Abstract
To investigate the significance of antibiotics for the producing organism(s) in the natural habitat, we screened a specimen of the fungicolous fungus Trichoderma phellinicola (syn. Hypocrea phellinicola) growing on its natural host Phellinus ferruginosus. Results revealed that a particular group of non-ribosomal antibiotic polypeptides, peptaibiotics, which contain the non-proteinogenic marker amino acid, α-aminoisobutyric acid, was biosynthesized in the natural habitat by the fungicolous producer and, consequently, released into the host. By means of liquid chromatography coupled to electrospray high-resolution time-of-flight mass spectrometry, we detected ten 20-residue peptaibols in the specimen. Sequences of peptaibiotics found in vivo were independently confirmed by analyzing the peptaibiome of an agar plate culture of T. phellinicola CBS 119283 (ex-type) grown under laboratory conditions. Notably, this strain could be identified as a potent producer of 39 new 17-, 18-, and 19-residue peptaibiotics, which display the same building scheme as the 20-residue peptaibols found in the specimen. Two of the 19-residue peptaibols are tentatively assigned to carry tyrosinol, a novel C-terminal residue, as deduced from high-resolution tandem mass-spectrometry data. For the new peptaibiotics produced by T. phellinicola, the name ‘hypophellin(s)’, based on the teleomorph name, is introduced.
1. Introduction
1.1. Fungi as a Prolific Source of Bioactive Natural Products
The current estimate of the total number of fungal species ranges between 1.0 and 1.5 million [1], whereas the number of those validly described should now exceed only 98,000 [2]. Of the 33,500 bioactive microbial metabolites known to date, the fungal kingdom contributes ca. 15,600. Approximately 10,000 of them were shown to display anti-infective, antitumor, and/or antiviral activities. Microbial-derived drugs on the market comprise ca. 400–500 active pharmaceutical agents [3], including therapeutically relevant antibiotics of fungal origin such as β-lactams, fusidic acid, and griseofulvin, as well as the two immunosuppressants mycophenolic acid and cyclosporin A [4].
Given that less than 1% of microorganisms visible under the microscope have been cultivated under laboratory conditions so far, microbial diversity provides an enormous, yet underestimated potential for future drug discovery [5] and in the search for new agricultural antibiotics [6].
1.2. The Potential of Trichoderma Species as Biological Control Agents (BCAs)
Species of the ubiquitous fungal genus Trichoderma and its Hypocrea teleomorphs have attracted considerable interest in the past two decades because of the pivotal role of their secondary metabolites in the antagonistic activities of biocontrol species [7–9]. Most of them occur as opportunistic, plant (endo)symbionts [10], some of which exhibit pronounced antimicrobial activity towards economically important plant pathogens. Recent examples include:
the hyperparasite Trichoderma stromaticum (syn. Hypocrea stromatica), the active agent of ‘Tricovab’ a commercial formulation against Crinipellis (syn. Moniliophthora) perniciosa, the Witches’ broom pathogen of cocoa (Theobroma cacao) [11] [12];
T paucisporum and T theobromicola, displaying in vitro-activities against frosty pod rot of cocoa, Moniliophthora roreri [13];
T martiale, which, in small-scale in situ field trials, proved highly effective against black pod rot of cocoa caused by Phytophthora palmivora [14].
The mode of action of phytoprotective Trichoderma species is considered rather complex. Depending on the species or even strains investigated, the following mechanisms may contribute to the antagonistic potential towards plant pathogenic fungi:
i) Competition for nutrients and/or space, ii) growth promotion of plants, especially colonization of roots, resulting in improved root and plant growth, iii) induction of localized and systemic resistance responses in plants, iv) mycoparasitism, v) increase of uptake and concentration of nutrients by the plant, including the production of siderophores, and vi) production of volatile and non-volatile antibiotics [10].
1.3. Peptaibiotics – Non-Ribosomally Biosynthesized Fungal Peptide Antibiotics Containing α,α-Dialkyl-α-amino Acids
During the past two decades, peptaibiotics have regained particular interest because of their unique bioactivities, resulting from their amphipathicity and helical conformations [15]. These are attributed to the presence of high proportions of peptide-bound α-aminoisobutyric acid (Aib), frequently accompanied by d- and/or l-isovaline (Iva) [16], and, in a few sequences, l-α-ethylnorvaline (EtNva), or 1-aminocyclopropane-1-carboxylic acid (Acc) [17]. The presence of these α,α-dialkyl-α-amino acids (Fig. 1,a) has been confirmed in acidic hydrolysates of more than 30 genera of fungi [18].
Fig. 1.
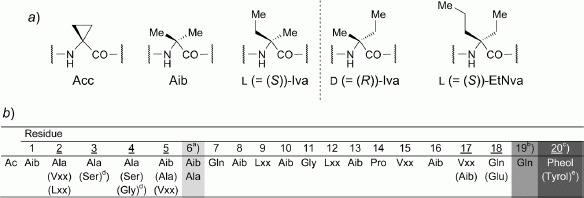
a) Structures and configurations of a,a-dialkylamino acids found in peptaibiotics. b) Building scheme of subfamily-1 (SF1) peptaibiotics, produced by Hypocrea phellinicola. Variable positions are underlined. Minor sequence variations are parenthesized. Deletions of certain amino acid positions are highlighted in different shades: C-terminal deletions are highlighted in dark, deletions of Gln in medium, and deletions of [Aib/Ala]6 in light gray. a) Deleted in 17-, 18-, and 19-residues hypophellins. b) Deleted in the 17-residue sequence 29. c) Deleted in 18-residue sequences 11, 12, and 28, and in the 17-residue sequence 29. d) Detected with DTU maXis gradient only. e) Detected with JLU micrOTOF-Q II gradient only.
Peptaibiotics are defined as non-ribosomally biosynthesised, linear or cyclic polypeptide antibiotics of exclusively fungal origin which i) have a molecular weight between 500 and 2,200 Da, thus containing 4–21 residues; ii) show a high content of the marker Aib, as well as further α,α-dialkylamino acids; iii) are characterized by the presence of other non-proteinogenic amino acids and/or lipoamino acids; iv) possess an acylated N-terminus, and v) in the case of linear peptides, have a C-terminal residue that, in most of them, consists of a free or O-acetylated, amide-bonded β-amino alcohol. The C-terminus might also be an amine, amide, sugar alcohol, 2,5-diketopiperazine, a heterocyclic residue, or an amino acid with free carboxy terminus. The majority of Aib-containing peptides carry a C-terminal residue representing a β-amino alcohol. Only this group is referred to as peptaibols sensu stricto, whereas for the others the comprehensive name peptaibiotics is used [17].
1.4. Detection of Peptaibiotics in T. phellinicola Growing on Its Natural Host
The genus Trichoderma, which currently consists of ca. 200 validly described species the number of which increases continually [19–28], is generally recognized as the most prolific source of peptaibiotics [17]. However, reports on the detection of peptaibiotics in samples collected in the natural habitat of the producer(s) are rare. Most of the ca. 1,000 individual sequences of peptaibiotics known to date have been sequenced in extracts of fungal cultures grown under artificial laboratory conditions.
The first example of peptaibiotics isolated from natural specimens were hypelcins A and B obtained from ca. 2 kg of dried, crushed stromata of Hypocrea peltata [29–31]. In 1997 and 1999, three reports were published on the isolation of peptaibiotics from fruiting bodies of Scleroderma texense, Tylopilus neofelleus, and Boletus sp., respectively; all being members of the Boletales [32–34]. However, in 2002, Kiet et al. [35] isolated chrysospermins A–D from the Vietnamese species Xerocomus langbianensis (Boletaceae, Boletales) and attributed the detection of these four 19-residue peptaibols [36] to an unrecognized infection of X. langbianensis with Sepedonium sp. This phenomenon was later commented on by Degenkolb et al. [37] [38]. Finally, Neuhof et al. [39] corroborated the assumption of Kiet et al. [35] by analyzing four fruiting bodies of members of the order Boletales infected by Sepedonium chrysospermum and S. microspermum, respectively. Notably, all samples were screened positive for peptaibiotics of the chrysospermin type. In 2006, Lehr et al. [40] demonstrated that 16-residue peptaibols, the antiamoebins, were solely responsible for antibiosis in herbivore dung naturally colonized by or artificially inoculated with Stilbella fimetaria (syn. S. erythrocephala).
1.5. Bioactivities of Peptaibiotics from Trichoderma
Peptaibiotics are thus assumed to play a key role in the infection process of a host by a fungicolous species because of their unique ability of forming voltage-gated ion channels. This phenomenon is best described by the dipole flip-flop gating model in planar lipid bilayers [41]. Their well-documented membrane activity, however, may also account for other striking bioactivities, such as neurolepsy [42], inhibition of amyloid β-peptide formation [43], inhibition of HIV-1 integrase [44], suppression of tumor cells, targeted calcium-mediated apoptosis, and autophagy in human hepatocellular carcinoma cells [45], as well as induction of defence responses and systemic resistance in tobacco against tobacco mosaic virus [46] and programmed cell death in fungal plant pathogens [47].
1.6. Choice of the Model Organism
Trichoderma phellinicola, a recently described polyporicolous species, which specifically occurs on effused basidiomes of Phellinus spp., was chosen as a model organism. Specimens of H. phellinicola have so far been recorded from Austria, Denmark, Germany [20], and the Czech Republic (see Exper. Part). This species is possibly specific for Phellinus ferruginosus [20].
To confirm the above hypothesis of peptaibiotic production under in vivo conditions, a specimen of Trichoderma phellinicola growing on its host Phellinus ferruginosus, was screened for peptaibiotics. For comparison, the ex-type culture of T. phellinicola, CBS 119283 (= C.P.K. 2137), was investigated. Both morphs were analyzed using a peptaibiomics approach as described in [48–50].
2. Results
2.1. General Considerations
All 17-, 18-, 19-, and 20-residue sequences discussed below were obtained from Trichoderma phellinicola [20]. The name ‘hypophellins’ (HPHs), which covers the entirety of long-chain peptaibiotics (>17 residues) produced by this species, is proposed. We base this name on the teleomorph name Hypocrea phellinicola, which used to be the valid name of the holomorph in dual nomenclature [20]. The introduction of a new name for peptaibiotics from a phylogenetically well-defined species is more favorable than earlier names for many of the 19- and 20-residue peptaibiotics mentioned below, viz. suzukacillins, trichocel-lins, trichokonins, and longibrachins, which were produced by phylogenetically undefined Trichoderma species with thus highly questionable names. The latter issue is further complicated by the fact that many of the peptaibiotic-producing Trichoderma strains reported in the literature have never been deposited in a public culture collection, or deposition was terminated [51].
Hypophellins are numbered consecutively with Arabic numbers as follows: i) sequences produced by the specimen; ii) sequences produced by the culture CBS 119283 grown and analyzed at JLU; iii) sequences produced by the culture CBS 119283 grown and analysed at DTU.
2.2. Peptaibiotic Pattern of the Teleomorph
Notably, the teleomorph of Trichoderma phellinicola proved to be a prolific source of ten 20-residue peptaibols, compounds 1-10, displaying the characteristic building scheme of subfamily 1 (SF1), one of the nine ‘peptaibol subfamilies’ (Fig. 1,b, and Tables 1 and 2), as introduced by Chugh and Wallace [52]4).
Table 1.
Sequences of 20-Residue Peptaibiotics Detected in the Specimen of Hypocrea phellinicola (micrOTOF-Q II screening)
| No. | tR [min] | [M+H]+ | Residue | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2a) | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||
| 1 | 37.8–38.1 | 1937.1209 | Ac | Aib | Ala | Aib | Ala | Aib | Ala | Gln | Aib | Vxx | Aib | Gly |
| 2 | 37.8–38.1 | 1938.1068 | Ac | Aib | Ala | Aib | Ala | Aib | Ala | Gln | Aib | Vxx | Aib | Gly |
| 3 | 39.1–39.3 | 1951.1358 | Ac | Aib | Ala | Aib | Ala | Aib | Ala | Gln | Aib | Vxx | Aib | Gly |
| 4 | 39.8–40.0 | 1952.1192 | Ac | Aib | Ala | Aib | Ala | Aib | Ala | Gln | Aib | Vxx | Aib | Gly |
| 5 | 40.2–40.4 | 1951.1416 | Ac | Aib | Ala | Aib | Ala | Aib | Aib | Gln | Aib | Vxx | Aib | Gly |
| 6 | 41.0–41.2 | 1952.1258 | Ac | Aib | Ala | Aib | Ala | Aib | Aib | Gln | Aib | Vxx | Aib | Gly |
| 7 | 41.3–41.7 | 1965.1615 | Ac | Aib | Ala | Aib | Ala | Aib | Aib | Gln | Aib | Vxx | Aib | Gly |
| 8 | 42.3–42.5 | 1966.1354 | Ac | Aib | Ala | Aib | Ala | Aib | Aib | Gln | Aib | Vxx | Aib | Gly |
| 9 | 43.0–43.2 | 1979.1718 | Ac | Aib | Aib | Aib | Ala | Aib | Aib | Gln | Aib | Vxx | Aib | Gly |
| 10 | 44.0–44.3 | 1980.1636 | Ac | Aib | Aib | Aib | Ala | Aib | Aib | Gln | Aib | Vxx | Aib | Gly |
| Compound identical or positionally isomeric with | Ref. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | Longibrachin A I | [53] |
| Trichoaureocin 3 | [54] | |||||||||
| Trichokonin VI (= gliodeliquescin A) | [55] [56] | |||||||||
| Trichobrachins II-5, II-6 | [57] | |||||||||
| Trichobrachin IIb A | [58] [59] | |||||||||
| Lxx | Aib | Pro | Vxx | Aib | Aib | Glu | Gln | Pheol | Longibrachin B II | [53] |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | Trichokonin VII | [55] |
| Trichoaureocin 4 | [54] | |||||||||
| Suzukacillin A-10a | [60] | |||||||||
| Trichobrachins II-7, II-8, II-9 | [57] | |||||||||
| Trichobrachin IIb B | [58] [59] | |||||||||
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Glu | Gln | Pheol | Longibrachin B III | [53] |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | Trichokonin VIII (= trichosporin B-IVc) | [55] [61] |
| Trichoaureocin V | [54] | |||||||||
| Trichobrachin IIb C | [58] [59] | |||||||||
| Lxx | Aib | Pro | Vxx | Aib | Aib | Glu | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | Longibrachin A IV | [53] |
| Trichoaureocin VI | [54] | |||||||||
| Trichobrachin IIb D | [58] [59] | |||||||||
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Glu | Gln | Pheol | New (longibrachin IV: [Gln]18→[Glu]18) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (homolog of 7) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Glu | Gln | Pheol | New (homolog of 8) | |
Table 2.
Diagnostic Fragment Ions of 20-Residue Peptaibiotics Detected in the Specimen of Hypocrea phellinicola (micrOTOF-Q II screening)
| Diagnostic fragment ions | Peaks [m/z]a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| tR [min] | 37.8–38.1 | 38.6–38.7 | 39.1–39.3 | 39.8–40.0 | 40.2–40.4 | 41.0–41.2 | 41.3–41.7 | 42.3–42.5 | 43.0–43.2 | 44.0–44.3 |
| [M + Na]+ | 1959.1047 | 1960.0872 | 1962.1376 | n.d. | 1973.1212 | 1974.1064 | 1987.1372 | 1988.1245 | 2001.1535 | 2002.1445 |
| [M + H]+ | 1937.1209 | 1938.1036 | 1951.1358 | 1952.1192 | 1951.1416 | 1952.1258 | 1965.1615 | 1966.1354 | 1979.1718 | 1980.1636 |
| a1 | 100.0808 | 100.0808 | 100.0808 | 100.0806 | 100.0809 | 100.0805 | 100.0808 | 100.0807 | n.d. | n.d. |
| a2 | 171.1181 | 171.1181 | 171.1197 | 171.1195 | 171.1188 | 171.1185 | 171.1191 | 171.1200 | 185.1315 | 185.1311 |
| a3 | 256.1657 | 256.1657 | 256.1662 | 256.1663 | 256.1666 | 256.1665 | 256.1669 | 256.1671 | 270.1811 | 270.1808 |
| a4 | 327.2121 | 327.2121 | 327.2155 | 327.2142 | 327.1992 | 327.2097 | 327.2102 | 327.2116 | 341.2238 | 341.2180 |
| a5 | n.d. | n.d. | 412.2739 | 412.2739 | 412.2572 | 412.2720 | 412.2776 | 412.2766 | n.d. | n.d. |
| b1 | 128.0758 | 128.0758 | 128.0762 | 128.0757 | 128.0763 | 128.0758 | 128.0765 | 128.0760 | 128.0758 | 128.0748 |
| b2 | 199.1102 | 199.1102 | 199.1109 | 199.1107 | 199.1111 | 199.1111 | 199.1113 | 199.1115 | 213.1251 | 213.1251 |
| b3 | 284.1604 | 284.1604 | 284.1615 | 284.1614 | 284.1618 | 284.1615 | 284.1623 | 284.1625 | 298.1845 | 298.1802 |
| b4 | 355.1982 | 355.1982 | 355.1973 | 355.1972 | 355.1981 | 355.1976 | 355.1986 | 355.1988 | 369.2109 | 369.2144 |
| b5 | 440.2479 | 440.2479 | 440.2494 | 440.2492 | 440.2502 | 440.1497 | 440.2508 | 440.2510 | 454.2669 | 454.2699 |
| b6 | 511.2839 | 511.2839 | 511.2850 | 511.2852 | 525.3019 | 525.3015 | 525.3023 | 525.3026 | 539.3231 | 539.3231 |
| b7 | 639.3431 | 639.3431 | 639.3455 | 639.3451 | 653.3661 | 653.3626 | 653.3690 | 653.3681 | 667.3870 | 667.3881 |
| b8 | 724.3937 | 724.3937 | 724.3961 | 724.3957 | 738.4118 | 738.4109 | 738.4130 | 738.4132 | 752.4381 | 752.4367 |
| b9 | 823.4750 | 823.4590 | 823.4611 | 823.4601 | 837.4777 | 837.4772 | 837.4790 | 837.4795 | 851.5058 | 851.5067 |
| b10 | 908.5298 | 908.5095 | 908.5131 | 908.5129 | 922.5302 | 922.5290 | 922.5314 | 922.5317 | 936.5594 | 936.5602 |
| b11 | 965.5490 | 965.5311 | 965.5470 | 965.5316 | 979.5501 | 979.5476 | 979.5508 | 979.5506 | 993.5822 | 993.5819 |
| b12 | 1078.6366 | 1078.6151 | 1078.6340 | 1078.6138 | 1092.6478 | 1092.6289 | 1092.6325 | 1092.6326 | 1106.6642 | 1106.6710 |
| b13 | 1163.6824 | 1163.6642 | 1163.6810 | 1163.6662 | 1177.6994 | 1177.6816 | 1177.6853 | 1177.6859 | 1191.7215 | 1191.7196 |
| y7 | 774.4598 | 775.4614 | 788.4742 | 789.4595 | 774.4586 | 775.4436 | 788.4750 | 789.4596 | 788.4750 | 789.4596 |
| y7 – H2O | 756.4445 | 757.4491 | n.d. | 771.4507 | n.d. | 757.4308 | n.d. | 771.4468 | n.d. | 771.4468 |
| y7 – AA (20) | 623.3556 | 624.3581 | 637.3711 | 638.3573 | 623.3563 | 624.3414 | 637.3722 | 638.3576 | 637.3722 | 638.3576 |
| y7 – AA (20-19) | 495.2979 | 496.2999 | 509.3124 | 510.2961 | 495.2955 | 496.2793 | 509.3120 | 510.2962 | 509.3120 | 510.2962 |
| y7 – AA (20-18) | 367.2385 | 367.2350 | 381.2515 | 381.2517 | 367.2364 | 367.2358 | 381.2519 | 381.2514 | 381.2519 | 381.2514 |
| y7 – AA (20-17) | 282.1900 | 282.1831 | 282.1850 | 282.1820 | 282.1853 | 282.1838 | 282.1839 | 282.1839 | 282.1839 | 282.1839 |
n.d., Not detected.
One Gln residue is found in position 7, and another one towards or at the C-terminus in position 18, whereas position 19 is either occupied by a third Gln or a Glu residue. A highly conserved Pro residue is located in position 14 of the peptide chain. All sequences have a Gly residue in position 11 and terminate in Pheol. At least seven, at most nine, residues are occupied by Aib. Variable amino acid residues are located in positions 2, 6, 17, and 18 (Fig. 1,b).
Most of the peptaibols sequenced resemble previously described compounds (Fig. 1,b, Table 1, and Fig. 2,a) such as longibrachins A and B [53], trichobrachins II [57], trichoaureocins [54], trichokonins [55] [62] [63], and suzukacillins A [60].
Fig. 2.
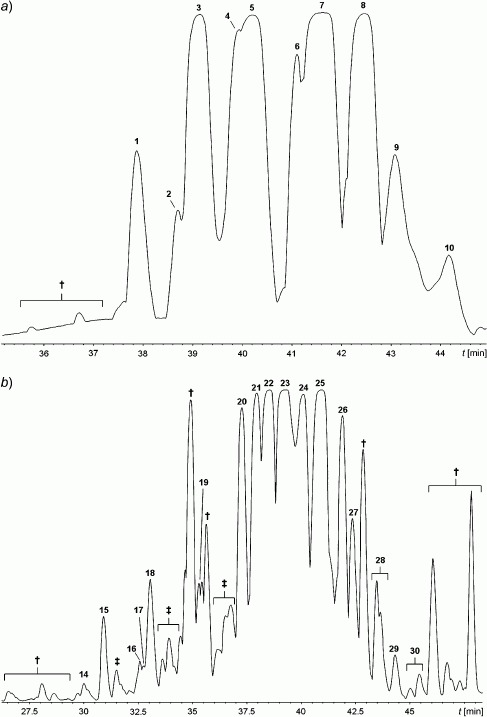
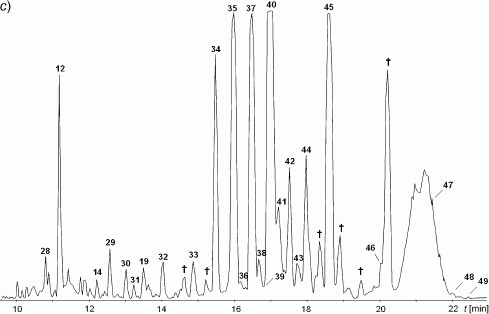
Base-peak chromatograms (BPCs) of a) the H. phellinicola specimen screened with the micrOTOF-Q II, b) the H. phellinicola ex-type plate culture screened with the micrOTOF-Q II, and c) the H. phellinicola specimen screened with the maXis. †, co-eluting peptaibiotics, not sequenced; ‡, non-peptaibiotic metabolite.
2.3. Peptaibiotic Pattern of the Culture
2.3.1. General Considerations
As observed before [20], ascospores of T phellinicola are unstable and die rapidly after collecting. This might have been the reason why no agar culture could be obtained from our specimen. As a substitute, the ex-type culture of T phellinicola CBS 119283 (= C.P.K. 2137) was provided, and its peptaibiotic pattern was analyzed. Except for the two lipopeptaibols 48 and 49, the remaining compounds 11–47 represent the characteristic building scheme of SF1, resembling the previously described 20-residue peptaibols suzukacillins A, trichosporins B, and trichocellins A [60] [61] [64–67].
2.3.2. micrOTOF-Q II Screening
In contrast to the specimen analyzed, the ex-type plate culture grown and analyzed at the Justus Liebig University of Giessen (JLU) produced two new 18- and fifteen new 19-residue peptaibols, compounds 11–27, which lacked the [Ala/Aib]6 residue of the 20-residue peptaibols found in the specimen (Tables 3 and 4, and Fig. 2,b). The two truncated 18-residue sequences, compounds 11 and 12, terminated in free Gln. Sequences 14 and 16–27 carry a C-terminal Pheol. For compounds 13 and 15, a C-terminal tyrosinol residue (abbreviated as ‘Tyrol’) was tentatively deduced from HR-ESI-MS/MS data (Tables 3 and 4, Fig. 3).
Table 3.
Sequences of 18- and 19-Residue Peptaibiotics Detected in the Ex-Type Culture (CBS 119283) of Hypocrea phellinicola (micrOTOF-Q II screening)
| No. | tR [min] | [M + H]+ | Residue | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||
| 11 | 30.9–31.1 | 1747.0135 | Ac | Aib | Ala | Aib | Ala | Ala | – | Gln | Aib | Lxx | Aib | Gly |
| 12 | 31.8–32.0 | 1761.0324 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 13 | 32.2–32.6 | 1896.0995 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 14 | 32.5–32.7 | 1910.1131 | Ac | Aib | Ala | Aib | Ser | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 15 | 32.8–33.1 | 1910.1140 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 16 | 35.1–35.3 | 1896.1035 | Ac | Aib | Ala | Ala | Ser | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 17 | 37.0–37.2 | 1866.0928 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 18 | 37.7–37.9 | 1880.1095 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 19 | 38.3–38.4 | 1880.1136 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 20 | 38.8–39.2 | 1894.1331 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 21 | 39.8–40.1 | 1895.1278 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 22 | 40.6–40.9 | 1908.1474 | Ac | Aib | Ala | Aib | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| 23 | 41.5–41.6 | 1909.1391 | Ac | Aib | Ala | Aib | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| 24 | 42.1–42.3 | 1922.1601 | Ac | Aib | [255] | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly | |
| 25 | 43.4–43.6 | 1936.1738 | Ac | Aib | [269] | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly | |
| 26 | 44.2–44.4 | 1936.1750 | Ac | Aib | Vxx | Aib | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| 27 | 45.0–45.6 | 1950.1894 | Ac | Aib | Lxx | Aib | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| Compound identical or positionally isomeric with | Ref. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | New (trichocellin A-VI – [Aib]5 – Pheol) | [67] | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | New (trichocellin A-VI – [Ala]6 – Pheol) | [67] | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Tyrol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Tyrol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New (trichosporin B IIIa – [Aib]6) | [64] [61] |
| New (trichobrachin IIb A – [Ala]6) | [58] [59] | |||||||||
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New (suzukacillin A-11a – [Ala]6) | [60] |
| New (trichosporin B-VIa – [Aib]6) | [61] | |||||||||
| New (trichosporin B-VIIa – [Aib]6) | [66] | |||||||||
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (trichosporin B-IVb – [Aib]6, trichosporin B-VIb – [Aib]6) | [61] |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (suzukacillin A-10b – [Ala]6) | [60] |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Glu | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Glu | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | – | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | – | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
Table 4.
Diagnostic Fragment Ions of 18- and 19-Residue Peptaibiotics Detected in the Ex-Type Culture (CBS 119283) of Hypocrea phellinicola (micrOTOF-Q II screening)
| Diagnostic fragment ions | Peaks [m/z]a) | |||||||
|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| tR [min] | 30.9–31.1 | 31.8–32.0 | 32.2–32.6 | 32.5–32.7 | 32.8–33.1 | 35.1–35.3 | 37.0–37.2 | 37.7–37.9 |
| [M + Na]+ | 1768.9850 | 1783.0115 | 1918.0846 | 1932.0976 | 1932.1017 | 1918.0877 | 1888.0616 | 1902.0891 |
| [M + H]+ | 1747.0135 | 1761.0324 | 1896.0995 | 1910.1131 | 1910.1140 | 1896.1035 | 1866.0928 | 1880.1095 |
| a1 | 100.0718 | n.d. | n.d. | n.d. | n.d. | 100.0720 | 100.0720 | n.d. |
| a3 | 256.1647 | 256.1624 | 242.1508 | 256.1641 | 256.1707 | 242.1511 | 242.1506 | 256.1675 |
| a4 | n.d. | n.d. | n.d. | n.d. | 327.1979 | n.d. | n.d. | 327.2046 |
| a5 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 398.2312 | n.d. |
| b1 | 128.0687 | 128.0658 | 128.0709 | 128.0708 | 128.0835 | 128.0715 | 128.0719 | 128.0721 |
| b2 | 199.1075 | 199.1076 | 199.1109 | 199.1110 | 199.1161 | 199.1118 | 199.1115 | 199.1081 |
| b3 | 284.1611 | 284.1617 | 270.1453 | 284.1622 | 284.1637 | 270.1434 | 270.1471 | 284.1634 |
| b4 | 355.1972 | 355.1977 | 341.1846 | 371.1863 | 355.2023 | 357.1765 | 341.1815 | 355.1988 |
| b4 – H2O | n.d. | n.d. | n.d. | 353.1758 | n.d. | 339.1676 | n.d. | n.d. |
| b5 | 426.2340 | 440.2546 | 426.2354 | 456.2441 | 440.2494 | 442.2277 | 426.2314 | 440.2546 |
| b6 | 554.2840 | 568.3175 | 554.2840 | 584.3226 | 568.3175 | 570.2870 | 554.2989 | 568.3023 |
| b7 | 639.3523 | 653.3691 | 639.3539 | 669.3625 | 653.3679 | 655.3443 | 639.3530 | 653.3685 |
| b8 | 752.4400 | 766.4531 | 752.4386 | 782.4408 | 766.4563 | 768.4296 | 752.4353 | 766.4519 |
| b9 | 837.4860 | 851.5024 | 837.4880 | 867.4961 | 851.5028 | 853.4825 | 837.4896 | 851.5066 |
| b10 | 894.5048 | 908.5271 | 894.5061 | 924.5223 | 908.5250 | 910.5022 | 894.5076 | 908.5242 |
| b11 | 1007.5856 | 1021.6063 | 1007.5967 | 1037.6039 | 1027.6073 | 1023.5862 | 1007.5917 | 1021.6085 |
| b12 | 1092.6441 | 1106.6573 | 1092.6442 | 1122.6523 | 1106.6575 | 1108.6413 | 1092.6474 | 1106.6629 |
| b12 – H2O | n.d. | n.d. | n.d. | n.d. | n.d. | 1090.6265 | 1074.6077 | 1088.6332 |
| y6 | 655.3841 | 655.3841 | – | – | – | – | – | – |
| y6 – AA (18) | 509.3130 | 509.3130 | – | – | – | – | – | – |
| y6 – AA (18-17) | 381.2540 | 381.2540 | – | – | – | – | – | – |
| y6 – AA (18-16) | 282.1709 | 282.1709 | – | – | – | – | – | – |
| y7 | – | – | 804.4624 | 788.4706 | 804.4669 | 788.4697 | 774.4592 | 774.4593 |
| y7 – H2O | – | – | 786.4472 | 770.4510 | n.d. | 770.4510 | 756.4383 | 756.4383 |
| y7 – AA (19) | – | – | 637.3680 | 637.3708 | 637.3725 | 637.3705 | 623.3566 | 623.3559 |
| y7 – AA (19-18) | – | – | 509.3068 | 509.3140 | 509.3085 | 509.3103 | 495.2961 | 495.2962 |
| y7 – AA (19-17) | – | – | 381.2489 | 381.2515 | 381.2545 | 381.2513 | 367.2370 | 367.2373 |
| y7 – AA (19-16) | – | – | n.d. | n.d. | 282.1814 | 282.1814 | 282.1815 | 282.1815 |
| 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 |
|---|---|---|---|---|---|---|---|---|
| 38.3–38.4 | 38.8–39.2 | 39.8–40.1 | 40.6–40.9 | 41.5–41.6 | 42.1–42.3 | 43.4–43.6 | 44.2–44.4 | 45.0–45.6 |
| 902.0921 | 1916.1081 | 1917.1085 | 1930.1235 | 1931.1236 | 1944.1425 | 1958.1599 | 1958.1548 | 1972.1635 |
| 1880.1136 | 1894.1331 | 1895.1278 | 1908.1474 | 1909.1391 | 1922.1601 | 1936.1738 | 1936.1750 | 1950.1894 |
| 100.0721 | 100.0721 | 100.0747 | 100.0722 | 100.0722 | 100.0722 | n.d. | n.d. | n.d. |
| 242.1514 | 256.1682 | 256.1682 | 256.1677 | 256.1649 | n.d. | n.d. | n.d. | n.d. |
| 313.1832 | 327.2048 | 327.2049 | 327.2042 | 327.2050 | n.d. | n.d. | n.d. | n.d. |
| n.d. | 412.2533 | 412.2564 | 426.2817 | n.d. | n.d. | n.d. | n.d. | n.d. |
| 128.0722 | 128.0724 | 128.0718 | 128.0720 | 128.0708 | 128.0712 | 128.0672 | 128.0701 | 128.0684 |
| 199.1121 | 199.1081 | 199.1118 | 199.1083 | 199.1141 | [255] | [269] | 227.1404 | 241.1564 |
| 270.1476 | 284.1608 | 284.1608 | 284.1641 | 284.1631 | 312.1955 | 326.2055 | ||
| 341.1814 | 355.1988 | 355.1973 | 355.1972 | 355.1972 | 383.2306 | 397.2427 | 383.2297 | 397.2477 |
| n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 426.2314 | 440.2531 | 440.2513 | 454.2685 | 454.2686 | 468.2836 | 482.3001 | 482.2988 | 496.3148 |
| 554.2989 | 568.3022 | 568.3119 | 582.3249 | 582.3249 | 596.3361 | 610.3531 | 610.3608 | 624.3766 |
| 639.3513 | 653.3673 | 653.3654 | 667.3841 | 667.3836 | 681.3976 | 695.4131 | 695.4110 | 709.4286 |
| 752.4346 | 766.4505 | 766.4489 | 780.4662 | 780.4659 | 794.4802 | 808.4949 | 808.4934 | 822.5109 |
| 837.4888 | 851.5044 | 851.5023 | 865.5205 | 865.5199 | 879.5335 | 893.5492 | 893.5457 | 907.5631 |
| 894.5075 | 908.5234 | 908.5216 | 922.5386 | 922.5395 | 936.5517 | 950.5713 | 950.5659 | 964.5813 |
| 1007.5920 | 1021.6065 | 1021.6039 | 1035.6228 | 1035.6231 | 1049.6347 | 1063.6526 | 1063.6516 | 1077.6661 |
| 1092.6463 | 1106.6606 | 1106.6578 | 1120.6786 | 1120.6785 | 1134.6898 | 1148.7069 | 1148.7051 | 1162.7188 |
| 1074.6284 | 1088.6331 | 1088.6424 | 1102.6441 | 1102.6440 | 1116.6595 | 1130.6997 | 1130.7031 | 1144.7051 |
| – | – | – | – | – | – | – | – | – |
| – | – | – | – | – | – | – | – | – |
| – | – | – | – | – | – | – | – | – |
| – | – | – | – | – | – | – | – | – |
| 788.4710 | 788.4710 | 789.4647 | 788.4718 | 789.4597 | 788.4710 | 788.4705 | 788.4678 | 788.4668 |
| 770.4509 | 770.4509 | 771.4475 | 770.4508 | 771.4390 | 770.4507 | 770.4507 | 770.1538 | 770.1538 |
| 637.3707 | 637.3707 | 638.3638 | 637.3705 | 638.3574 | 637.3721 | 637.3678 | 637.3649 | 637.3676 |
| 509.3096 | 509.3096 | 510.3014 | 509.3108 | 510.2964 | 509.3105 | 509.3113 | 509.3093 | 509.3082 |
| 381.2513 | 381.2513 | 381.2483 | 381.2524 | 381.2520 | 381.2505 | 381.2508 | 381.2506 | 381.2492 |
| n.d. | n.d. | 282.1837 | 282.1813 | 282.1813 | 282.1813 | 282.1920 | 282.1781 | 282.1917 |
n.d., Not detected.
Fig. 3.
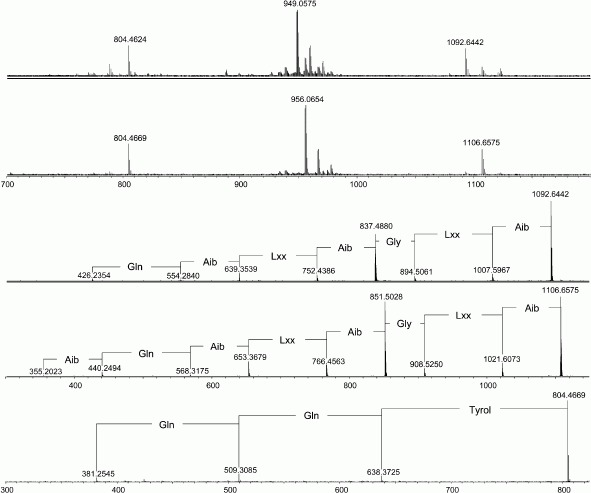
Sequencing of compounds 13 and 15 ocontaining a new C-terminal residue with a peak at m/z 804.46, tentatively assigned as tyrosinol (Tyrol)
2.3.3. maXis Screening
All SF1 peptaibiotics, compounds 12, 14, 19, 28–47, of the ex-type plate culture grown and analyzed at DTU (Tables 5 and 6, and Fig. 2,c) exhibit the characteristic deletion of the Ala/Aib residue in position 6. However, different positional isomers and homologues were found, e.g., the 17-residue deletion sequence 29, lacking the C-terminal dipeptide [Gln18–Pheol19]. In compound 31, a Ser-residue was found in position 3, whereas compound 30 exhibited a Gly residue in position 4. Overall, the structural diversity of peptaibiotics produced by the two cultures was much higher as compared to the specimen: variable amino acid residues were in positions 2,3, 4, 5, 6, 17, 18, and 20 (Fig. 1,b).
Table 5.
Sequences of 10-, 11-, 17-, 18-, and 19-Residue Peptaibiotics Detected in the Ex-Type Culture (CBS 119283) of Hypocrea phellinicola (maXis screening)
| No. | tR [min] | [M + H]+ | Residue | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||
| 28 | 10.8 | 1747.0131 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 12 | 11.2 | 1761.0273 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 14 | 12.2 | 1911.1213 | Ac | Aib | Ala | Aib | Ser | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 29 | 12.6 | 1632.9708 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 30 | 13.0 | 1880.1000 | Ac | Aib | Ala | Aib | Gly | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 31 | 13.2 | 1882.0784 | Ac | Aib | Ala | Ser | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 19 | 13.5 | 1880.1008 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 32 | 14.1 | 1896.0964 | Ac | Aib | Ala | Ser | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 33 | 14.9 | 1880.1035 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 34 | 15.5 | 1866.0863 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 35 | 15.9 | 1880.1012 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 36 | 16.2 | 1867.0706 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 37 | 16.4 | 1880.1007 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 38 | 16.7 | n.d. | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 39 | 16.8 | 1880.1009 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 40 | 17.0 | n.d. | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 41 | 17.2 | 1880.0997 | Ac | Aib | Ala | Ala | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 42 | 17.5 | 1894.1210 | Ac | Aib | Ala | Aib | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| 43 | 17.7 | 1895.1007 | Ac | Aib | Ala | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly |
| 44 | 18.0 | 1894.1177 | Ac | Aib | Ala | Ala | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| 45 | 18.6 | 1908.1341 | Ac | Aib | Ala | Aib | Ala | Vxx | – | Gln | Aib | Lxx | Aib | Gly |
| 46 | 20.0 | 1922.1467 | [227]a) | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly | ||
| 47 | 21.5 | 1936.1660 | [241]b) | Aib | Ala | Aib | – | Gln | Aib | Lxx | Aib | Gly | ||
| 48 | 22.0 | 1009.7031 | Occ) | Aib | Gly | Lxx | Aib | – | Gly | Lxx | Aib | Gly | Lxx | Lxxol |
| 49 | 22.1–22.2 | 1066.7242 | Oc | Aib | Gly | Lxx | Aib | Gly | Gly | Lxx | Aib | Gly | Lxx | Lxxol |
| Compound identical or positionally isomeric with | Ref. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | New (trichocellin A-V–[Ala]6 – Pheol) | [67] | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | New (trichocellin A-VI – [Ala]6 – Pheol) | [67] | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | New (12 – [Gln]18) | |||
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (17: [Ala]4→[Gly]4) | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (trichosporin B-IVb – [Aib]6, trichosporin B-VIb – [Aib]6) | [61] |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (positional isomer of 19, 37, and 41) | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New (positional isomer of 17) | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New (trichosporin B-VIa – [Aib]6, trichosporin B-VIIb – [Aib]6) | [61] [66] |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Glu | Gln | Pheol | New (35: [Gln]17→[Glu]17) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (positional isomer of 19, 33, and 41) | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Glu | Gln | Pheol | New (39: [Gln]17→[Glu]17) | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New (positional isomer of 35) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (trichosporin B-VIIa – [Aib]6) | [66] |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (positional isomer of 19, 33, and 37) | |
| Lxx | Aib | Pro | Vxx | Aib | Aib | Gln | Gln | Pheol | New | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Glu | Gln | Pheol | New (positional isomer of 40) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (positional isomer of 45) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | New (positional isomer of 44) | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | – | |
| Lxx | Aib | Pro | Vxx | Aib | Vxx | Gln | Gln | Pheol | – | |
| New | ||||||||||
| Trichogin A IV | [68] | |||||||||
| Sequence 13 or 14 from Trichoderma cf. strigosum CBS 119777 | [49] | |||||||||
| Partial sequence 4 from Hypocrea citrina CBS 853.70 | [48] | |||||||||
| Partial sequence 4 from Hypocrea vinosa CBS 247.63 | [48] | |||||||||
The N-terminal sequence of compound 46, which is represented by a mass difference of 227 Da, could not be assigned.
The N-terminal sequence of compound 47, which is represented by a mass difference of 241 Da, could not be assigned.
Oc, Tentatively assigned as n-octanoyl residue.
Table 6.
Diagnostic Fragment Ions of 10-, 11-, 17-, 18-, and 19-Residue Peptaibiotics Detected in the Ex-Type Culture (CBS 119283) of Hypocrea phellinicola (maXis screening)
| Diagnostic fragment ions | Peaks [m/z]a) | |||||
|---|---|---|---|---|---|---|
| 28 | 12 | 14 | 28 | 30 | 31 | |
| tR [min] | 10.8 | 11.2 | 12.2 | 12.6 | 13.0 | 13.2 |
| [M + H]+ | 1747.0131 | 1761.0273 | 1911.1213 | 1632.9708 | 1880.1000 | 1882.0784 |
| b1 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| b2 | n.d. | 199.1093 | n.d. | 199.1087 | 199.1087 | 199.1123 |
| b3 | 284.1601 | 284.1607 | 284.1613 | 284.1609 | 284.1604 | 286.1389 |
| b4 | 355.1989 | 355.1980 | 371.1938 | 355.1975 | 341.1819 | 357.1760 |
| b4 – H2O | n.d. | n.d. | 438.2353 | n.d. | 412.2541 | 424.2167 |
| b5 | 440.2512 | 440.2509 | 456.2470 | 440.2506 | 426.2347 | 442.2296 |
| b6 | 568.3097 | 568.3098 | 584.3039 | 568.3096 | 554.2926 | 570.2869 |
| b7 | 653.3615 | 653.3626 | 669.3571 | 653.3619 | 639.3458 | 655.3404 |
| b8 | 766.4456 | 766.4471 | 782.4415 | 766.4461 | 752.4294 | 768.4257 |
| b9 | 851.5003 | 851.4996 | 867.4953 | 851.4987 | 837.4826 | 853.4789 |
| b10 | 908.5192 | 908.5208 | 924.5190 | 908.5199 | 894.5026 | 910.4971 |
| b11 | 1021.6077 | 1021.6046 | 1038.5981 | 1021.6053 | 1007.5901 | 1023.5860 |
| b12 | 1106.6578 | 1106.6578 | 1122.6537 | 1106.6590 | 1092.6412 | 1108.6356 |
| b12 – H2O | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| y5 | – | – | – | 527.3191 | – | – |
| y5 – AA (17) | – | – | – | 381.2497 | – | – |
| y5 – AA (17-16) | – | – | – | 282.1814 | – | – |
| y5 – AA (17-15) | – | – | – | 197.1287 | – | – |
| y6 | 641.3626 | 655.3768 | – | – | – | – |
| y6 – AA (18) | 495.2923 | 509.3095 | – | – | – | – |
| y6 – AA (18-17) | 367.2353 | 381.2500 | – | – | – | – |
| y6 – AA (18-16) | 282.1812 | 282.1816 | – | – | – | – |
| y6 – AA (18-15) | 197.1274 | 197.1288 | – | – | – | – |
| y7 | – | – | 788.4676 | – | 788.4676 | 774.4501 |
| y7 – H2O | – | – | 637.3673 | – | 637.3673 | 623.3515 |
| y7 – AA (19) | – | – | 509.3117 | – | 509.3117 | 495.2926 |
| y7 – AA (19-18) | – | – | 381.2509 | – | 381.2509 | 367.2344 |
| y7 – AA (19-17) | – | – | 282.1814 | – | 282.1814 | 282.1813 |
| y7 – AA (19-16) | – | – | 197.1284 | – | 197.1284 | 197.1270 |
| 19 | 32 | 33 | 34 | 35 | 36 |
|---|---|---|---|---|---|
| 13.5 | 14.1 | 14.9 | 15.5 | 15.9 | 16.2 |
| 1880.1008 | 1896.0964 | 1880.1035 | 1866.0863 | 1880.1012 | 1867.0706 |
| n.d. | n.d. | n.d. | 128.0697 | n.d. | n.d. |
| 199.1123 | 199.1123 | 199.1123 | 199.1074 | 199.1078 | 199.1078 |
| 270.1449 | 286.1389 | 270.1449 | 270.1449 | 284.1605 | 270.1438 |
| 341.1826 | 357.1760 | 341.1826 | 341.1819 | 355.1975 | 341.1816 |
| n.d. | 424.2191 | 408.2242 | 408.2280 | 422.2402 | n.d. |
| 426.2349 | 442.2296 | 426.2349 | 426.2349 | 440.2506 | 426.2354 |
| 554.2934 | 570.2869 | 554.2934 | 554.2933 | 568.3087 | 554.2932 |
| 639.3463 | 655.3404 | 639.3463 | 639.3465 | 653.3621 | 639.3461 |
| 752.4301 | 768.4257 | 752.4301 | 752.4303 | 766.4461 | 752.4295 |
| 837.4813 | 853.4789 | 837.4813 | 837.4833 | 851.4992 | 837.4824 |
| 894.5075 | 910.4971 | 894.5075 | 894.5044 | 908.5203 | 894.5037 |
| 1007.5825 | 1023.5860 | 1007.5825 | 1007.5891 | 1021.6041 | 1007.5911 |
| 1092.6420 | 1108.6370 | 1092.6440 | 1092.6432 | 1106.6582 | 1092.6413 |
| n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| – | – | – | – | – | – |
| 788.4661 | 788.4667 | 788.4668 | 774.4504 | 774.4503 | 775.4366 |
| 637.3669 | 637.3683 | 637.3683 | 623.3499 | 623.3499 | 624.3356 |
| 509.3092 | 509.3078 | 509.3078 | 495.2931 | 495.2931 | 496.2769 |
| 381.2498 | 381.2500 | 381.2500 | 367.2337 | 367.2337 | 367.2337 |
| 282.1806 | 282.1815 | 282.1815 | 282.1812 | 282.1812 | 282.1814 |
| 197.1288 | 197.1286 | 197.1286 | 197.1284 | 197.1284 | 197.1287 |
| Diagnostic fragment ions | Peaks [m/z]a) | |||||
|---|---|---|---|---|---|---|
| 37 | 38 | 39 | 40 | 41 | 42 | |
| tR [min] | 16.4 | 16.7 | 16.8 | 17.0 | 17.2 | 17.5 |
| [M + H]+ | 1880.1007 | n.d. | 1880.1009 | n.d. | 1880.0997 | 1894.1210 |
| b1 | n.d. | n.d. | 128.0701 | 128.0713 | n.d. | n.d. |
| b2 | 199.1075 | 199.1075 | 199.1077 | 199.1075 | 199.1075 | 199.1080 |
| b3 | 270.1444 | 284.1603 | 284.1602 | 284.1599 | 270.1444 | 284.1604 |
| b4 | 341.1819 | 355.1974 | 355.1975 | 355.1973 | 341.1819 | 355.1974 |
| b4 – H2O | n.d. | n.d. | 422.2399 | n.d. | n.d. | 436.2493 |
| b5 | 426.2350 | 440.2504 | 440.2499 | 440.2504 | 426.2350 | 454.2659 |
| b6 | 554.2935 | 568.3091 | 568.3080 | 568.3086 | 554.2935 | 582.3240 |
| b7 | 639.3462 | 653.3619 | 653.3613 | 653.3615 | 639.3462 | 667.3770 |
| b8 | 752.4307 | 766.4459 | 766.4450 | 766.4452 | 752.4307 | 780.4612 |
| b9 | 837.4843 | 851.4983 | 851.4983 | 851.4987 | 837.4843 | 865.5140 |
| b10 | 894.5019 | 908.5197 | 908.5205 | 908.5230 | 894.5019 | 922.5363 |
| b11 | 1007.5901 | 1021.6066 | 1021.6041 | 1021.6054 | 1007.5901 | 1035.6190 |
| b12 | 1092.6420 | 1106.6569 | 1106.6577 | 1106.6577 | 1092.6420 | 1120.6761 |
| b12 – H2O | n.d. | n.d. | n.d. | 1088.6517 | n.d. | 1103.6621 |
| y5 | – | – | – | – | – | – |
| y5 – AA (17) | – | – | – | – | – | – |
| y5 – AA (17-16) | – | – | – | – | – | – |
| y5 – AA (17-15) | – | – | – | – | – | – |
| y6 | – | – | – | – | – | – |
| y6 – AA (18) | – | – | – | – | – | – |
| y6 – AA (18-17) | – | – | – | – | – | – |
| y6 – AA (18-16) | – | – | – | – | – | – |
| y6 – AA (18-15) | – | – | – | – | – | – |
| y7 | 788.4660 | 775.4348 | 774.4505 | 788.4664 | 788.4664 | 774.4522 |
| y7 – H2O | 637.3704 | 624.3348 | 623.3515 | 637.3670 | 637.3670 | 623.3499 |
| y7 – AA (19) | 509.3084 | 469.2766 | 495.2929 | 509.3079 | 509.3079 | 495.2931 |
| y7 – AA (19-18) | 381.2504 | 367.2338 | 367.2338 | 381.2493 | 381.2493 | 367.2337 |
| y7 – AA (19-17) | 282.1806 | 282.1808 | 282.1808 | 282.1807 | 282.1807 | 282.1812 |
| y7 – AA (19-16) | 197.1288 | 197.1283 | 197.1274 | 197.1282 | 197.1282 | 197.1284 |
| 43 | 44 | 45 | 46 | 47 | 48 | 49 |
|---|---|---|---|---|---|---|
| 17.7 | 18.0 | 18.6 | 20.0 | 21.5 | 22.0 | 22.1–22.2 |
| 1895.1007 | 1894.1177 | 1908.1341 | 1922.1467 | 1936.1660 | 1009.7031 | 1066.7242 |
| n.d. | 128.0684 | 128.0684 | n.d. | n.d. | n.d. | n.d. |
| 199.1084 | 199.1074 | 199.1080 | 227.1386 | 241.1536 | 212.1663 | 212.1644 |
| 284.1606 | 270.1440 | 284.1604 | 312.1916 | 326.2076 | 269.1858 | 269.1850 |
| 355.1969 | 341.1818 | 355.1974 | 383.2288 | 397.2443 | 382.2698 | 382.2695 |
| n.d. | 422.2401 | 436.2550 | n.d. | n.d. | – | – |
| 440.2499 | 440.2501 | 454.2659 | 468.2807 | 482.2975 | 467.3234 | 467.3230 |
| 568.3077 | 568.3087 | 582.3240 | 596.3410 | 610.3540 | 524.3442 | 524.3428 |
| 653.3609 | 653.3614 | 667.3770 | 681.3925 | 695.4084 | 637.4289 | 581.3654 |
| 766.4466 | 766.4453 | 780.4612 | 794.4774 | 808.4926 | 722.4814 | 694.4498 |
| 851.4985 | 851.4983 | 865.5140 | 879.5284 | 893.5450 | 779.5027 | 779.5029 |
| 908.5184 | 908.5202 | 922.5363 | 936.5518 | 950.5672 | 892.5860 | 836.5243 |
| 1021.6039 | 1021.6067 | 1035.6190 | 1049.6372 | 1063.6524 | – | 949.6064 |
| 1106.6577 | 1106.6590 | 1120.6744 | 1134.6878 | 1148.7083 | – | – |
| 1088.6389 | n.d. | 1102.6586 | n.d. | n.d. | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| – | – | – | – | – | – | – |
| 789.4503 | 788.4660 | 788.4670 | 788.4660 | 788.4650 | – | – |
| 638.3516 | 637.3677 | 637.3670 | 637.3677 | 637.3678 | – | – |
| 510.2927 | 509.3076 | 509.3079 | 509.3076 | 509.3077 | – | – |
| 381.2498 | 381.2495 | 381.2493 | 381.2495 | 381.2492 | – | – |
| 282.1814 | 282.1807 | 282.1807 | 282.1807 | 282.1814 | – | – |
| 197.1292 | 197.1284 | 197.1282 | 197.1284 | 197.1277 | – | – |
n.d., Not detected.
2.4. Lipopeptaibols as Trace Components in the Plate Cultures
Two lipopeptaibols, compounds 48 and 49, were produced as trace components in the DTU plate culture. Compound 49 probably represents trichogin A IV [68] [69] or a positional isomer thereof. The new positionally isomeric compound 48, named ‘lipophellin 1’, is characterized by the deletion of [Gly]5 of compound 49 (Tables 5 and 6, and Fig. 2,c).
3. Discussion
3.1. Hypophellins, Novel Long-Chain Peptaibiotics from T. phellinicola
The most notable result of this investigation is, indeed, the unequivocal confirmation of peptaibiotic biosynthesis in the natural habitat of T phellinicola growing on its host Phellinus ferruginosus, commonly known as the Rusty Porecrust. We here describe for the first time the in vivo detection of non-ribosomal peptide antibiotics5), which may significantly contribute to the complex interaction of a fungicolous ascomycete growing on its basidiomycetous host.
3.2. The Peptaibiome of the Specimen
The teleomorph produced a microheteroge-neous mixture of ten 20-residue HPHs, four of which, 6, 8, 9, and 10, are new (Table 1). Compared to smaller sequences consisting of less than 17 residues, long-chain peptaibiotics display a higher membrane-pore-formation activity by several orders of magnitude [71].
Depending on the individual sequence, seven to nine Aib residues are present, which strongly promote the formation of helical structures. i.e., α- or 310-helices, and even mixed forms [72–74], which is due to the steric constraints imposed by the geminal Me groups of the Cα-atom [75]. All of them exhibit the structurally important features, which are required for the formation of transmembrane ion channels in artificial lipid bilayer membranes, as compiled by Duclohier [76], and Duclohier and Wróblewski [77]. A multitude of bioactivities has been described for 20-residue peptaibols of similar structure, which are compiled in Table 7.
Table 7.
Biological Activities of Selected 20-Residue Peptaibols Structurally Closely Related to Hypophellins
| Peptaibols | Bioactivities reported | Ref. |
|---|---|---|
| Longibrachins | Ion-channel formation in BLM, antimycoplasmic | [53] |
| Suzukacillins | Antibacterial, antifungal | [78] |
| Ion-channel formation in BLM | [79] | |
| Haemolysis of human erythrocytes | [80] | |
| Trichoaureocins | Haemolysis of sheep erythrocytes, antibacterial (g+) | [54] |
| Trichobrachins | Antibacterial (g+ ) | [57] |
| Trichocellins | Induction of Ca2+-dependent catecholamine secretion from bovine adrenal medullary chromaffin cells | [67] |
| Ion-channel formation in BLM | [81] | |
| Trichokonins | Agonist towards Ca2+-channels in bullfrog cardiac myocytes | [55] [82] |
| Antibacterial (g+ ), antifungal | [83] | |
| Induction of defense responses and systemic resistance in tobacco against tobacco mosaic virus | [46] | |
| Induction of apoptotic programmed cell death in fungal plant pathogens | [47] | |
| Trichosporins B | Uncoupling of the respiratory activity of rat liver mitochondria | [64] [84] |
| Induction of Ca2+-dependent catecholamine secretion from bovine adrenal medullary chromaffin cells | [85–87] | |
| Ion-channel formation in BLM | [88] | |
| Antitrypanosomal | [66] | |
| Paracelsins | Antibacterial (g+ ) | [89] |
| Increasing digestibility of starch and cellulose in ruminants; haemolysis of human erythrocytes; acutely toxic in mice (LD50 5 mg/kg, i.p.) | [90] | |
| Mosquitocidal (larvae of Culex pipiens) | [91] | |
| Toxic against aquatic invertebrates (Daphnia magna, Artemisia salina) | [92] [93] | |
| Ion-channel formation in BLM | [71] | |
| Antifungal | [93] | |
3.3. The Peptaibiome of the Ex-Type Plate Culture
In contrast to what has been observed for the specimen, 20-residue peptaibols could not be detected. Instead, fifteen 19-residue peptaibols were detected in the micrOTOF-Q II screening and another eighteen in the maXis screening. Although sequences of 11–47 still exhibit the characteristic building scheme of SF1, they are distinguished from the 20-residue peptaibols of the teleomorph specimen by a deletion of the Aib/Ala residue in position 6 (Δ Ala/Aib6) of the peptide chain. This deletion, however, is predicted not to negatively influence the bioactivity of these long-chain peptaibols, as all important structural features are still present, which comply with the requirements for the formation of transmembrane ion channels in artificial lipid bilayer membranes [76] [77]. The three 18-residue sequences, 11, 12, and 28, exhibit a deletion of the C-terminal amino alcohol, whereas the dipeptide [Gln18–Pheol19] is deleted in 29, a 17-residue sequence. Truncated versions of SF1 peptaibols lacking the C-terminal amino alcohol or even the adjacent Gln residue have been reported before.
The ten 19-residue peptaibiotics, trichobrachins I (TB I), lacking the C-terminal Pheol residue, as well as the two 18-residue trichobrachins II-1 and -2 (TB II), which exhibit a deletion of the C-terminal dipeptide [Gln19–Pheol20], were shown to originate from 20-residue trichobrachins II (TB II) by enzymatic degradation [57]. Two minor desPheol compounds F30, representing 1.3% of the alamethicin (ALM) mixture investigated, have been detected by non-aqueous capillary electrophoresis (NACE) coupled to electrospray mass spectrometry [94].
3.4. l-Phenylalaninol as Constituent of Natural Products
C-Terminal l-Pheol is commonly found in peptaibiotics [17] [18] but has also been infrequently reported as a constituent of other plant and fungal secondary metabolites such as N-benzoyl-l-phenylalaninol from Catharanthus pusillus [95] and Diospyros quaesita [96], O-acetyl-N-(N′-benzoyl-l-phenylalanyl)-l-phenylalaninol from Euphorbia fischeriana and E. kansui [97], and N-benzoyl-O-[N′-benzoyl-l-phenylalanyl]-l-phenylalaninol from Penicillium arenicola (syn. P. canadense) [98].
3.5. l-Tyrosinol as a Constituent of Natural Products
To the best of our knowledge, neither d- nor l-tyrosinol6) has ever been reported as constituent of either linear or cyclic peptides of microbial origin, including peptaibiotics. However, l-tyrosinol is a ‘cryptic’ building block of the following natural products:
3.6. The Lifestyle of Trichoderma phellinicola: Findings and Thoughts
Taken these findings together, we dare predict a mycoparasitic lifestyle of the host-specific polyporicolous Trichoderma phellinicola:
It has been demonstrated by in vitro studies that chitinases and β-1,3-glucanases act synergistically with peptaibiotics in inhibiting spore germination and hyphal elongation of Botrytis cinerea. Parallel formation of hydrolytic enzymes and 19-residue antifungal trichorzianins A and B by the potent mycoparasite Trichoderma atroviride7) is triggered in the presence of cell walls of plant-pathogenic fungi [106]. Trichorzianins have previously been shown to form voltage-gated ion channels in planar lipid bilayers [107] and to modify the membrane permeability of liposomes, and they are active against Rhizoctonia solani and Phythophthora cactorum [108]. Based on these findings, a model of how peptaibiotics such as trichorzianins and hydrolases interact synergistically was proposed.
First, the host cell wall is digested enzymatically; thereafter, peptaibiotics will penetrate the cell membrane to form ion channels. Cell leakage reduces the ability of the host to effectively repair its cell wall. Eventually, inhibition of chitin and β-glucan synthesis further amplifies the destructive effect of chitinases and β-1,3-glucanases [108]. These mechanisms, however, may also account for the recently published induction of programmed cell death in plant fungal pathogens [47] caused by the 20-residue peptaibol trichokonin VI (= gliodeliquescin A [56])8), from T. koningii, T. pseudokoningii, and T. deliquescens (syn. Gliocladium deliquescens) [20]. The presence of peptaibiotics was also shown to play a role in the induction of plant defence responses [110].
3.7 Remarks on Non-Ribosomal Biosynthesis and Module Skipping by T. phellinicola
The exclusive production of 20-residue peptaibols by the T. phellinicola teleomorph indicates the presence of a 20-module NRPS. As the culture CBS 119283 has been shown to produce 17-, 18-, and 19-residue peptaibiotics only, it is likely to contain a 19-module NRPS, lacking the 6th module activating Ala or Aib. In addition, modules 3 and 4 show differing substrate specificities, as compared to the teleomorph, thus permitting the incorporation of Ala or Ser in position 3 and of Gly, Ala, or Ser in position 4, respectively. These findings indicate substantial variations in the sequences of the SF1-type peptaibol synthetases of both strains. As has been discussed in the case of SF4-type peptaibols, genes involved in secondary-metabolite products show a much broader sequential variety than housekeeping genes [50]. We here, indeed, find evidence for a significant structural variation within a large gene.
Experimental Part
Chemicals. All solvents used, MeCN (99.9%), MeOH (99.9%), CH2Cl2 (99.8%), and HCOOH (98%), were of LC/MS grade from Sigma-Aldrich (D-Steinheim). Water was purified by a Merck-Millipore Milli-Q Synthesis A10 system (D-Schwalbach/Ts.).
Origin of Specimen. The teleomorphic specimen of Trichoderma phellinicola growing on its host Phellinus ferruginosus was collected in the ‘Národni park Podyjí’ (Czech Republic, Moravia), near Hardegg at the bridge across the River Thaya, just across the border between Austria and the Czech Republic.
Origin of Trichoderma phellinicola CBS 119283 (ex-type). All details concerning this new species were given by Jaklitsch [20].
Extraction of Specimens. The teleomorph was extracted with CH2Cl2/MeOH 1:1 (v/v), the solvent was evaporated in vacuo (Rotavapor R-215, Biichi, D-Essen), and the extract was cleaned up over Sep-Pak Classic C18 cartridges (Waters, D-Eschborn) as described by Krause et al. [48].
Cultivation and Extraction of Pure Cultures. Cultures of the specimen were grown on potato dextrose agar (PDA; Becton Dickinson, D-Heidelberg) at 23° for 6 d. These subcultures were used for inoculation of the main cultures. After 10 d of cultivation at 23° in the dark, main cultures were extracted as described for the teleomorph.
LC/MS Analysis. Two QTOF systems, both from Bruker Daltonic (D-Bremen) controlled by HyStar v. 3.2 were used. Both instruments were equipped with an orthogonal ESI source and coupled to a Dionex UltiMate 3000 UHPLC (Dionex, D-Idstein).
System 1: high-resolution micrOTOF Q-II mass spectrometer. For separation, an Acclaim 120 C8, 3 μm, 2.1 × 150 mm, column (Dionex, D-Idstein) at a flow rate of 0.25 ml/min−1 and a temp. of 35° was used. Eluent A consisted of H2O + 0.1% HCOOH and eluent B of 95% MeCN + 0.1% HCOOH. Subsamples of 10 μl were injected. The column was held at 80% A/20% B for 5 min, then a gradient from 20% B to 100% over 55 min was applied. Thereafter, the column was held at 100% B for 15 min, returned to the start conditions in 1 min, and finally equilibrated for 14 min.
Samples were screened for peptaibiotics in the positive-ion mode using the following three-step routine procedure: first a full scan was recorded from m/z 50 to 3000. In System 1, this was followed by CID measurements from m/z 50 to 2000, recorded at energy of 150 eV. Finally, results of CID-MS were verified by MS/MS experiments on selected precursor ions. For precursors of m/z < 1000, a collision energy of 30 eV was applied, precursor ions in the m/z range from 1000 to 1500 were fragmented at a collision energy of 35 eV and precursor ions of m/z > 1500 at a collision energy of 40 eV. The isolation width for MS/MS experiments was set to ± 1 Da.
System 2: The maXis 3G QTOF mass spectrometer operated at a resolution of 40,000 FWHM. An Acquity BEH300 C18,1.7 μm, 2.1 × 150 mm, column (Waters, D-Eschborn) was used for separation, using H2O + 0.1% HCOOH (eluent A) and 100% MeCN + 0.1% HCOOH(eluent B). The flow rate was set to 0.3 ml/min and the temp. to 40°. The gradient started with 90% A/10% B and was changed to 50% A/50% B at 7 min, then to 30% A/70 % B at 25 min, then raised to 100% B at 38 min, and held at 100% B until 41 min before setting to starting conditions from time 42 min to 46 min. Three μl were injected. MS were scanned in the m/z range of 100–2,000. Auto MS with precursor ion-dependent collision energy optimization was used for fragmentation in the range of 10–65 eV.
Data interpretation was performed using the DataAnalysis v. 4.0 software (Bruker Daltonic, D-Bremen). Use of high-resolution (HR)ESI-MS allowed the unequivocal sequencing of fragment-ion series according to the Roepstorff/Fohlman–Biemann nomenclature. In cases where the isomeric amino acids (Leu/Ile and Val/Iva, resp.) or the corresponding amino alcohols (Leuol/Ileol) with the same elemental composition could not be distinguished, the abbreviations Lxx, Vxx, and Lxxol were used instead [48–50].
This study was supported by the Hessian Ministry for Science and Art by a grant from the LOEWE-Schwerpunkt program ‘Insect Biotechnology’ to A. V. DTU acknowledges the grant from the Danish Research Council (FI 2136-08-0023) for the maXis QTOF system, and MYCORED (EC KBBE-2007-222690-2) for supporting A. I. Support by the Austrian Science Fund (FWF; project P22081-B17) is acknowledged by W. M. J. The authors are indebted to Prof. Dr. Hartmut Laatsch (Institute of Organic and Biomolecular Chemistry, University of Göttingen, Germany) for his valuable comments on the occurrence of tyrosinol as a constituent of natural products.
Footnotes
These subfamilies were introduced at a time when the total number of peptaibiotics described did not exceed 200 sequences. As of October 2012, ca. 1,000 individual sequences are known, which also exhibit new building schemes and constituents. Consequently, there is an urgent need to reconsider this classification.
Hypophellins were simultaneously detected in an LC/MS/MS screening of 15 specimens belonging to nine Hypocrea species, which have been collected in their natural habitat. Recently, a manuscript on the in vivo detection of hypopulvins, novel peptaibiotics from the polyporicolous fungus H. pulvinata, has been published. The results therein corroborate that peptaibiotics are produced by a fungicolous fungus during infection of its natural hosts [70].
C-Terminal β-amino alcohols with the d-configuration have not yet been reported for peptaibiotics.
The trichorzianin-producing strain ATCC 36042 (= CBS 391.92) was originally identified as T. harzianum [104] but later shown to belong to T. atroviride [105]. The high degree of misidentification of Trichoderma species prior to introduction of phylogenetic analysis is still regarded a major problem, unless authors describe how their cultures were identified [17].
Gliodeliquescin A has been isolated from Gliocladium deliquescens NRRL 1086 [109] and not from NRRL 3091 [56]. According to phylogenetic data (18S-rRNA, and ITS 1 and 2), G. deliquescens NRRL 1086 (= CBS 228.48 = ATCC 10097) was re-identified as G. viride (http://www.straininfo.net/strains/260309).
References
- 1.Hawksworth DL. Mycol. Res. 2001;105:1422. [Google Scholar]
- 2.Kirk PM, Cannon PF, Minter DW, Stalpers JA, editors. 10th edn. Wallingford, Oxon: CABI Europe; 2008. ‘Dictionary of Fungi’. [Google Scholar]
- 3.Bérdy J. J. Antibiot. 2012;65:385. doi: 10.1038/ja.2012.27. corrigendum in J. Antibiot201265, 441. [DOI] [PubMed] [Google Scholar]
- 4.Laatsch H. ‘Antibase 2012 SciDex – The Natural Compounds Identifier’, Wiley-VCH, Weinheim, 2012.
- 5.Cragg GM, Grothaus PG, Newman DJ. in ‘Plant Bioactives and Drug Discovery: Principles, Practice, and Perspectives’, 4th edn., Ed. V. Cechinel-Filho, John Wiley & Sons, Hoboken, 2012, p. 1.
- 6.Bérdy J. J. Antibiot. 2005;58:1. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 7.Vinale F, Sivasithamparam K, Ghisalberti EL, Marra R, Woo SL, Lorito M. Soil Biol. Biochem. 2008;40:1. [Google Scholar]
- 8.Vinale F, Sivasithamparam K, Ghisalberti EL, Marra R, Barbetti MJ, Li H, Woo SL, Lorito M. Physiol. Mol. Plant Pathol. 2008;72:80. [Google Scholar]
- 9.Harman GE, Obregón MA, Samuels GJ, Lorito M. Plant Dis. 2010;94:928. doi: 10.1094/PDIS-94-8-0928. [DOI] [PubMed] [Google Scholar]
- 10.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Nat. Rev. Microbiol. 2004;2:43. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 11.Pomella AWV, de Souza JT, Niella GR, Bateman RP, Hebbar PK, Loguercio LL, Lumsden DR. in ‘Biological control: a global perspective’, Eds. C. Vincent, M. S. Goettel, G. Lazarovits, CAB International, Wallingford/AAFC, Oxon, 2007, Chapt. 23, p. 210.
- 12. Ministério da Agricultura, Pecuária e Abastecimento (MAPA)/Comissão Executiva do Plano da Lavoura Cacaueira (CEPLAC), J. Cacau2011/2012, 6(Nov./Feb.), 5.
- 13.Samuels GJ, Suarez C, Solis K, Holmes KA, Thomas SE, Ismaiel AA, Evans HC. Mycol. Res. 2006;110:381. doi: 10.1016/j.mycres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Hanada RE, Pomella AVW, Soberanis W, Loguercio LL, Pereira JO. Biol. Control. 2009;50:143. [Google Scholar]
- 15.Gessmann R, Brückner H, Petratos C. J. Pept. Sci. 2003;9:753. doi: 10.1002/psc.490. [DOI] [PubMed] [Google Scholar]
- 16.De Zotti M, Schievano E, Mammi S, Kaptein B, Broxterman QB, Singh SB, Brückner H, Toniolo C. Chem. Biodiversity. 2010;7:1612. doi: 10.1002/cbdv.200900287. [DOI] [PubMed] [Google Scholar]
- 17.Degenkolb T, Brückner H. Chem. Biodiversity. 2008;5:1817. doi: 10.1002/cbdv.200890171. [DOI] [PubMed] [Google Scholar]
- 18.Brückner H, Becker D, Gams W, Degenkolb T. Chem. Biodiversity. 2009;6:38. doi: 10.1002/cbdv.200800331. [DOI] [PubMed] [Google Scholar]
- 19.Jaklitsch WM. Stud. Mycol. 2009;63:1. doi: 10.3114/sim.2009.63.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaklitsch WM. Fungal Diversity. 2011;48:1. doi: 10.1007/s13225-011-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaklitsch WM, Voglmayr H. Mycologia. 2012;104:1213. doi: 10.3852/11-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaklitsch WM, Stadler M, Voglmayr H. Mycologia. 2012;104:925. doi: 10.3852/11-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuels GJ, Ismaiel A. Mycologia. 2011;103:616. doi: 10.3852/10-227. [DOI] [PubMed] [Google Scholar]
- 24.Samuels GJ, Ismaiel A, de Souza J, Chaverri P. Mycol. Prog. 2012;11:215. [Google Scholar]
- 25.Samuels GJ, Ismaiel A, Mulaw TB, Szakacs G, Druzhinina IS, Kubicek CP, Jaklitsch WM. Fungal Diversity. 2012;55:77. doi: 10.1007/s13225-012-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi K, Tsurumi Y, Suzuki R, Chuaseeharonnachai C, Sri-Indrasutdhi V, Boonyuen N, Okane I, Suzuki K-I, Nakagiri A. Mycologia. 2012;104:1109. doi: 10.3852/11-253. [DOI] [PubMed] [Google Scholar]
- 27.Kim CS, Shirouzu T, Nakagiri A, Sotome K, Nagasawa E, Maekawa N. Antonie van Leeuwenhoek. 2012;102:629. doi: 10.1007/s10482-012-9758-3. [DOI] [PubMed] [Google Scholar]
- 28.Li Q-R, Tan P, Yiang Y-L, Hyde KD, Mckenzie EHC, Bahkali AH, Kang J-C, Wang Y. Mycol. Prog. 2013;12:167. [Google Scholar]
- 29.Fujita T, Takaishi Y, Moritoki H, Ogawa T, Tokimoto K. Chem. Pharm. Bull. 1984;32:1822. [Google Scholar]
- 30.Matsuura K, Yesilada A, Iida A, Takaishi Y, Kanai M, Fujita T. J. Chem. Soc., Perkin Trans. I. 1993:381. [Google Scholar]
- 31.Matsuura K, Shima O, Takeda Y, Takaishi Y, Nagaoka Y, Fujita T. Chem. Pharm. Bull. 1994;42:106. doi: 10.1248/cpb.42.1063. [DOI] [PubMed] [Google Scholar]
- 32.Aretz W, Knauf M, Kogler H, Stahl W, Stump H, Vértesy L, Wink J. in ‘Abstracts of the 9th Dechema Meeting on Natural Products’, Irsee Monastry, Germany, poster 18, 1997.
- 33.Lee S-J, Yeo W-H, Yun B-S, I.-D.Yoo J. Pept. Sci. 1999;5:374. doi: 10.1002/(SICI)1099-1387(199908)5:8<374::AID-PSC211>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 34.Lee S-J, Yun B-S, Cho D-H, Yoo I-D. J. Antibiot. 1999;52:998. doi: 10.7164/antibiotics.52.998. [DOI] [PubMed] [Google Scholar]
- 35.Kiet TT, Gräfe U, Saluz H-P, Schlegel B. Di Truyên Hoc Vá Úng Dųng (Genet. Appl., Hanoi)2002 62, Special Issue on Biotechnology.
- 36.Dornberger K, Ihn W, Ritzau M, Gräfe U, Schlegel B, Fleck WF, Metzger JW. J. Antibiot. 1995;48:977. doi: 10.7164/antibiotics.48.977. [DOI] [PubMed] [Google Scholar]
- 37.Degenkolb T, Berg A, Gams W, Schlegel B, Gräfe U. J. Pept. Sci. 2003;9:666. doi: 10.1002/psc.497. [DOI] [PubMed] [Google Scholar]
- 38.Degenkolb T, Kirschbaum J, Brückner H. Chem. Biodiversity. 2007;4:1052. doi: 10.1002/cbdv.200790096. [DOI] [PubMed] [Google Scholar]
- 39.Neuhof T, Berg A, Besl H, Schwecke T, Dieckmann R, von Döhren H. Chem. Biodiversity. 2007;4:1103. doi: 10.1002/cbdv.200790099. [DOI] [PubMed] [Google Scholar]
- 40.Lehr N-A, Meffert A, Antelo L, Sterner O, Anke H, Weber RWS. FEMS Microbiol. Ecol. 2006;55:106. doi: 10.1111/j.1574-6941.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- 41.Menestrina G, Voges K-P, Jung G, Boheim G. J. Membr. Biol. 1986;93:111. doi: 10.1007/BF01870804. [DOI] [PubMed] [Google Scholar]
- 42.Berek I, Becker A, Schröder H, Härtl A, Höllt V, Grecksch G. Behav. Brain Res. 2009;203:232. doi: 10.1016/j.bbr.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Hosotani N, Kumagai K, Honda S, Ito A, Shimatani T, Saji I. J. Antibiot. 2007;60:184. doi: 10.1038/ja.2007.20. [DOI] [PubMed] [Google Scholar]
- 44.Singh SB, Herath K, Guan Z, Zink DL, Dombrowski AW, Polishook JD, Silverman KC, Lingham RB, Felock PJ, Hazuda DJ. Org. Lett. 2002;4:1431. doi: 10.1021/ol025540a. [DOI] [PubMed] [Google Scholar]
- 45.Shi M, Wang H-N, Xie S-T, Luo Y, Sun C-Y, Chen X-L, Zhang Y-Z. Mol. Cancer. 2010;9:26. doi: 10.1186/1476-4598-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y, Zhang D-D, Dong X-W, Zhao P-B, Chen L-L, Song X-Y, Wang X-J, Chen X-L, Shi M, Zhang Y-Z. FEMS Microbiol. Lett. 2010;313:120. doi: 10.1111/j.1574-6968.2010.02135.x. [DOI] [PubMed] [Google Scholar]
- 47.Shi M, Chen L, Wang X-W, Zhang T, Zhao P-B, Song X-Y, Sun C-Y, Chen X-L, Zhou B-C, Zhang Y-Z. Microbiology. 2012;158:166. doi: 10.1099/mic.0.052670-0. [DOI] [PubMed] [Google Scholar]
- 48.Krause C, Kirschbaum J, Brückner H. Amino Acids. 2006;30:435. doi: 10.1007/s00726-005-0275-9. [DOI] [PubMed] [Google Scholar]
- 49.Degenkolb T, Gräfenhan T, Berg A, Nirenberg HI, Gams W, Brückner H. Chem. Biodiversity. 2006;3:593. doi: 10.1002/cbdv.200690063. [DOI] [PubMed] [Google Scholar]
- 50.Degenkolb T, Karimi Aghcheh R, Dieckmann R, Neuhof T, Baker SE, Druzhinina IS, Kubicek CP, Brückner H, von Döhren H. Chem. Biodiversity. 2012;9:499. doi: 10.1002/cbdv.201100212. [DOI] [PubMed] [Google Scholar]
- 51.Neuhof T, Dieckmann R, Druzhinina IS, Kubicek CP, von Döhren H. Microbiology. 2007;153:3417. doi: 10.1099/mic.0.2007/006692-0. [DOI] [PubMed] [Google Scholar]
- 52.Chugh JK, Wallace BA. Biochem. Soc. Trans. 2001;29:565. doi: 10.1042/bst0290565. [DOI] [PubMed] [Google Scholar]
- 53.Leclerc G, Goulard C, Prigent Y, Bodo B, Wróblewski H, Rebuffat S. J. Nat. Prod. 2001;64:164. doi: 10.1021/np000240s. [DOI] [PubMed] [Google Scholar]
- 54.Jaworski A, Brückner H. Amino Acids. 2001;21:1. this abstract is found on p. 6/7. [Google Scholar]
- 55.Huang Q, Tezuka Y, Kikuchi T, Nishi A, Tubaki K, Tanaka K. Chem. Pharm. Bull. 1995;43:223. doi: 10.1248/cpb.43.223. [DOI] [PubMed] [Google Scholar]
- 56.Brückner H, Przybylski M. Chromatographia. 1984;19:188. [Google Scholar]
- 57.Krause C, Kirschbaum J, Brückner H. Chem. Biodiversity. 2007;4:1083. doi: 10.1002/cbdv.200790098. [DOI] [PubMed] [Google Scholar]
- 58.Brückner H, Kripp T, Kieß M. in ‘Peptides 1990, Proceedings of the 21st European Peptide Symposium. Platja d'Aro, Spain’, Eds. E. Giralt, D. Andreu, ESCOM, Leiden, 1991, p. 347.
- 59.Brückner H, Kripp T, Kieß M. in ‘Chemistry of Peptides and Proteins. Proceedings of the 7th USSR-FRG Symposium Chemistry of Peptides and Proteins, Dilizhan, 1989’ and in ‘Chemistry of Peptides and Proteins. Proceedings of the 8th USSR-FRG Symposium Chemistry of Peptides and Proteins, Aachen, 1991’, Eds. D. Brandenburg, V. Ivanov, W. Voelter, Mainz Verlag, Aachen, 1993, DWI Reports, Vol. 112A+B, p. 357.
- 60.Krause C, Kirschbaum J, Jung G, Brückner H. J. Pept. Sci. 2006;12:321. doi: 10.1002/psc.728. [DOI] [PubMed] [Google Scholar]
- 61.Iida A, Okuda M, Uesato S, Takaishi Y, Shingu T, Saito M, Morita M, Fujita T. J. Chem. Soc., Perkin Trans. I. 1990:3249. [Google Scholar]
- 62.Huang Q, Tezuka Y, Hatanaka Y, Kikuchi T, Nishi A, Tubaki K. Chem. Pharm. Bull. 1995;43:1663. doi: 10.1248/cpb.43.1663. [DOI] [PubMed] [Google Scholar]
- 63.Huang Q, Tezuka Y, Hatanaka Y, Kikuchi T, Nishi A, Tubaki K. Chem. Pharm. Bull. 1996;44:590. doi: 10.1248/cpb.44.590. [DOI] [PubMed] [Google Scholar]
- 64.Fujita T, Iida A, Uesato S, Takaishi Y, Shingu T, Saito M, Morita M. J. Antibiot. 1988;41:814. doi: 10.7164/antibiotics.41.814. [DOI] [PubMed] [Google Scholar]
- 65.Iida J, Iida A, Takahashi Y, Takaishi Y, Nagaoka Y, Fujita T. J. Chem. Soc., Perkin Trans. I. 1993:357. [Google Scholar]
- 66.Iwatsuki M, Kinoshita Y, Niitsuma M, Hashida J, Mori M, Ishiyama A, Namatame M, Nishihara-Tsukashima A, Nonaka K, Masuma R, Otoguro K, Yamada H, Shiomi K, Ōmura S. J. Antibiot. 2010;63:331. doi: 10.1038/ja.2010.41. [DOI] [PubMed] [Google Scholar]
- 67.Wada S-I, Nishimura T, Iida A, Toyama N, Fujita T. Tetrahedron Lett. 1994;35:3095. [Google Scholar]
- 68.Auvin-Guette C, Rebuffat S, Prigent Y, Bodo B. J. Am. Chem. Soc. 1992;114:2170. [Google Scholar]
- 69.Peggion C, Formaggio F, Crisma M, Epand RF, Epand RM, Toniolo C. J. Pept. Sci. 2003;9:679. doi: 10.1002/psc.500. [DOI] [PubMed] [Google Scholar]
- 70.Röhrich CR, Iversen A, Jaklitsch WM, Voglmayr H, Berg A, Dörfelt H, Thrane U, Vilcinskas A, Nielsen KF, von Döhren H, Brückner H, Degenkolb T. Fungal Biol. 2012;116:1219. doi: 10.1016/j.funbio.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grigoriev PA, Schlegel B, Kronen M, Berg A, Härtl A, Gräfe U. J. Pept. Sci. 2003;9:763. doi: 10.1002/psc.502. [DOI] [PubMed] [Google Scholar]
- 72.De Zotti M, Damato F, Formaggio F, Crisma M, Schievano E, Mammi S, Kaptein B, Broxterman QB, Felock PJ, Hazuda DJ, Singh SB, Kirschbaum J, Brückner H, Toniolo C. Chem. – Eur. J. 2010;16:316. doi: 10.1002/chem.200900945. [DOI] [PubMed] [Google Scholar]
- 73.Gessmann R, Axford D, Owen RL, Brückner H, Petratos K. Acta Crystallogr., Sect. D. 2012;68:109. doi: 10.1107/S090744491105133X. [DOI] [PubMed] [Google Scholar]
- 74.Gessmann R, Axford D, Evans G, Brückner H, Petratos K. J. Pept. Sci. 2012;18:678. doi: 10.1002/psc.2454. [DOI] [PubMed] [Google Scholar]
- 75.De Zotti M, Biondi B, Crisma M, Hjørringgaard CU, Berg A, Brückner H, Toniolo C. Biopolymers (Pept. Sci.) 2012;98:36. doi: 10.1002/bip.21679. [DOI] [PubMed] [Google Scholar]
- 76.Duclohier H. Eur. Biophys. J. 2004;33:169. doi: 10.1007/s00249-003-0383-y. [DOI] [PubMed] [Google Scholar]
- 77.Duclohier H, Wróblewski H. J. Membr. Biol. 2001;184:1. doi: 10.1007/s00232-001-0077-2. [DOI] [PubMed] [Google Scholar]
- 78.Ooka T, Takeda I. Agric. Biol. Chem. 1972;36:112. [Google Scholar]
- 79.Boheim G, Janko K, Leibfritz D, Ooka T, König WA, Jung G. Biochim. Biophys. Acta. 1976;433:182. doi: 10.1016/0005-2736(76)90186-3. [DOI] [PubMed] [Google Scholar]
- 80.Irmscher G, Jung G. Eur. J. Biochem. 1977;80:165. doi: 10.1111/j.1432-1033.1977.tb11868.x. [DOI] [PubMed] [Google Scholar]
- 81.Wada S-I, Iida A, Asami K, Tachikawa E, Fujita T. Biochim. Biophys. Acta. 1997;1325:209. doi: 10.1016/s0005-2736(96)00260-x. [DOI] [PubMed] [Google Scholar]
- 82.Huang Q, Tezuka Y, Kikuchi T, Mosome Y. Eur. J. Pharmacol. 1994;271 doi: 10.1016/0014-2999(94)90290-9. R5. [DOI] [PubMed] [Google Scholar]
- 83.Song X-Y, Shen Q-T, Xie S-T, Chen X-L, Sun C-Y, Zhang Y-Z. FEMS Microbiol. Lett. 2006;260:119. [Google Scholar]
- 84.Okuda M, Iida A, Uesato S, Nagaoka Y, Tujita T, Takaishi Y, Terada H. Biol. Pharm. Bull. 1994;17:482. doi: 10.1248/bpb.17.482. [DOI] [PubMed] [Google Scholar]
- 85.Tachikawa E, Takahashi S, Furumachi K, Kashimoto T, Iida A, Nagaoka Y, Fujita T, Takaishi Y. Mol. Pharmacol. 1991;40:790. [PubMed] [Google Scholar]
- 86.Tachikawa E, Takahashi S, Mizuma K, Kashimoto T, Nagaoka Y, Iida A, Fujita T. Biol. Pharm. Bull. 1995;18:1165. doi: 10.1248/bpb.18.1165. [DOI] [PubMed] [Google Scholar]
- 87.Tachikawa E, Nogimori K, Takahashi S, Mizuma K, Itoh K, Kashimoto T, Nagaoka Y, Iida A, Fujita T. Biochim. Biophys. Acta. 1996;1282:140. doi: 10.1016/0005-2736(96)00052-1. [DOI] [PubMed] [Google Scholar]
- 88.Nagaoka Y, Iida A, Kambara T, Tachikawa E, Asami K, Fujita T. Biol. Pharm. Bull. 1995;18:640. doi: 10.1248/bpb.18.640. [DOI] [PubMed] [Google Scholar]
- 89.Brückner H, Graf H. Experientia. 1983;39:528. doi: 10.1007/BF01965190. [DOI] [PubMed] [Google Scholar]
- 90.Brückner H, Graf H, Bokel M. Experientia. 1984;40:1189. doi: 10.1007/BF01946646. [DOI] [PubMed] [Google Scholar]
- 91.Mat'ha V, Jegorov A, Kieß M, Brückner H. Tissue Cell. 1992;24:559. doi: 10.1016/0040-8166(92)90071-e. [DOI] [PubMed] [Google Scholar]
- 92.Favilla M, Macchia L, Gallo A, Altomare C. Food Chem. Toxicol. 2006;44:1922. doi: 10.1016/j.fct.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 93.Maddau L, Cabras A, Franceschini A, Linaldeddu BT, Crobu S, Roggio T, Pagnozzi D. Microbiology. 2009;155:3371. doi: 10.1099/mic.0.030916-0. [DOI] [PubMed] [Google Scholar]
- 94.Psurek A, Neusüß C, Degenkolb T, Brückner H, Balaguer E, Imhof D, Scriba GKE. J. Pept. Sci. 2006;12:279. doi: 10.1002/psc.720. [DOI] [PubMed] [Google Scholar]
- 95.Battersby AR, Kapil RS. Tetrahedron Lett. 1965;39:3529. doi: 10.1016/s0040-4039(01)89337-0. [DOI] [PubMed] [Google Scholar]
- 96.Ma C-Y, Musoke SF, Tan GT, Sydara K, Bouamanivong S, Southavong B, Soejarto DD, Fong HHS, Zhang H-J. Chem. Biodiversity. 2008;5:2442. doi: 10.1002/cbdv.200890209. [DOI] [PubMed] [Google Scholar]
- 97.Uemura D, Sugiura K, Hirata Y. Chem. Lett. 1975;6:537. [Google Scholar]
- 98.McCorkindale NJ, Baxter RL, Roy TP, Shields HS, Stewart RM, Hutchinson SA. Tetrahedron. 1978;34:2791. [Google Scholar]
- 99.Cheng Y, Schneider B, Riese U, Schubert B, Li Z, Hamburger M. J. Nat. Prod. 2004;67:1854. doi: 10.1021/np049761w. [DOI] [PubMed] [Google Scholar]
- 100.Jia J-M, Tao H-H, Feng B-M. Chem. Pharm. Bull. 2009;57:99. doi: 10.1248/cpb.57.99. [DOI] [PubMed] [Google Scholar]
- 101.Dethe DH, Ranjan A, Pardeshi VH. Org. Biomol. Chem. 2011;9:7990. doi: 10.1039/c1ob06320k. [DOI] [PubMed] [Google Scholar]
- 102.Ciminiello P, Dell'Aversano C, Fattorusso E, Forino M, Magno S, Ianaro A, Di Rosa M. Eur. J. Org. Chem. 2001:49. doi: 10.1021/jo001437s. [DOI] [PubMed] [Google Scholar]
- 103.Ciminiello P, Dell'Aversano C, Fattorusso E, Forino M, Grauso L, Santelia FU, Tartaglione L, Moutsos VI, Pitsinos EN, Couladouros EA. Eur. J. Org. Chem. 2007:5434. [Google Scholar]
- 104.El Hajji M, Rebuffat S, Lecommandeur D, Bodo B. Int. J. Pept. Protein Res. 1987;29:207. doi: 10.1111/j.1399-3011.1987.tb02247.x. [DOI] [PubMed] [Google Scholar]
- 105.Kuhls K, Lieckfeldt E, Samuels GJ, Kovacs W, Meyer W, Petrini O, Gams W, Börner T, Kubicek CP. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7755. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schirmböck M, Lorito M, Wang YL, Hayes CK, Arisan-Atac C, Scala F, Harman GE, Kubicek CP. Appl. Environ. Microbiol. 1994;60:4364. doi: 10.1128/aem.60.12.4364-4370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Molle G, Duclohier H, Spach G. FEBS Lett. 1987;224:208. doi: 10.1016/0014-5793(87)80449-0. [DOI] [PubMed] [Google Scholar]
- 108.Lorito M, Varkas V, Rebuffat S, Bodo B, Kubicek CP. J. Bacteriol. 1996;178:6382. doi: 10.1128/jb.178.21.6382-6385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brückner H, Wunsch P, Kussin C. in ‘Second forum on peptides. Vol. 174’, Eds. A. Aubry, M. Marraud M, B. Vitoux, Colloque INSERM/John Libbey Eurotext, London, p. 103.
- 110.Viterbo A, Wiest A, Brotman Y, Chet I, Kenerley C. Mol. Plant Pathol. 2007;8:737. doi: 10.1111/j.1364-3703.2007.00430.x. [DOI] [PubMed] [Google Scholar]


