ng of mRNA per reaction (Ambion). Real-time PCR was performed on the reverse-transcribed samples using SYBR Green PCR Master Mix (Applied Biosystems). RT reactions in which no reverse transcriptase had been added served as a monitor for the efficiency of the DNaseI digestion. All reactions were carried out in triplicate. The fold difference in various transcripts was calculated by the ΔΔCT method using GAPDH as the internal control, and we assigned wild-type values as 1. For every reaction, we observed a single peak on the dissociation curve plot. Primer sequences were as follows: cyclin E-F, 5′-CAGAGCAGCGAGCAGGAGA-3′; cyclin E-R, 5′-CAGCTGCTTCCACACCACTG; Notch1-F, 5′-CTCAGCATGTGCAGAGTCTACTACC; Notch1-R, 5′-GAGAAGTACTCCCAAGGCCCA; Hes1-F, 5′-CTCCTCGCTCACTTCGGACT; Hes1-R, 5′-TAGCAGTGGCCTGAGGCTCT-3′; Herp1-F, 5′-CCGCAGCCTCCAGTCCT-3′; Herp1-R, 5′-TGTCCCCCAGGGTTGGT-3′; Herp2-F, 5′-GCCTGCACTTAGCAGCCCT-3′; Herp2-R, 5′-ACGCAGGTCGACACTGGTG-3′; GAPDH-F, 5′-TGTACCGTCTAGCATATCTCCGAC-3′; GAPDH-R, 5′-ATGATGTGCTCTAGCTCTGGGTG-3′.
Western Blotting. Embryo and placenta protein at ED10.0 was prepared and immunoblotted as described (60). Antibodies used to detect blotted proteins were as follows: anti-cyclin E (M20, Santa Cruz Biotechnology), Notch1 ICD (Cell Signaling Technology, Beverly, MA), Notch3 ICD (Upstate Biotechnology, Lake Placid, NY), p107 (Santa Cruz Biotechnology) and Notch4 ICD (Upstate Biotechnology). Quantification was carried out using nih image 1.61 software, normalizing all values for cyclin E intensity to the respective indicated loading controls.
Results
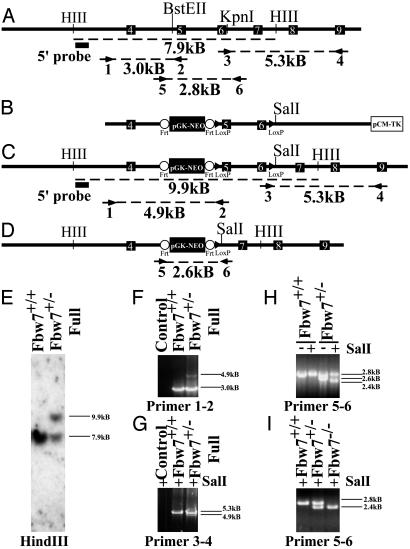
Generation of Fbw7 Targeting Construct and Fbw7+/- Mice. We used the Recombination Cloning System to generate the mouse Fbw7 targeting construct (56). We selected exons 5 and 6 for deletion (Fig. 1 A and B). Because these exons contain the F-box of Fbw7, this mutation would compromise all F-box-dependent functions in vivo. We electroporated this construct into ES cells. Of 192 clones examined by Southern blot hybridization and PCR, 22 were found to include the homologously targeted integrand, including the 3′ distal lox site and the 5′ organization of the genomic locus intact (Fig. 1 A, C, E, F, and G). Two were chosen for subsequent electroporation with either a Cre- or a Flp-encoding plasmid. Cre-mediated recombination was expected to delete the region between the lox sites and generate a loss of function allele for Fbw7 (Fbw7-) (Fig. 1D), whereas Flp-mediated recombination should delete the neomycin reporter and render a conditional “floxed” allele. We did not observe any Flp-mediated recombination in 192 lines screened after electroporation and thus were unable to generate a conditional allele. Of 192 lines screened from the Cre electroporation, 22 exhibited the predicted recombination between the lox sites (8.7%), and four were injected into C57BL/6 blastocysts (Fig. 1H, primers 5–6). Fbw7+/- animals appear normal and are fertile. No obvious phenotypes have been observed in these mice, and they do not develop spontaneous tumors up to 1 year of age.
Fig. 1.
Targeted disruption of the mouse Fbw7 locus. (A) Restriction map of the Fbw7 genomic locus, showing intron–exon boundaries and location for exons 4–9 (indicated by white numbered black boxes) of mouse Fbw7. Primer pairs (arrows) used for PCR analysis are indicated numerically beneath the primer, and the size of the predicted product is indicated beneath dashed line. The probe used for Southern hybridization is indicated by a filled black box. (B) Map of targeting construct. Frt sites are indicated by open circles, and loxP sites are indicated by filled triangles. (C) The restriction map of the Fbw7 locus after homologous integration of the construct in B. Predicted PCR fragment sizes are shown. (D) The restriction map of the Fbw7 locus after Cre-mediated recombination. (E) Southern hybridization using the probe indicated by the filled black box in A and C after HindIII digestion of ES cell DNA. (F) PCR analysis of the 5′ end of the targeted Fbw7 locus by using primer pair 1–2 shows single homologous integration of frt-NEO-frt-loxP cassette into ES cells. (G) PCR analysis of the 3′ end of the targeted locus in ES cells by using primer pair 3–4 confirming the presence of the distal loxP site, indicated by the presence of a unique SalI restriction site engineered into the loxP site. (H) PCR genotyping of ES cells using primers 56 following Cre-mediated recombination, compared with wild-type cell line. (I) PCR genotyping of embryos from yolk sac DNA with primer pair 5–6. The “full” designation describes the Fbw7 mutant allele shown in C before Cre recombination whereas a minus sign alone denotes that allele after Cre-mediated recombination (D).
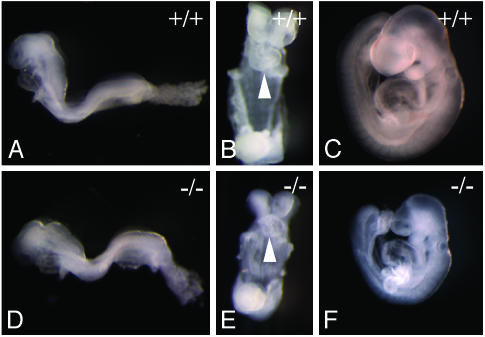
Loss of Fbw7 Results in Midgestational Embryonic Lethality. We intercrossed Fbw7+/- mice to produce Fbw7-/- offspring. Of 159 offspring, no Fbw7-/- mice were detected, suggesting that homozygous loss of Fbw7 causes embryonic lethality (Table 1). To ascertain the timing and nature of the lethality, we studied the morphology of embryos from timed Fbw7+/- intercrosses. Genotypes were confirmed by PCR of yolk sac DNA preparations (Table 1 and Fig. 1I). Embryos harvested on ED8.5 or earlier are indistinguishable on gross inspection from their wild-type littermates (Fig. 2 A and B versus D and E). Gross inspection confirmed that the allantois connects to the chorion (data not shown). By ED9.5, however, Fbw7-/- embryos appeared appreciably smaller and developmentally delayed. In many cases, neural tube closure is incomplete. All Fbw7-/- embryos at ED9.5 displayed grossly underdeveloped fore-, mid-, and hind-brain regions (Fig. 2 C and F).
Table 1. Genotypes of offspring from Fbw7-/+ intercross at the indicated times.
| Fbw7
|
|||
|---|---|---|---|
| +/+ | +/- | -/- | |
| Weaning | 57 | 108 | 0 |
| E8.5 | 8 | 22 | 11 |
| E9.5 | 28 | 40 | 25 |
| E10.5 | 11 | 10 | 10 |
| E13.5 | 2 | 4 | 0 |
Fig. 2.
Morphology of Fbw7-/- embryos. Shown are embryos from timed Fbw7+/- intercrosses dissected away from maternal decidua and yolk sacs. Genotyping was confirmed by PCR genotyping of yolk sacs by using primer pair 5–6 as shown in Fig. 1I. (A) Left-lateral view of wild-type embryo at ED8.5. (B) Ventral view of wild-type embryo ED8.5. The white arrowhead identifies rightward-looping heart tube. (C) Left lateral view of wild-type embryo at ED9.5. (D) Left-lateral view of Fbw7-/- embryo at ED8.5. (E) Ventral view of Fbw7-/- embryo at ED8.5. The white arrowhead identifies rightward-looping heart tube. (F) Left-lateral view of Fbw7-/- embryo at ED9.5.
Fbw7-/- embryos do complete axial rotation and initiate branchial arch and anterior limb bud development successfully (Fig. 2 B and C). At ED10.5, Fbw7-/- embryos were ≈60% the size of their normal littermates, indicating a substantial developmental delay (data not shown). Because no live embryos were observed past ED10.5, we infer that Fbw7-/- embryos die around this time.
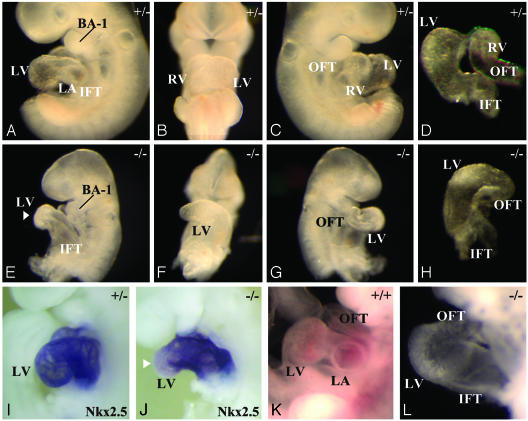
Fbw7 Is Required for Normal Heart Morphogenesis. The heart is the first organ to form during embryogenesis. During normal heart development, the linear, symmetric heart tube undergoes a left to right looping at ED8.0, which establishes the positions of the future heart chambers. Subsequent to looping, the respective heart chambers form by ballooning out from the linear heart tube along the outer curvature (61, 62). Although Fbw7-/- embryos formed a looped, contractile, linear heart tube by ED8.5, there were subtle delays in ventricular maturation (Fig. 2 B and E, white arrowheads). At ED9.5, chamber formation along the entire length of the linear heart tube appeared delayed (Fig. 3 A–D versus E–H). In Fbw7+/+ embryos, the maturation of the left ventricle typically resulted in a thick bulbous chamber at the ventral curvature of the looped heart tube. In Fbw7-/- hearts, only a poorly demarcated ventricular structure is produced (Fig. 3 A and B versus E and F). Furthermore, when normal hearts have completed a clear morphological demarcation between atrium and ventricle, Fbw7-/- linear hearts appeared thickened along the entire length of their in-flow tract, and distinct atrial and ventricular chambers were not evident (Fig. 3 E and F). Finally, in half of the mutant embryos, we observed delayed right ventricular maturation (Fig. 3 C and D versus G and H). The bulbo-ventricular groove between the right and left ventricle was absent, and outflow tract looping and growth were affected. At ED10.5, these differences were more pronounced (Fig. 3 K and L). Although the wild-type heart has all four future heart chambers and is ready to engage in septation, the Fbw7-/- animals never differentiate ventricular or atrial structures and exist as thickened looped tubes, devoid of chamber formation. Still, the hearts of Fbw7-/- embryos maintain contractility at ED10.5.
Fig. 3.
Heart defects in Fbw7-/- embryos. (A–D) A wild-type embryo at ED9.5 illustrating normal heart morphogenesis. (A) Left-lateral view shows chamber formation of the future left ventricle (LV) and left atrium (LA) following the inflow tract (IFT). (B) Ventral view of wild-type heart illustrating the differentiation of the future right ventricle (RV) and LV along the linear heart tube. (C) Right-lateral view shows the RV and the appearance of a mature outflow tract (OFT). (D) Cranial view of the mature hearts displays normal organization of the presumptive chambers (LV and RV) with respect to IFT and OFT. (E–H) Fbw7-/- hearts at ED9.5 illustrate defective development. (E) Left-lateral view shows delayed chamber formation. IFT leads directly to primitive LV structure, indicated by white arrowhead. (F) Ventral view shows that the linear heart develops only a primitive LV, with no apparent RV development. (G) Right-lateral view depicts absence of RV chamber formation. (H) Cranial view of mutant heart. (I and J) Left-lateral view of Nkx2.5 in situ hybridization on embryos at ED9.5. (I) Expression pattern of Nkx2.5 in all cells of the developing heart tube. (J) Reduced expression of Nkx2.5 at ventral-most portion of the heart curvature, the future left ventricle of the Fbw7-/- heart (white arrowhead). (K and L) Embryos at ED10.5. (K) Wild-type heart with well differentiated chambers. (L) Fbw7-/- heart remains a linear heart tube with very little chamber formation. BA-1, first branchial arch.
To further characterize the specificity of the chamber formation defects, we performed whole-mount in situ hybridization using the markers cardiac actin, Nkx2.5, and atrial natriuretic factor. We observed no difference in the expression pattern or intensity of cardiac actin in hearts at ED9.5, indicating an intact commitment to cardiomyocyte cell lineage in the absence of Fbw7 (data not shown). The expression of Nkx2.5, a core cardiac transcription factor, appeared during early cardiogenesis in Fbw7-/- embryos, but reduced Nkx2.5 expression was evident in the future left ventricle region, (Fig. 3 A and I versus E and J). In parallel with this, we observed reduced expression levels of the Nkx2.5-dependent ventricular atrial natriuretic factor transcripts in Fbw7-/- hearts (data not shown). These results suggest that, whereas the cellular commitment to cardiogenesis occurs normally in Fbw7-/- hearts, specific molecular and morphogenetic events contributing to the formation of the respective chambers do not occur.
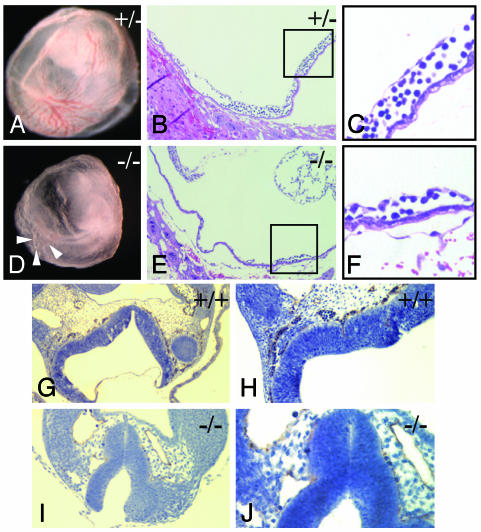
Fbw7 Is Required for the Formation of a Functional Vasculature. Fbw7-/- yolk sacs are indistinguishable from their wild-type littermates at ED8.5 (data not shown). At ED9.5, however, significant differences emerge. Fbw7+/+ and Fbw7+/- embryos at ED9.5 form a mature yolk sac vasculature replete with both large and small vessels and dense blood islands (Fig. 4A). The Fbw7-/- yolk sac appeared pale, and, although a primary vascular plexus forms and partial remodeling in some areas does occur, it is incomplete and disorganized (Fig. 4D). In general, the Fbw7-/- yolk sac vasculature appears as a homogeneous meshwork of vessels of uniform diameter (Fig. 4D, white arrowheads). Furthermore, we observed a paucity of blood cells in both the Fbw7-/- embryos and their yolk sacs (Fig. 4 A versus D). We confirmed these results histologically. Whereas wild-type animals had a rich vascular network with both large and small vessels, Fbw7-/- animals had reduced vessels and nucleated blood cells (compare Fig. 4 B and C with E and F).
Fig. 4.
Vascular defects in Fbw7-/- yolk sacs and embryos. (A–C) Fbw7+/+ yolk sac at ED9.5. (A) Gross appearance of yolk sac. (B, C, E, and F) Images stained with hematoxylin (blue) and eosin (red). (B) Histologic analysis of yolk sac demonstrates abundant large and small vessels with rich blood islands. (C) High magnification of boxed area from B shows mature vessel with intact blood island. (D–F) Fbw7-/- yolk sac at ED9.5. (D) Gross appearance of yolk sac. (E) Histologic appearance of Fbw7-/- yolk sac confirms the absence of a mature vasculature. (F) Higher magnification of the area indicated in the box in E. (G–J) Platelet endothelial cell adhesion marker 1 immunocytohistochemistry (stained in brown and counterstained with hematoxylin in blue) reveals neural tube vasculature at ED9.5. (G and H) Fbw7+/+ embryo at ×10 and ×20 magnification, respectively. (I and J) Fbw7-/- embryo at ×20 and ×40 magnification, respectively.
These vascular and hematopoietic defects extended to the embryo as well. We observe a reduction not only in the number of blood cells in the embryo but also in the density of well organized small-diameter vasculature. This difference was most evident in the capillary beds underlying the neural tube. Here, platelet endothelial cell adhesion marker 1 staining in Fbw7+/+ embryos reveals a rich vascular network bathing this tissue (Fig. 4 G and H). In contrast, Fbw7-/- embryos lack such well organized vessels along the entire length of their neural tubes (Fig. 4 I and J). Although they did possess remnants of microvasculature (Fig. 4J), these vessels appeared neither well organized nor well developed, and they did not contain any substantial nucleated blood cells compared with their wild-type counterparts. Thus, the loss of Fbw7 results in a wide array of hematopoietic and vascular defects in both the extraembryonic yolk sac and the embryo proper.
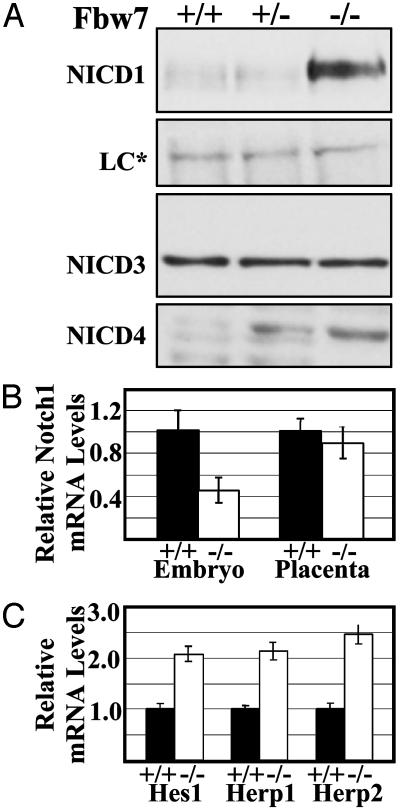
Increased Expression of NICD and Notch-Dependent Targets in Fbw7-/- Embryos. A number of studies directly demonstrated the involvement of the SCFFbw7 complex and its homologues in C. elegans in mediating the ubiquitination-dependent turnover of the NICD (12, 16). To examine these in vivo during development, we analyzed embryos at ED10.0 for the expression of NICD family members. In blots from whole embryo lysates, we observed a substantial increase in the amount of the NICD1 as well as NICD4 in Fbw7-/- embryos compared with their wild-type littermates. In contrast, Notch 3 did not appear to be affected (Fig. 5A). NICD4 appears to be increased even in the Fbw7-/- embryos, yet Fbw7+/- animals are phenotypically indistinguishable from Fbw7+/+ animals. Similar results were obtained for lysates from embryos at ED9.5 (data not shown).
Fig. 5.
NICD protein and NICD-dependent transcripts are more abundant in Fbw7-/- embryos. (A) Western blots using indicated antibodies against whole embryo lysates. The loading control (LC*) is a cross-reactive band. (B) Relative levels of Notch 1 mRNA by real-time PCR using SYBR-green detection of mRNA prepared from embryos and placenta of the indicated genotypes. (C) Relative levels of the Notch-dependent transcripts Hes1, Herp1, and Herp2 in Fbw7+/+ and Fbw7-/- embryos by using quantitative real-time PCR analysis as in B.
To determine whether the increase in NICD protein levels in Fbw7-/- embryos was due to increased transcription of Notch 1, we used quantitative, real-time RT-PCR on analogous samples. This analysis confirmed that the differences observed for NICD1 was not due to higher Notch1 mRNA, which was actually 2-fold lower in Fbw7-/- embryos than in Fbw7+/- littermates. NICD1 protein and mRNA levels were not affected in Fbw7-/- placental lysates (data not shown and Fig. 5B). We further asked whether the apparent stabilization of NICD1 in Fbw7-/- embryos exerted any biological consequence by assessing the levels of the known NICD1 targets Hes1, Herp1, and Herp2 (32). Hes1, Herp1, and Herp2 mRNAs were 2.1-, 2.1-, and 2.5-fold more abundant, respectively, in Fbw7-/- embryos at ED9.5 than in their wild-type littermates (Fig. 5C). This finding is consistent with our hypothesis that the increased levels of NICD in Fbw7-/- embryos profoundly impacts developmental pathways in the form of deregulated expression of critical developmental regulators Hes1, Herp1, and Herp2.
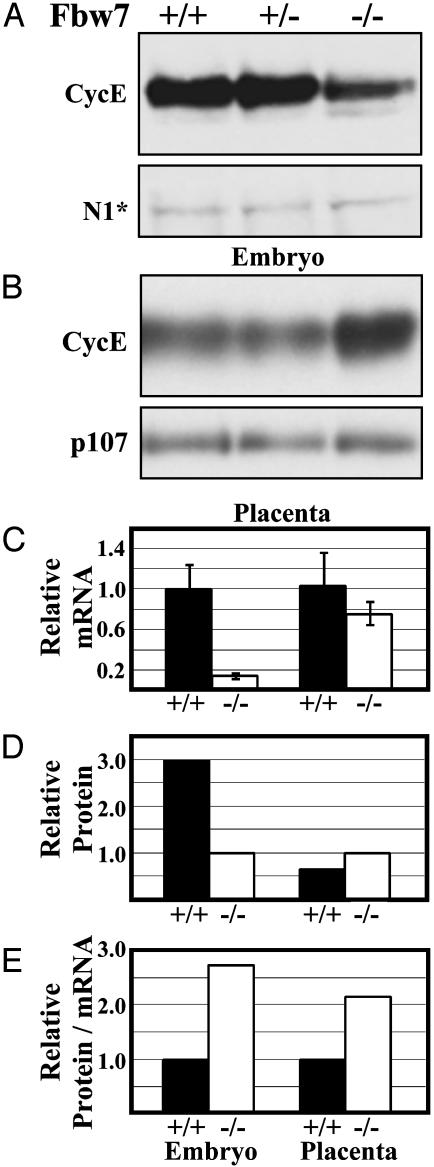
Cyclin E Levels Are Increased in Fbw7-/- Placentas. The SCF has been implicated in the in vivo destruction of cyclin E (14, 15, 63–65). To test whether these relationships held during mammalian development, we analyzed protein extracts from embryos at ED10.0 and their placenta for levels of cyclin E. There was a 2- to 3-fold increase in cyclin E abundance in Fbw7-/- placental lysates (Fig. 6 B and D). Real-time, quantitative RT-PCR analyses of analogous Fbw7-/- and wild-type littermates showed no significant change in the abundance of cyclin E mRNA in Fbw7-/- placentas (Fig. 6C). In contrast, Fbw7-/- embryos displayed a 3-fold reduction in cyclin E protein levels (Fig. 6 A and D); however, they showed an 8-fold reduction in the abundance of cyclin E mRNA in Fbw7-/- embryos (Fig. 6C). Quantification of the amount of cyclin E protein versus mRNA shows a 2.7-fold increase in Fbw7-/- versus Fbw7+/+ embryos and a 2.2-fold increase in Fbw7-/- versus Fbw7+/+ placentas (Fig. 6E). These results suggest an in vivo stabilization of cyclin E in the absence of Fbw7 (Fig. 6E).
Fig. 6.
Cyclin E is more abundant in Fbw7-/- placenta and embryo lysates relative to mRNA levels. Western blots using anti-cyclin E antibodies against whole-embryo (A) and whole-placenta (B) protein extracts are shown. NICD1 (N1*) cross-reactive band is the loading control in A. P107 is the loading control in B. (C) Quantitative, real-time PCR analysis of cyclin E transcripts in Fbw7-/- versus Fbw7+/+. (D) Relative cyclin E band intensities from blots in A and B normalized to loading controls. (E) Relative cyclin E protein/mRNA ratios calculated from the data in B and C.
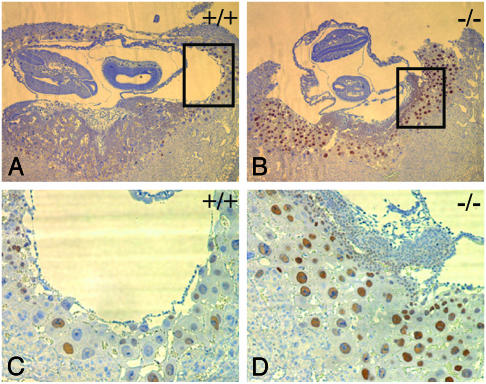
To assess which cells might contain increased cyclin E, we used immunocytohistochemistry to analyze the placentas of wild-type and Fbw7-/- embryos. We observed cyclin E expression in >90% of the Fbw7-/- trophoblast giant cells versus 50–60% of these cells in their wild-type littermates (Fig. 7 A and C versus 7 B and D). This correlated with an increased number of giant cells undergoing DNA synthesis (data not shown).
Fig. 7.
Fbw7-/- trophoblast giant cells contain more cyclin E and increased DNA synthesis. Placenta stained with antibodies against cyclin E (brown) counterstained with hematoxylin (blue). (A and C) Fbw7+/+ at ×4 and ×10 magnification. (B and D) Fbw7-/- ×4 and ×10 magnification.
Discussion
The mammalian SCFFbw7 is an E3 ubiquitin ligase that previously has been implicated in the control of cyclin E and Notch ubiquitination in vitro. Although biochemical evidence and cell culture studies supported these implications, no mammalian studies had been performed in vivo to establish the organismal role of Fbw7. Here, we investigated the in vivo role of Fbw7 through targeted disruption of the mouse Fbw7 locus. We find that Fbw7 is essential for mouse development, establishing a critical role for the control of Fbw7 substrate levels in development.
Loss of Fbw7 results in midgestational embryonic lethality, likely arising from defects in cardiovascular and hematopoietic development. Hematopoiesis is compromised in Fbw7-/- embryos, because few nucleated blood cells were detected in Fbw7-/- yolk sacs and embryos. Additionally, small vessel development in the yolk sac as well as the embryo proper was impaired. Furthermore, Fbw7-/- embryos demonstrated consistent defects in the formation of their left atrium and left ventricle, and, with less penetrance, they exhibited defects in the maturation of their right ventricle and outflow tract. Finally, Fbw7-/- embryos often showed defective somitogenesis.
We think it likely that the defects in hematopoietic and vascular development are the primary consequence of Fbw7 deletion and compromise the function of the yolk sac placenta, the effects of which are further exacerbated by the vascular defects in the embryo proper. Mice lacking various elements in the Notch signaling pathway show severely impaired vascular and hematopoietic development (31, 38, 50, 51, 66). Although Fbw7 mutants exhibit increased Notch signaling, it is possible that correct development requires the proper balance of Notch signaling.
Although not apparent until ED9.0, the defects in hematopoietic and vascular development impose nutritional stress on the embryo. Any developmental defects that occur later may be intrinsic to Fbw7 loss or indirect due to nutritional stress. Although we cannot rule out that the observed heart defects occur secondarily to nutritional stress, we feel that the defects in cardiac chamber formation are likely to be directly due to Fbw7 loss. First, we observed defects in left-sided heart development in every mutant embryo at ED9.5 we analyzed. Additionally, we observed subtle delays in heart chamber formation even by ED8.5, at which point no other obvious indication of nutritional stress existed. The formation of the right ventricular chamber occurred slightly later than that of the left chamber and thus could be more susceptible to nutritional stresses. Interestingly, there were many occasions in which the left ventricle failed to mature but the right side and outflow tract appeared normal. We believe that if the nutritional stress were of sufficient magnitude to block the slightly earlier development of the left heart, one might expect that the subsequent development of the right side of the heart would invariably be impaired. Because this is not always the case, we feel this further argues that the left-sided heart defects are a direct consequence of Fbw7 impairment. Finally, both Fbw7 (11) and its substrate Notch 1 (refs. 11, 66, and 67 and data not shown) are expressed in the mouse heart. This is consistent with the idea that the reduced capacity of Fbw7-/- embryos to temporally control Notch signaling affects normal cardiac development. In accordance with this is the finding that both Notch and its cognate binding partner recombination signal binding protein Jκ are capable of suppressing cardiomyogenesis (68, 69).
Fbw7 Is Required for Proper Control of Notch Signaling. Fwb7 has been implicated in Notch signaling in C. elegans. sel10, the Fbw7 homologue, acts antagonistically to Notch (34). Furthermore, SCF ligases containing recombinant Sel10 are capable of ubiquitinating Notch in vitro (12, 16). Although it is not possible to demonstrate stabilization of NICD in the absence of Fbw7 in vivo, we have shown an increased abundance of the NICD1 and NICD4 proteins without increased transcription of their respective genes. Thus, our results are consistent with the stabilization of the NICD by means of reduced ubiquitination. In support of the observed increase in NICD levels, we observed increases in the expression of Notch-activated transcripts Hes1, Herp1, and Herp2 in Fbw7-/- embryos. Thus, the increased NICD1 signals serves to alter the levels of gene expression in Fbw7-/- embryos. A number of studies have shown that unchecked Notch signaling wields deleterious effects in the developing embryos (40, 70). Given this critical role played by Notch signaling during development, the observed increase in signaling in Fbw7 mutants is likely to contribute in a causative manner to the vascular defects observed in Fbw7-/- embryos and extraembryonic tissues.
Fbw7 Negatively Regulates Cyclin E During Development. In Fbw7-/- placental lysates, we observe an almost 2-fold increase in cyclin E levels that cannot be accounted for by increased transcription of cyclin E. Previous studies implicating the SCF complex in cyclin E destruction in which Cul1 was deleted demonstrated an increased abundance of cyclin E and increased DNA synthesis in trophoblast giant cells of the placenta (64, 65). Our results are consistent with these findings, because Fbw7-/- giant cells exhibit elevated levels of cyclin E and increased DNA synthesis. Overexpression of cyclin E in cell culture has been shown to enhance the rate of transit into S phase (71, 72). It is unclear whether the alterations in the cyclin E levels of Fbw7-/- giant cells affect their biology to the extent that placental function is compromised. Placentation appears otherwise normal in Fbw7-/- embryos.
Fbw7-/- embryos did not demonstrate a gross increase in cyclin E. Instead, we observed a consistent 2- to 3-fold decrease in the protein levels in these embryos. However, cyclin E mRNA levels are almost 8-fold lower than their normal counterparts. Thus the ratio of cyclin E protein to its mRNA is elevated 2.7-fold in the mutant versus the wild-type embryo (Fig. 6E). Taken together, these data are consistent with the stabilization of cyclin E, masked by the overall reduction in cyclin E transcription in Fbw7-/- embryos. It is unclear why cyclin E mRNA is expressed at lower levels in Fbw7-/- embryos. It is possible that constitutive Notch signaling, Notch downstream targets, or some other unidentified substrate of the SCFFbw7 affects the transcription of cyclin E. Nonetheless, given the differences between relative protein levels and relative mRNA levels, we favor the interpretation that, as in Fbw7-/- placental tissue, stabilization of cyclin E occurs in Fbw7-/- embryos.
Fbw7 has been proposed to be a tumor suppressor. A number of studies have implicated the unregulated expression of both cyclin E and the NICDs in tumors (17, 18, 42, 53, 55). However, we did not observe tumor formation in Fbw7+/- animals up to 12 months of age. It is possible that these animals will display increased tumorigenesis later in life, or loss of both alleles of Fbw7 in adult tissues may be required to reveal a role in tumorigenesis. The ablation of Fbw7 in adult tissues could be accomplished by a conditional allele of Fbw7 or small interfering RNA. Given the established role of Fbw7 in control of known oncogenes combined with the relatively strong data implicating it in human tumorigenesis, we suspect that loss of Fbw7 will contribute to cancer in mice under the proper conditions, such as combining it with known oncogenes or carcinogen treatments.
These studies have demonstrated that Fbw7 plays an essential in vivo role in the control of the levels of critical developmental regulators. We demonstrated Fbw7 control of three such regulators, cyclin E, Notch 1, and Notch 4. Their deregulation may explain several of the phenotypes observed in Fbw7-/- animals. Furthermore, it is likely that other substrates will be found that might also contribute to the developmental defects observed in these mice. Because the embryos from these mice persist long enough to obtain proteins, the validity of future Fbw7 substrates can be tested using these animals. It is our hope that, in the future, these mice will be able to contribute to a mouse model for cancer caused by loss of human Fbw7.
Acknowledgments
We thank D. Killen for help with sectioning, A. Major for Ki67 stains, G. Nepala for valued help with real-time PCR analysis, and Dr. D. Craig Allred and Carlos Genty for help with platelet endothelial cell adhesion marker 1 stains. M.T.T. thanks A. Osborn, D. Schmucker, J. T. Winston, D. Cortez, D. Killen, and T. Westbrook for helpful discussions throughout the course of this work. M.T.T. was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Training Grant T32DK07696. This work was supported by National Institute of Health Grants AG11085 and CA58024 (to J.W.H. and S.J.E.), P01-HL49953 (to R.J.S.), and DK57693 (to K.M.). S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Abbreviations: SCF, Skp1 Cul1 F-box protein E3 ubiquitin ligase; NICD, Notch intracellular domain; ES, embryonic stem; EDn, n days postcoitus.
See accompanying Biography on page 3336.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
References
- 1.Baumeister, W., Walz, J., Zuhl, F. & Seemuller, E. (1998) Cell 92, 367-380. [DOI] [PubMed] [Google Scholar]
- 2.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- 3.Hershko, A., Heller, H., Elias, S. & Ciechanover, A. (1983) J. Biol. Chem. 258, 8206-8214. [PubMed] [Google Scholar]
- 4.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86, 263-274. [DOI] [PubMed] [Google Scholar]
- 5.Feldman, R. M., Correll, C. C., Kaplan, K. B. & Deshaies, R. J. (1997) Cell 91, 221-230. [DOI] [PubMed] [Google Scholar]
- 6.Kamura, T., Koepp, D. M., Conrad, M. N., Skowyra, D., Moreland, R. J., Iliopoulos, O., Lane, W. S., Kaelin, W. G., Jr., Elledge, S. J., Conaway, R. C., et al. (1999) Science 284, 657-661. [DOI] [PubMed] [Google Scholar]
- 7.Patton, E. E., Willems, A. R., Sa, D., Kuras, L., Thomas, D., Craig, K. L. & Tyers, M. (1998) Genes Dev. 12, 692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209-219. [DOI] [PubMed] [Google Scholar]
- 9.Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J. & Harper, J. W. (1999) Science 284, 662-665. [DOI] [PubMed] [Google Scholar]
- 10.Winston, J. T., Koepp, D. M., Zhu, C., Elledge, S. J. & Harper, J. W. (1999) Curr. Biol. 9, 1180-1182. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama, S., Hatakeyama, S., Nakayama, K., Ishida, N. & Kawakami, K. (2001) Genomics 78, 214-222. [DOI] [PubMed] [Google Scholar]
- 12.Gupta-Rossi, N., Le Bail, O., Gonen, H., Brou, C., Logeat, F., Six, E., Ciechanover, A. & Israel, A. (2001) J. Biol. Chem. 276, 34371-34378. [DOI] [PubMed] [Google Scholar]
- 13.Moberg, K. H., Bell, D. W., Wahrer, D. C., Haber, D. A. & Hariharan, I. K. (2001) Nature 413, 311-316. [DOI] [PubMed] [Google Scholar]
- 14.Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W. & Elledge, S. J. (2001) Science 294, 173-177. [DOI] [PubMed] [Google Scholar]
- 15.Strohmaier, H., Spruck, C. H., Kaiser, P., Won, K. A., Sangfelt, O. & Reed, S. I. (2001) Nature 413, 316-322. [DOI] [PubMed] [Google Scholar]
- 16.Wu, G., Lyapina, S., Das, I., Li, J., Gurney, M., Pauley, A., Chui, I., Deshaies, R. J. & Kitajewski, J. (2001) Mol. Cell. Biol. 21, 7403-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bortner, D. M. & Rosenberg, M. P. (1997) Mol. Cell. Biol. 17, 453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enders, G. H. (2002) Breast Cancer Res. 4, 145-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed, S. I. (1997) Cancer Surv. 29, 7-23. [PubMed] [Google Scholar]
- 20.Spruck, C. H., Strohmaier, H., Sangfelt, O., Muller, H. M., Hubalek, M., Muller-Holzner, E., Marth, C., Widschwendter, M. & Reed, S. I. (2002) Cancer Res. 62, 4535-4539. [PubMed] [Google Scholar]
- 21.Spruck, C. H., Won, K. A. & Reed, S. I. (1999) Nature 401, 297-300. [DOI] [PubMed] [Google Scholar]
- 22.Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. (1999) Science 284, 770-776. [DOI] [PubMed] [Google Scholar]
- 23.Logeat, F., Bessia, C., Brou, C., LeBail, O., Jarriault, S., Seidah, N. G. & Israel, A. (1998) Proc. Natl. Acad. Sci. USA 95, 8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaumueller, C. M., Qi, H., Zagouras, P. & Artavanis-Tsakonas, S. (1997) Cell 90, 281-291. [DOI] [PubMed] [Google Scholar]
- 25.Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., Ray, W. J. & Kopan, R. (2000) Mol. Cell 5, 197-206. [DOI] [PubMed] [Google Scholar]
- 26.Brou, C., Logeat, F., Gupta, N., Bessia, C., LeBail, O., Doedens, J. R., Cumano, A., Roux, P., Black, R. A. & Israel, A. (2000) Mol. Cell 5, 207-216. [DOI] [PubMed] [Google Scholar]
- 27.Schroeter, E. H., Kisslinger, J. A. & Kopan, R. (1998) Nature 393, 382-386. [DOI] [PubMed] [Google Scholar]
- 28.Struhl, G. & Adachi, A. (1998) Cell 93, 649-660. [DOI] [PubMed] [Google Scholar]
- 29.Struhl, G. & Greenwald, I. (1999) Nature 398, 522-525. [DOI] [PubMed] [Google Scholar]
- 30.Kageyama, R. & Ohtsuka, T. (1999) Cell Res. 9, 179-188. [DOI] [PubMed] [Google Scholar]
- 31.Kojika, S. & Griffin, J. D. (2001) Exp. Hematol. (Charlottesville, Va) 29, 1041-1052. [DOI] [PubMed] [Google Scholar]
- 32.Iso, T., Kedes, L. & Hamamori, Y. (2003) J. Cell. Physiol. 194, 237-255. [DOI] [PubMed] [Google Scholar]
- 33.Beatus, P. & Lendahl, U. (1998) J. Neurosci. Res. 54, 125-136. [DOI] [PubMed] [Google Scholar]
- 34.Hubbard, E. J., Wu, G., Kitajewski, J. & Greenwald, I. (1997) Genes Dev. 11, 3182-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.del Amo, F. F., Gendron-Maguire, M., Swiatek, P. J., Jenkins, N. A., Copeland, N. G. & Gridley, T. (1993) Genomics 15, 259-264. [DOI] [PubMed] [Google Scholar]
- 36.Del Amo, F. F., Smith, D. E., Swiatek, P. J., Gendron-Maguire, M., Greenspan, R. J., McMahon, A. P. & Gridley, T. (1992) Development (Cambridge, U.K.) 115, 737-744. [DOI] [PubMed] [Google Scholar]
- 37.Higuchi, M., Kiyama, H., Hayakawa, T., Hamada, Y. & Tsujimoto, Y. (1995) Brain Res. Mol. Brain Res. 29, 263-272. [DOI] [PubMed] [Google Scholar]
- 38.Joutel, A., Corpechot, C., Ducros, A., Vahedi, K., Chabriat, H., Mouton, P., Alamowitch, S., Domenga, V., Cecillion, M., Marechal, E., et al. (1996) Nature 383, 707-710. [DOI] [PubMed] [Google Scholar]
- 39.Lardelli, M., Dahlstrand, J. & Lendahl, U. (1994) Mech. Dev. 46, 123-136. [DOI] [PubMed] [Google Scholar]
- 40.Lardelli, M., Williams, R., Mitsiadis, T. & Lendahl, U. (1996) Mech. Dev. 59, 177-190. [DOI] [PubMed] [Google Scholar]
- 41.Reaume, A. G., Conlon, R. A., Zirngibl, R., Yamaguchi, T. P. & Rossant, J. (1992) Dev. Biol. 154, 377-387. [DOI] [PubMed] [Google Scholar]
- 42.Uyttendaele, H., Marazzi, G., Wu, G., Yan, Q., Sassoon, D. & Kitajewski, J. (1996) Development (Cambridge, U.K.) 122, 2251-2259. [DOI] [PubMed] [Google Scholar]
- 43.Weinmaster, G., Roberts, V. J. & Lemke, G. (1991) Development (Cambridge, U.K.) 113, 199-205. [DOI] [PubMed] [Google Scholar]
- 44.Weinmaster, G., Roberts, V. J. & Lemke, G. (1992) Development (Cambridge, U.K.) 116, 931-941. [DOI] [PubMed] [Google Scholar]
- 45.Williams, R., Lendahl, U. & Lardelli, M. (1995) Mech. Dev. 53, 357-368. [DOI] [PubMed] [Google Scholar]
- 46.Lardelli, M. & Lendahl, U. (1993) Exp. Cell Res. 204, 364-372. [DOI] [PubMed] [Google Scholar]
- 47.Conlon, R. A., Reaume, A. G. & Rossant, J. (1995) Development (Cambridge, U.K.) 121, 1533-1545. [DOI] [PubMed] [Google Scholar]
- 48.Hamada, Y., Kadokawa, Y., Okabe, M., Ikawa, M., Coleman, J. R. & Tsujimoto, Y. (1999) Development (Cambridge, U.K.) 126, 3415-3424. [DOI] [PubMed] [Google Scholar]
- 49.Huppert, S. S., Le, A., Schroeter, E. H., Mumm, J. S., Saxena, M. T., Milner, L. A. & Kopan, R. (2000) Nature 405, 966-970. [DOI] [PubMed] [Google Scholar]
- 50.Kumano, K., Chiba, S., Kunisato, A., Sata, M., Saito, T., Nakagami-Yamaguchi, E., Yamaguchi, T., Masuda, S., Shimizu, K., Takahashi, T., et al. (2003) Immunity 18, 699-711. [DOI] [PubMed] [Google Scholar]
- 51.Krebs, L. T., Xue, Y., Norton, C. R., Shutter, J. R., Maguire, M., Sundberg, J. P., Gallahan, D., Closson, V., Kitajewski, J., Callahan, R., et al. (2000) Genes Dev. 14, 1343-1352. [PMC free article] [PubMed] [Google Scholar]
- 52.Swiatek, P. J., Lindsell, C. E., del Amo, F. F., Weinmaster, G. & Gridley, T. (1994) Genes Dev. 8, 707-719. [DOI] [PubMed] [Google Scholar]
- 53.Robbins, J., Blondel, B. J., Gallahan, D. & Callahan, R. (1992) J. Virol. 66, 2594-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellisen, L. W., Bird, J., West, D. C., Soreng, A. L., Reynolds, T. C., Smith, S. D. & Sklar, J. (1991) Cell 66, 649-661. [DOI] [PubMed] [Google Scholar]
- 55.Zagouras, P., Stifani, S., Blaumueller, C. M., Carcangiu, M. L. & Artavanis-Tsakonas, S. (1995) Proc. Natl. Acad. Sci. USA 92, 6414-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, P., Li, M. Z. & Elledge, S. J. (2002) Nat. Genet. 30, 31-39. [DOI] [PubMed] [Google Scholar]
- 57.McMahon, A. P. & Bradley, A. (1990) Cell 62, 1073-1085. [DOI] [PubMed] [Google Scholar]
- 58.Wei, L., Imanaka-Yoshida, K., Wang, L., Zhan, S., Schneider, M. D., DeMayo, F. J. & Schwartz, R. J. (2002) Development (Cambridge, U.K.) 129, 1705-1714. [DOI] [PubMed] [Google Scholar]
- 59.Reecy, J. M., Li, X., Yamada, M., DeMayo, F. J., Newman, C. S., Harvey, R. P. & Schwartz, R. J. (1999) Development (Cambridge, U.K.) 126, 839-849. [DOI] [PubMed] [Google Scholar]
- 60.Winston, J. T., Strack, P., Beer-Romero, P., Chu, C. Y., Elledge, S. J. & Harper, J. W. (1999) Genes Dev. 13, 270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christoffels, V. M., Habets, P. E., Franco, D., Campione, M., de Jong, F., Lamers, W. H., Bao, Z. Z., Palmer, S., Biben, C., Harvey, R. P. & Moorman, A. F. (2000) Dev. Biol. 223, 266-278. [DOI] [PubMed] [Google Scholar]
- 62.Olson, E. N. & Srivastava, D. (1996) Science 272, 671-676. [DOI] [PubMed] [Google Scholar]
- 63.Singer, J. D., Gurian-West, M., Clurman, B. & Roberts, J. M. (1999) Genes Dev. 13, 2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Y., Penfold, S., Tang, X., Hattori, N., Riley, P., Harper, J. W., Cross, J. C. & Tyers, M. (1999) Curr. Biol. 9, 1191-1194. [DOI] [PubMed] [Google Scholar]
- 65.Dealy, M. J., Nguyen, K. V., Lo, J., Gstaiger, M., Krek, W., Elson, D., Arbeit, J., Kipreos, E. T. & Johnson, R. S. (1999) Nat. Genet. 23, 245-248. [DOI] [PubMed] [Google Scholar]
- 66.Loomes, K. M., Underkoffler, L. A., Morabito, J., Gottlieb, S., Piccoli, D. A., Spinner, N. B., Baldwin, H. S. & Oakey, R. J. (1999) Hum. Mol. Genet. 8, 2443-2449. [DOI] [PubMed] [Google Scholar]
- 67.Loomes, K. M., Taichman, D. B., Glover, C. L., Williams, P. T., Markowitz, J. E., Piccoli, D. A., Baldwin, H. S. & Oakey, R. J. (2002) Am. J. Med. Genet. 112, 181-189. [DOI] [PubMed] [Google Scholar]
- 68.Schroeder, T., Fraser, S. T., Ogawa, M., Nishikawa, S., Oka, C., Bornkamm, G. W., Honjo, T. & Just, U. (2003) Proc. Natl. Acad. Sci. USA 100, 4018-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rones, M. S., McLaughlin, K. A., Raffin, M. & Mercola, M. (2000) Development (Cambridge, U.K.) 127, 3865-3876. [DOI] [PubMed] [Google Scholar]
- 70.Henderson, A. M., Wang, S. J., Taylor, A. C., Aitkenhead, M. & Hughes, C. C. (2001) J. Biol. Chem. 276, 6169-6176. [DOI] [PubMed] [Google Scholar]
- 71.Ohtsubo, M. & Roberts, J. M. (1993) Science 259, 1908-1912. [DOI] [PubMed] [Google Scholar]
- 72.Resnitzky, D., Gossen, M., Bujard, H. & Reed, S. I. (1994) Mol. Cell. Biol. 14, 1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winston, J. T., Chu, C. & Harper, J. W. (1999) Genes Dev. 13, 2751-2757. [DOI] [PubMed] [Google Scholar]
- 74.Geng, Y., Yu, Q., Sicinska, E., Das, M., Schneider, J. E., Bhattacharya, S., Rideout, W. M., Bronson, R. T., Gardner, H. & Sicinski, P. (2003) Cell 114, 431-443. [DOI] [PubMed] [Google Scholar]