Abstract
The molecular structure of the pepstatin-insensitive carboxyl peptidase from Scytalidium lignicolum, formerly known as scytalidopepsin B, was solved by multiple isomorphous replacement phasing methods and refined to an R factor of 0.230 (Rfree = 0.246) at 2.1-Å resolution. In addition to the structure of the unbound peptidase, the structure of a product complex of cleaved angiotensin II bound in the active site of the enzyme was also determined. We propose the name scytalidocarboxyl peptidase B (SCP-B) for this enzyme. On the basis of conserved, catalytic residues identified at the active site, we suggest the name Eqolisin for the enzyme family. The previously uninvestigated SCP-B fold is that of a β-sandwich; each sheet has seven antiparallel strands. A tripeptide product, Ala-Ile-His, bound in the active site of SCP-B has allowed for identification of the catalytic residues and the residues in subsites S1, S2, and S3, which are important for substrate binding. The most likely hydrolytic mechanism involves nucleophilic attack of a general base (Glu-136)-activated water (OH-) on the si-face of the scissile peptide carbonylcarbon atom to form a tetrahedral intermediate. Electrophilic assistance and oxyanion stabilization is provided by the side-chain amide of Gln-53. Protonation of the leaving-group nitrogen is accomplished by the general acid function of the protonated carboxyl group of Glu-136.
Pepstatin-insensitive carboxyl peptidases (1–8) originally found by Murao and Oda can be classified into two groups, bacterial and fungal carboxyl peptidases. The 3D structures of the two members of the bacterial carboxyl peptidases have recently been determined (9, 10). From the tertiary folds, it was clear that these enzymes are members of the subtilisin family. Each enzyme has a glutamate residue acting as the general base instead of the histidine present in the subtilisins. The environments of the carboxylates contribute to the low pH optima of these enzymes.
The fungal pepstatin-insensitive carboxyl peptidases (5) are represented by several enzymes, among them the enzyme that is the subject of this article. Formerly, this enzyme has been called Scytalidopepsin-B because of its low pH optimum and preponderance of acidic residues (8). It is isolated from Scytalidium lignicolum ATCC24568. We have chosen to rename this enzyme scytalidocarboxyl peptidase-B (SCP-B) to reflect its fungal source and active-site carboxyl groups. SCP-B is synthesized as a precursor consisting of two regions: an amino-terminal preprosegment of 54 amino acid residues and a mature enzyme consisting of 206 amino acid residues (11). The mature enzyme has no sequence similarity to the well known pepsin-like or retroviral aspartic peptidases, but it has significant similarity to the other fungal pepstatin-insensitive carboxyl peptidases (Fig. 1).
Fig. 1.
clustalw multiple-sequence alignment of the eqolisins. The sequence numbering for each mature enzyme is indicated. The enzymes in this alignment are SCP-B (11), Aspergillus niger carboxyl peptidase A (AnCPA) (12), Sclerotinia sclerotiorum carboxyl peptidase (SSCP) (13), Cryphonectria parasitica peptidases B and C (EAPB and EAPC) (14), and Talaromyces emersonii carboxyl peptidase (TECP) (GenBank accession no. AF439998). The β-strands constituting the secondary structure of SCP-B determined herein are indicated by numbered arrows above the sequence of SCP-B. Amino acid color-coding is as follows: green, neutral hydrophilic side chains; red, hydrophobic side chains; blue, negatively charged side chains; and magenta, positively charged side chains. The catalytic residues Gln-53 and Glu-136 are highlighted in black. Asterisks denote residues that are identical in all six sequences; colons or periods denote highly conserved residues. The 11 residues in the box in the AnCPA sequence are removed in the mature enzyme (12).
SCP-B and the other carboxyl peptidases described here have been classified in family A4 of the aspartic endopeptidases in the MEROPS (http://merops.sanger.ac.uk). database. On the basis of our structural results, however, we prefer to name this family the Eqolisins (pronounced “echo”-lisin) derived from the active-site residues, glutamic acid (E) and glutamine (Q). We suggest that the eqolisins constitute a new peptidase family that is not part of the aspartic peptidases.
SCP-B works best in acidic conditions, having an optimal pH of 2.0 with casein as substrate, suggesting that carboxyl groups are intimately involved in the catalytic function. SCP-B is not inhibited by pepstatin, acetyl-pepstatin, nor by diazoacetyl-dl-norleucine methyl ester, but it is inhibited by 1,2-epoxy-3-(p-nitrophenoxy)propane. SCP-B cleaves the oxidized insulin B chain at Tyr-26–Thr-27, Phe-24–Phe-25, and other positions after Tyr and Glu. It has also been shown to cleave the His-6–Pro-7 bond of angiotensin II and, to a lesser extent, the bond between Tyr-4 and Ile-5 of this octapeptide (15).
Several studies have been carried out to identify the catalytic residues of the eqolisins. Tsuru and colleagues (16, 17) reported that Glu-53 and Asp-98, residues in SCP-B, were candidates for its catalytic residues. The nucleotide sequence of the SCP-B gene, elucidated after these studies, however, revealed that Glu-53 was in fact a glutamine residue (18). Furthermore, it turned out that Asp-98 is not conserved in any of the other members of this peptidase family. Takahashi and colleagues (19) reported from mutational data that Glu-219 and Asp-123 (sequence numbers in the mature enzyme, Glu-149 and Asp-53) in AnCPA were involved in the catalytic function. These residues are conserved in the SCP-B molecule as Glu-136 and Asp-43, respectively. Thus, without structural data, the catalytic mechanism for this unique family of peptidases is still shrouded in mystery.
We have solved the crystal structures of SCP-B in the unbound form and in a form complexed with hydrolytic products of angiotensin II. On the basis of these 3D structures and of sequence and biochemical data, it was determined that SCP-B has a catalytic dyad consisting of residues Gln-53 and Glu-136. These two residues and the surrounding segments of polypeptide chain are highly conserved in all fungal pepstatin-insensitive carboxyl peptidases of the eqolisin family (Fig. 1). From the results derived here, the eqolisins should occupy a more distinct position in the overall classification of peptidases (20).
Methods
Crystallization and Data Collection. Lyophilized SCP-B was dissolved in water at 20 mg/ml. Crystals were grown at room temperature by the hanging-drop method from 42% saturated ammonium sulfate/0.1 M sodium acetate buffer (pH 4.0)/10% (vol/vol) ethylene glycol. Before data collection, crystals of SCP-B were transferred for several seconds into a cryosolution containing 30% glycerol, 45% saturated ammonium sulfate, and 0.1 M sodium acetate (pH 4.0) then frozen in the cryostream. Heavy-atom derivative crystals were obtained by soaking native crystals in 1.5 mM mersalyl for 4 h or in 3 mM uranyl acetate overnight before freezing and data collection. For soaking a potential substrate into the crystals, a drop consisting of 3 μl of 10 mM angiotensin II in water and 3 μl of the crystallization solution was equilibrated for several hours and an SCP-B crystal was soaked in it overnight. The diffraction data were collected the next day from a flash-frozen crystal.
All data except for the native set were collected on a Rigaku RAXIS-IV++ area detector with CuKα radiation generated by a Rigaku RU-H3R rotating anode generator (Table 1). The native data were collected on beam line 9-1 at the Stanford Synchrotron Radiation Laboratory. All data were processed with the hkl suite of programs (21).
Table 1. Summary of crystallographic data.
| Native | Uranyl | Mercury | Angiotensin II | |
|---|---|---|---|---|
| Crystals | ||||
| Space group | P6322 | P6322 | P6322 | P6322 |
| Cell parameters | ||||
| a, b | 108.553 | 108.601 | 108.648 | 109.369 |
| c | 114.149 | 113.880 | 114.312 | 113.830 |
| X-ray | SSRL BL 9-1,* | Cu Kα | Cu Kα | Cu Kα |
| λ = 0.979 Å | ||||
| Resolution, Å | 20–1.9 (1.97–1.90) | 47–2.4 (2.49–2.40) | 36–2.4 (2.49–2.40) | 27–2.0 (2.07–2.0) |
| Rmerge† | 5.8 (48.2) | 9.3 (42.6) | 9.3 (43.7) | 5.0 (41.3) |
| Completeness, % | 96.5 (88.8) | 93.9 (89.3) | 99.9 (100) | 99.9 (100) |
| Mean I/σ(I) | 8.1 | 14.2 | 13.6 | 19.2 |
| Refinement | ||||
| Resolution, Å | 20–2.1 | 27–2.0 | ||
| No. of protein atoms | 1,450 | 1,450 | ||
| No. of water molecules | 167 | 121 | ||
| No. of ligand atoms | — | 37 | ||
| R factor† (Rfree)*§ | 22.97 (24.57) | 23.35 (25.58) | ||
| No. of reflections | 18,539 (1,032) | 26,254 (1,422) | ||
| rms deviation bond, Å | 0.006 | 0.006 | ||
| rms deviation angle, ° | 1.4 | 1.4 |
SSRL BL 9-1, Stanford Synchrotron Radiation Laboratory (Stanford University, Stanford, CA) beam line 9-1
Rmerge = 100 Σ |Ih – 〈I〉|/ΣIh
R factor = 100 Σ ||Fobs |–| Fcalc ||/Σ |Fobs| calculated for all observed data
Rfree = R factor for ≈5% of the randomly chosen unique reflections not used in the refinement
Structure Determination and Refinement. The structure was solved by the multiple isomorphous replacement method from data collected from the mersalyl and uranyl heavy-atom derivatives by using the program solve (22). solve assigned one unique heavy-atom site for each of the derivatives. The phases derived from the multiple isomorphous replacement procedure resulted in a figure- of-merit (FOM) of 0.35. Subsequently, density modification carried out by the program resolve (23, 24) increased the FOM to 0.68. Examination of the electron density computed from the phase set at this stage showed clearly interpretable molecular features. Thus, these phases and the corresponding electron density map were used for automated chain tracing by the program warpntrace (25). The program was able to fit 168 residues in nine chain segments. Substitution of the correct sequence and additional model building was done manually with xtalview (26). The program cns (27) was used for refinement of both the native and product-bound structures (Table 1). The refined coordinates have been deposited in the Protein Data Bank with ID codes 1S2B and 1S2K.
The crystal quality of SCP-B was variable. Despite the relatively high resolution recorded for the native data set, the quality of the data in the high-resolution shells was poor. Overall the mean I/σ(I) is 8.1; it is much worse (1.3) for the data in the range from 2.1 Å to 1.9 Å. The value of Rmerge is also large for the same range of data; overall, the data are only 89% complete. For these reasons, the data were truncated to 2.1 Å for use in the refinement. Very likely the poor data quality is the cause for the disappointingly high R factors for the unbound enzyme (Table 1). The data for the product-bound form of SCP-B was not truncated in resolution, as the I/σ(I) was much larger in the high-resolution shell (2.07 to 2.0 Å) than that of the native data set.
Results
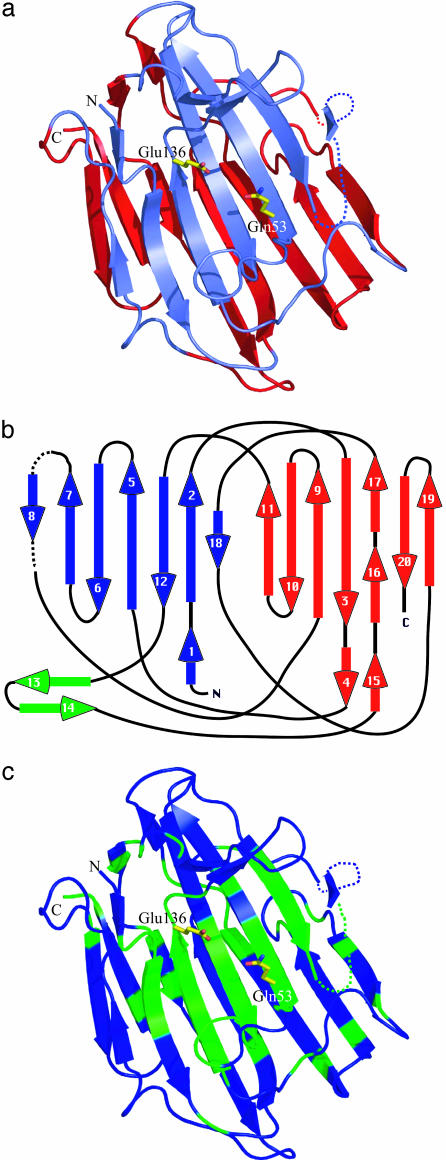
SCP-B is the founding member of the eqolisins, a new family of proteolytic enzymes (Fig. 1). The overall structure of the enzyme is that of a β-sandwich formed from two antiparallel β-sheets; each sheet consists of seven strands (Fig. 2 a and b). A blast search with the SCP-B sequence has identified five additional members of the eqolisins (Fig. 1). Each of these enzymes is synthesized as a preproprecursor protein. The preprosegments are ≈55 amino acids in length, and the prosegments are rich in positively charged residues. This observation is very similar to the charge distribution in the zymogens of the aspartic peptidases (28). The mature eqolisins have a preponderance of negatively charged residues (e.g., SCP-B has 32 Asp plus Glu residues) over positively charged residues (SCP-B has only 2 Lys and 1 Arg). Clearly, this distribution is one of the determining factors for the low pI of SCP-B (3.0).
Fig. 2.
(a) Ribbon representation of SCP-B. The secondary structure assignment was determined by Database of Secondary Structure of Proteins (31). This drawing and Figs. 2c, 3, 4, 5, and 6b were produced by the program pymol (32). The active-site residues, Gln-53 and Glu-136, are shown in stick representation. The upper sheet is slate, and the lower sheet is red. (b) A topological diagram representing the secondary structure in SCP-B. The upper sheet harboring the active site is blue, and the lower sheet is red. The loop unique to SCP-B that is adjacent to the active site is shown in green. Seven loops cross from the upper to the lower sheet or from the lower to the upper sheet in this topography. (c) A representation of the regions of SCP-B that are highly conserved among the six members of the eqolisin family. Those segments of the chain corresponding to these highly conserved regions of SCP-B (asterisks in Fig. 1) are green.
The mature enzymes have several highly conserved segments of polypeptide chain (in SCP-B numbering: residues 4–10, 38–46, 50–58, 63–73, 81–89, 130–140, and 150–157; Fig. 1). From Fig. 2c, it can be seen that many of these regions of conserved residues are clustered on the upper β-sheet surrounding the active site and the substrate-binding site. The high level of sequence identity and the clustering of the conserved regions on the upper β-sheet that comprises the active site and substrate-binding region confirm that these enzymes are all related members of the eqolisin family.
SCP-B has three disulfide bridges, not all of which are conserved in the other members of the eqolisins. The most highly conserved disulfide is that between Cys-47 and Cys-127 (absent in the C enzyme from C. parasitica). In SCP-B this disulfide surrounds the buried Asp-43, a residue that is highly conserved in all members of the family. Another disulfide is unique to SCP-B (Cys-141 to Cys-148). It is at the base of the hairpin loop (green arrows in Fig. 2b) that is present only in SCP-B. This loop has been deleted in the other carboxyl peptidases of the eqolisin family (residues Asn-144 to Glu-149) (see Figs. 1 and 2c).
The isoelectric point of SCP-B is in the acidic range (pI = 3.0). Most of the carboxylate groups in SCP-B are solvent-exposed, so they would not contribute to such a low pI. However, several carboxylate residues have depressed pKa values either as a result of being involved in carboxyl–carboxylate interactions (29) (i.e., the sharing of one proton by two carboxyl groups) or as a result of being buried in the core of the enzyme. The carboxyl groups of Asp-57 and Asp-65 share a hydrogen bond (O—O distance of 2.5 Å) and likely have one of the two pKa values of ≈1.5. The carboxylate of Asp-43 is buried and negatively charged, because it is the recipient of four hydrogen bonds from the neighboring main-chain NH groups from residues Cys-127 to Asn-130, inclusive. The environments of these two carboxyl groups would contribute strongly to the low pI value of SCP-B.
The Unbound Active Site of SCP-B. From the results of the biochemical, kinetic, and mutagenic studies that have been carried out on SCP-B and AnCPA, it is most likely that residues Glu-136 and Gln-53 are key catalytic residues. These residues are located on the concave surface of the upper β-sheet (Fig. 2a) and are separated by a closest approach of 5.03 Å between atoms Glu-136 Oε2 and Gln-53 Oε1. A water molecule, HOH169, forms a hydrogen bond to each of these oxygen atoms in the native enzyme (Table 2 and Fig. 3). By analogy with the aspartic peptidases' active sites (30), we propose that this water is the nucleophilic water in the hydrolytic mechanism of the eqolisins. For the side-chain carboxylate of Glu-136 to act as a general base, it must be negatively charged. The negative charge on the carboxyl group of Glu-136 is likely, because it is the recipient of three hydrogen bonds (Fig. 3b), one from HOH169 (3.0 Å), one from the Nε1 of Trp-39 (2.9 Å), and one from the Nδ2 atom of Asn-5 (3.0 Å). Glu-136 is only partially solvent-accessible. Therefore, Glu-136 is unlikely to enter into a covalent acyl-enzyme intermediate (an acid anhydride) by direct nucleophilic attack on the carbonyl-carbon of the scissile peptide. As with the aspartic peptidases, no space exists for such a close substrate approach.
Table 2. Hydrogen bonding in the active site of SCP-B.
| Atoms | Distance, Å | Atoms | Distance, Å |
|---|---|---|---|
| Glu136 Oε2...HOH169 | 3.0 | Trp6 Nε1...HOH212 | 3.0 |
| Gln53 Oε1...HOH169 | 2.9 | Asp57 Oδ2...Asp65 Oδ2 | 2.6 |
| Glu136 Oε2...Trp39 Nε1 | 2.9 | Asp57 Oδ1...Thr37 Oγ1 | 2.8 |
| Glu136 Oε1...Asn5 Nδ2 | 3.0 | Trp67 Nε1...Asp57 Oδ1 | 2.8 |
| Gln53 Nε2...Glu69 Oε1 | 2.9 | Asp65 Oδ1...Tyr59 OH | 2.7 |
| Gln53 Nε2...HOH304 | 2.7 | HOH169...HOH304 | 2.8 |
Fig. 3.
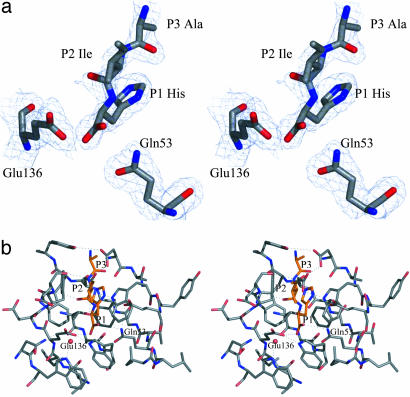
(a) Electron density associated with residues that constitute the active site of SCP-B. The electron density was calculated by using coefficients (2m|Fo|-D|Fc|), and calculated phases (αc) (33); the map is contoured at 1σ within 3.5 Å of the active-site residues and solvent molecules. (b) Residues of the active site of the unbound form of SCP-B are represented in stereo; oxygen atoms are red, nitrogen atoms are blue, and carbon atoms are slate. The water molecules are shown as red spheres; hydrogen bonds between polar side-chain atoms and to waters are shown as dotted lines. The hydrogen bond distances are given in Table 2.
The side chain of Gln-53 is wedged between the indole ring of Trp-67 and the side chain of Ile-51 (Fig. 3b). The amide Nε2 of Gln-53 donates a hydrogen bond to the Oε1 of Glu-69 (2.9 Å). The Oε1 carbonyl oxygen of Gln-53 receives a hydrogen bond from the nucleophilic water, HOH169 (2.9 Å). In addition, the amide nitrogen Nε2 of Gln-53 makes a hydrogen bond to another water, HOH304 (2.7 Å). This latter site is postulated as the binding site for the carbonyl-oxygen atom of the scissile peptide bond of a substrate, thereby dictating the Ψ conformational angle of the P1 residue of a bound substrate.
The carboxyl–carboxylate interaction between Asp-65 and Asp-57 is at the distal end of the S1 pocket; it provides a net negative charge for positively charged P1 side chains (Fig. 3b). The side chain Oδ1 of Asp-57 receives two other hydrogen bonds, one from the Oγ1 atom of Thr-37 (2.8 Å) and the other from the Nε1 atom of Trp-67 (2.8 Å). All the above-mentioned hydrogen bonds are between residues that are conserved in all six known members of the eqolisin family (Fig. 1).
The Binding of Hydrolytic Products of Angiotensin II to SCP-B. To identify the active site of SCP-B and to visualize the key residues that are involved in catalysis, angiotensin II (Asp-Arg-Val-Tyr-Ile-His-Pro-Phe) was soaked into pregrown crystals of SCP-B; diffraction data were collected to 2.0-Å resolution (Table 1). A difference electron density map (Fig. 4a) showed two separate regions of positive density in the substrate-binding cleft of SCP-B. One was interpreted as a tripeptide, Ala-Ile-His (Fig. 4), and the other was interpreted as a single tyrosine (not shown). One would have expected a hexapeptide to remain as a product complex in the enzymes' active site; however, the interpretable regions of difference density corresponded to a tripeptide Tyr-Ile-His with the Tyr side chain of this tripeptide not visible beyond Cβ. It is not at all clear from where the single tyrosine derived, but a tyrosine is in angiotensin II that with appropriate enzyme-catalyzed cleavages could be the source.
Fig. 4.
(a) Electron density associated with the product Ala-Ile-His-COO- and the two catalytic residues Gln-53 and Glu-136. The details of the contour levels are as given in Fig. 3a. (b) The environment of the angiotensin II hydrolysis product Ala-Ile-His (orange) is shown. The C terminus of the tripeptide corresponds to a cleavage site in Angiotensin II at the His–Pro bond, and the tripeptide indicates the binding mode of three residues of the N-terminal segment of the substrate. Hydrogen bonds between groups on the enzyme and on the tripeptide are indicated by dotted lines.
The atomic model of the refined product complex included the tripeptide, Ala-Ile-His, the single tyrosine, and 200 residues of SCP-B (six of the residues in the segment Tyr-71 to Gly-80 are disordered). The resulting electron density for the P1 His of the product is very clear (Fig. 4a), but the side chain of the Ile in the P2 position is in weak electron density and the atoms have B factors in the range of 60 Å2. The tripeptide binds in an extended conformation with the NH and CO of the P2 Ile, making antiparallel hydrogen-bonding interactions with the CO and NH of Glu-139 (Fig. 4b). The OXT atom of the P1 His carboxyl group at the C terminus of the tripeptide forms a hydrogen bond with the side-chain carboxyl group of Glu-136 (2.6 Å). The distance from Gln-53 Oε1 to P1 His OXT is 3.4 Å and is likely a van der Waals contact rather than a hydrogen bond. Thus, the simplest interpretation for these interactions would have the side-chain carboxyl group of Glu-136 protonated and a second hydrogen bond to the P1 His O donated by Gln-53 Nε2 (3.18 Å). The side-chain imidazolium group of the P1 His packs between the phenyl side chain of Phe-138 and the indole ring of Trp-67 (Fig. 4b). Both residues are highly conserved among the six known members of this family. In addition, the Nε2 atom of the P1 His imidazolium group forms a hydrogen bond to the carboxylate of Asp-57 (3.1 Å).
The protein inhibitors of the serine peptidases [e.g., the third domains of avian ovomucoids (34, 35)] bind to their cognate enzymes in the manner of good substrates. The P1 residues of these inhibitors have average main-chain conformational angles of φ ≈ -118°, Ψ ≈+35°, and side chain χ1 ≈-65°. The nucleophilic attack of the serine Oγ on the carbonyl-carbon atom of the substrate takes place on the re-face of the scissile peptide in those enzymes. The resulting tetrahedral intermediate would have a Ψ angle of ≈+60°, a staggered conformation. In the eqolisins, the direction of nucleophilic attack of water HOH169 is on the si-face of the scissile peptide (see below). In the product complex with SCP-B, the conformational angles for the P1 His are φ = -112°, Ψ (pseudo) = -6°, and side chains χ1 =-49° and χ2 =+77°. The values of χ1 and χ2 are well within the range of values observed for histidine side chains.
From a series of fluorescence resonance energy transfer experiments on oligopeptide substrates, the preference for the P1 residue was determined to be tyrosine followed closely by histidine (K.O. and K. Takada, unpublished results). In addition, these substrates showed preferences at the P2 and P3 positions for tyrosine and lysine or arginine, respectively. The preference for positively charged residues at P2 and P3 can be understood from the overall net negative charge on SCP-B due to the preponderance of Asp and Glu residues in the molecule. The most favored substrate was cleaved at the Tyr–Ala bond in the tetrapeptide sequence, Arg-Tyr-Tyr-Ala.
Two main reasons exist for why pepstatin is a poor inhibitor of the eqolisins. Modeling a leucine side chain at the P1 position indicates that Leu would not fit well sterically or electrostatically. Also, the 3-OH group of a statine residue in pepstatin has the wrong stereochemical configuration.
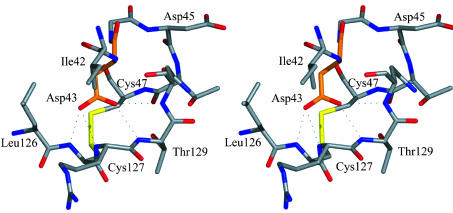
Mutational data from the closely related aspergillopepsin II (A. niger carboxyl proteinase-A or AnCPA) suggested that Asp-123 and Glu-219 (mature numbering, Asp-53 and Glu-149) are the catalytic residues (20, 36). The suggestion that Asp-53 (Asp-43 in SCP-B) is a catalytic residue is better interpreted as a disruption of the folding pathway of the enzyme caused by the mutation of Asp-53. As pointed out, in SCP-B, Asp-43 is in a highly conserved region (Fig. 1), but it is distant from the active site and buried in the core of the enzyme. In addition, the carboxylate of Asp-43 is most likely negatively charged, because it is the recipient of four hydrogen bonds from successive main-chain NH groups in the segment of Cys-127 to Asn-130, inclusive (Fig. 5). Any mutation of Asp-53 (Asp-43) would therefore have a severely detrimental effect on the folding of AnCPA or any of the members of this family.
Fig. 5.
The carboxylate group of Asp-43 in SCP-B is buried and is the recipient of four H bonds from the main-chain NH groups from residues Cys-127 to Asn-130, inclusively.
Proposed Hydrolytic Mechanism for the Eqolisins. We have developed a plausible catalytic mechanism (Fig. 6a) for SCP-B that is based on the experimentally determined structures of the native unbound SCP-B (Fig. 3b) and the bound tripeptide product complexed to SCP-B (Fig. 4b). This mechanism should be applicable to all the members of the eqolisin family. The mechanism proposed in Fig. 6a has similarities to those of the aspartic and metallopeptidases in that it has a general baseactivated water molecule as the nucleophile, but it is unique in many its features.
Fig. 6.
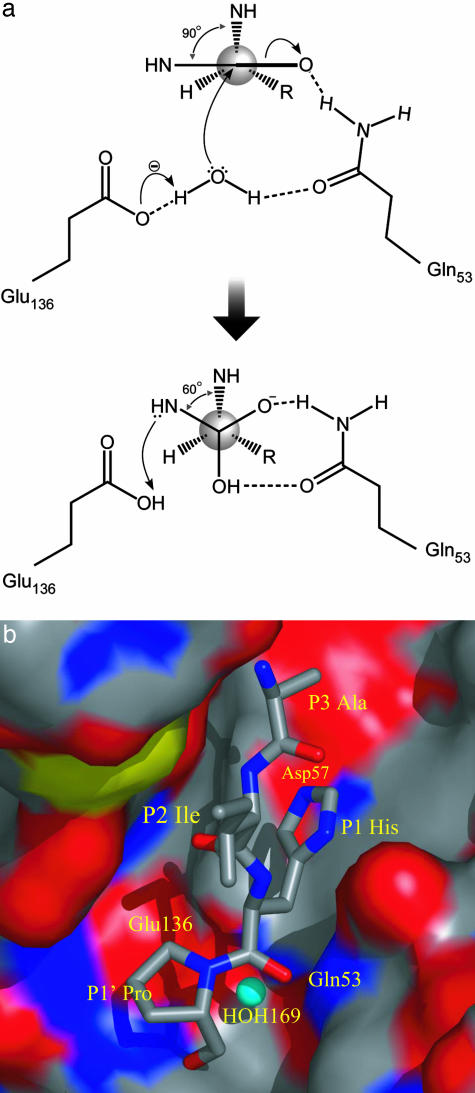
(a) The proposed catalytic mechanism of SCP-B. The water molecule hydrogen-bonded to both Glu-136 and Gln-53 is the nucleophile. The general base is the carboxylate of Glu-136. The side-chain amide of Gln-53 assists in the nucleophilic attack and stabilizes the tetrahedral intermediate by hydrogen bonding. (b) A model of a substrate Ala-Ile-His-Pro bound in a productive mode in the active site of SCP-B. The nucleophilic attack by the OH- ion (blue sphere) is on the si-face of the scissile peptide. The surface of SCP-B is represented and colored according to the underlying atoms (slate, carbon; blue, nitrogen; red, oxygen).
In our mechanistic proposal for the eqolisins, the water (HOH169) bound between Glu-136 and Gln-53 (Fig. 3b) acts as the nucleophile that is activated to a hydroxide ion by the general base functionality of the carboxylate of Glu-136 (Fig. 6a). Electrophilic assistance by polarizing the carbonyl bond of the scissile peptide is provided by the side-chain amide of Gln-53. The hydrogen-bonded interaction from Gln-53 Nε2 to the substrate carbonyl oxygen is bifunctional; it assists in the formation of the tetrahedral intermediate, and it provides stabilization of the oxyanion that results in that intermediate.
We propose that a productive enzyme–substrate complex would have P1 conformational angles of φ ≈ -112° and Ψ ≈ 90° as indicated in Fig. 6. As the nucleophilic attack takes place (the formation of the covalent bond from the OH- ion to the carbonylcarbon atom of the scissile peptide), the carbonyl-carbon atom becomes tetrahedral in character and the Ψ angle decreases to ≈60°, thus adopting a fully staggered conformation for the tetrahedral intermediate (Fig. 6a). Gln-53 provides for stabilization of the tetrahedral intermediate by donating a hydrogen bond from Gln-53 Nε2 to the oxyanion of the tetrahedral intermediate and by receiving a hydrogen bond (to Gln-53 Oε1) from the OH of the attacking water molecule. The nucleophilic attack takes place on the si-face of the scissile peptide. This direction of attack is seen only in the papain-like cysteine peptidase family; it is the re-face attack that takes place in the serine, aspartic, and metallopeptidases.
Protonation of the leaving-group nitrogen is an obligatory step in any hydrolytic mechanism for an amide. In our mechanism, it is likely that the proton donor will be the protonated Glu-136 (Fig. 6a) by analogy with the aspartic and metallopeptidases. The tetrahedral character of the nitrogen atom in the intermediate will place the lone-pair electrons on the nitrogen in an appropriate position to abstract the proton from the carboxyl group of Glu-136. The distance from the proton-donor oxygen atom of the Glu-136 carboxyl group to the leaving-group nitrogen atom of the tetrahedral intermediate is ≈3.2 Å (estimated from the equivalent distance in the metalloenzymes).
Conclusions
Several reasons exist to place the eqolisins into a new family of peptidases. First, they have a unique fold not previously observed for proteolytic enzymes. No other known peptidase has a β-sandwich as a tertiary fold. Second, they have a unique catalytic dyad of a glutamate and a glutamine to activate the nucleophilic water and to stabilize the tetrahedral intermediate on the hydrolytic pathway, respectively. Third, the nucleophilic attack on the carbonyl-carbon atom is from the si-face of the scissile peptide. This feature is in common with the papain-like peptidases but distinct from the serine, aspartic, and metalloproteinases. Last, the conformational angle Ψ of the P1 residue of ecolisin substrates is unique among peptidase families. In the eqolisins, substrates approach the catalytic residues with the P1 residue having φ,Ψ ≈ -115°, +90°. The serine and aspartic peptidases bind substrates with the main-chain conformational angles of the P1 residues of φ,Ψ ≈ -120°, +35°, respectively; the Ψ angle for the P1 residues of substrates of the metallopeptidases is ≈150° [estimated from the coordinates, 5TMN (37)]. The cysteine peptidases of the papain family have φ,Ψ angles of ≈ -120°, -30° for their substrate P1 residues. Nature has used common features of proteolysis but has mixed these with unusual and different folds and different modes of substrate binding. The chemistry, however, that of hydrolysis of a peptide bond, is common to all peptidolytic enzymes.
Acknowledgments
We thank Ken Ng and Ernst Bergmann for collecting the native data set of SCP-B on the synchrotron at SSRL; M.N.G.J. thanks Daniel Bur for some important discussions at a recent conference ICAPI-03 in Japan; and we thank Jason Maynes and Perry d'Obrenan for help with the figures. This research was supported by grants from the Canadian Institutes of Health Research to the Canadian Institutes of Health Research Group in Protein Structure and Function, the Alberta Synchrotron Institute, and the Alberta Heritage Foundation for Medical Research.
Abbreviations: SCP-B, scytalidocarboxyl peptidase B; AnCPA, Aspergillus niger carboxyl peptidase A.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1S2B and 1S2K).
References
- 1.Murao, S., Oda, K. & Matsushita, Y. (1973) Agric. Biol. Chem. 37, 1417-1421. [Google Scholar]
- 2.Oda, K. & Murao, S. (1974) Agric. Biol. Chem. 38, 2435-2444. [Google Scholar]
- 3.Oda, K. & Murao, S. (1976) Agric. Biol. Chem. 40, 1221-1225. [Google Scholar]
- 4.Majima, E., Murao, S., Oda, K. & Ichishima, E. (1988) Agric. Biol. Chem. 52, 787-793. [Google Scholar]
- 5.Murao, S. & Oda, K. (1985) in Aspartic Proteinases and their Inhibitors, ed. Kostka, V. (Walter de Gruyter, Berlin), pp. 379-399.
- 6.Oda, K. & Murao, S. (1991) in Structure and Function of the Aspartic Proteinases: Genetics, Structures, and Mechanisms, ed. Dunn, B. M. (Plenum, New York), pp. 185-201.
- 7.Oda, K., Takahashi, S., Shin, T. & Murao, S. (1995) in Aspartic Proteinases: Structure, Function, Biology, and Biochemical Implications, ed. Takahashi, K. (Plenum, New York), pp. 529-542.
- 8.Oda, K., Takahashi, S., Ito, M. & Dunn, B. M. (1998) in Aspartic Proteinases: Retroviral and Cellular Enzymes, ed. James, M. N. G. (Plenum, New York), pp. 349-353.
- 9.Wlodawer, A., Li, M., Dauter, Z., Gustchina, A., Uchida, K., Oyama, H., Dunn, B. M. & Oda, K. (2001) Nat. Struct. Biol. 8, 442-446. [DOI] [PubMed] [Google Scholar]
- 10.Comellas-Bigler, M., Fuentes-Prior, P., Maskos, K., Huber, R., Oyama, H., Uchida K., Dunn, B. M., Oda, K. & Bode, W. (2002) Structure (London) 10, 865-876. [DOI] [PubMed] [Google Scholar]
- 11.Oda, N., Gotoh, Y., Oyama, H., Murao, S., Oda, K. & Tsuru, D. (1998) Biosci. Biotechnol. Biochem. 62, 1637-1639. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, H., Kimura, T., Makabe, O. & Takahashi, K. (1991) J. Biol. Chem. 266, 19484-19489. [PubMed] [Google Scholar]
- 13.Jara, P., Gilbert, S., Delmas, P., Guillemot, J. C., Jaghad, M., Ferrara, P. & Loison, G. (1996) Mol. Gen. Genet. 250, 97-105. [DOI] [PubMed] [Google Scholar]
- 14.Poussereau, N., Creton, S., Billon-Grant, G., Rascle, C. & Fevre, M. (2001) Microbiology 147, 717-726. [DOI] [PubMed] [Google Scholar]
- 15.Oda, K. (1998) in Handbook of Proteolytic Enzymes, eds. Barrett, A. J., Rawlings, N. D. & Woessner, J. F. (Academic, San Diego), pp. 969-970.
- 16.Tsuru, D., Shimada, S., Maruta, S., Yoshimoto, T., Oda, K., Murao, S., Miyata, T. & Iwanaga, S. (1986) J. Biochem. (Tokyo) 99, 1537-1539. [DOI] [PubMed] [Google Scholar]
- 17.Tsuru, D., Kobayashi, R., Nakagawa, N. & Yoshimoto, T. (1989) Agric. Biol. Chem. 53, 1305-1312. [Google Scholar]
- 18.Kakimori, T., Yoshimoto, T., Oyama, H., Oda, N., Gotoh, Y., Oda, K., Murao, S. & Tsuru, D. (1996) Biosci. Biotechnol. Biochem. 60, 1210-1211. [DOI] [PubMed] [Google Scholar]
- 19.Huang, X. P., Kagami, N., Inoue, H., Kojima, M., Kimura, T., Makabe, O., Suzuki, K. & Takahashi, K. (2000) J. Biol. Chem. 275, 26607-26614. [DOI] [PubMed] [Google Scholar]
- 20.Rawling, N. D. (1998) in Handbook of Proteolytic Enzymes, eds. Barrett, A. J., Rawlings, N. D. & Woessner, J. F. (Academic, San Diego), pp. 963-967.
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger, T. C. (1999) Acta Crystallogr. D 55, 1863-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458-463. [DOI] [PubMed] [Google Scholar]
- 26.McRee, D. E. (1999) J. Struc. Biol. 125, 156-165. [DOI] [PubMed] [Google Scholar]
- 27.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 28.James, M. N. G. & Sielecki, A. R. (1986) Nature 319, 33-38. [DOI] [PubMed] [Google Scholar]
- 29.Sawyer, L. & James, M. N. G. (1982) Nature 295, 79-80. [DOI] [PubMed] [Google Scholar]
- 30.James, M. N. G., Sielecki, A. R., Hayakawa, K. & Gelb, M. H. (1992) Biochemistry 31, 3872-3886. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577-2637. [DOI] [PubMed] [Google Scholar]
- 32.Delano, W. (2002) The pymol Molecular Graphics System (Delano Scientific, San Carlos, CA).
- 33.Read, R. J. (1986) Acta Crystallogr. A 42, 140-149. [Google Scholar]
- 34.Laskowski, M., Jr., & Qasim M. A. (2000) Biochim. Biophys. Acta 1477, 324-327. [DOI] [PubMed] [Google Scholar]
- 35.Bode, W. & Huber, R. (1992) Eur. J. Biochem. 204, 433-451. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi, K., Kagami, N., Huang, X.-P., Kojima, M. & Inoue, H. (1998) in Aspartic Proteinases: Retroviral and Cellular Enzymes, ed. James, M. N. G. (Plenum, New York), pp. 275-282.
- 37.Holden, H. M., Tronrud, D. E., Monzingo, A. F., Weaver, L. H. & Matthews, B. W. (1987) Biochemistry 26, 8542-8553. [DOI] [PubMed] [Google Scholar]