Abstract
The yeast Mediator complex is required for transcriptional regulation both in vivo and in vitro, and its function is conserved in all eukaryotes. Mediator interacts with both transcriptional activators and RNA polymerase II, but little is known about the mechanisms by which it operates at the molecular level. Here, we show that the cyclin-dependent kinase Srb10 interacts with, and phosphorylates, the Med2 subunit of Mediator both in vivo and in vitro. A point mutation of the single phosphorylation site in Med2 results in a strongly reduced expression of the REP1, REP2, FLP1, and RAF1 genes, which are all located on the endogenous 2-μm plasmid. Combined with previous studies on the effects of SRB10/SRB11 deletions, our data suggest that posttranslational modifications of Mediator subunits are important for regulation of gene expression.
Keywords: transcriptional regulation, Srb11, RNA polymerase II
The Mediator complex was originally identified in Saccharomyces cerevisiae as an activity required for transcriptional activation in an in vitro transcription system reconstituted from highly purified RNA polymerase II (pol II) and general transcription factors (1, 2). Mediator was later purified to homogeneity and shown to be a complex composed of 20 subunits that interacts with the C-terminal domain of the largest pol II subunit (3, 4). More recent work has described several subunit–subunit interactions and subdomains within Mediator (5–8), and low-resolution structures of Mediator alone or in complex with pol II have been determined by three-dimensional reconstruction from electron micrographs of single particles (9, 10). These structures indicate a division of Mediator into distinct domains.
In parallel with the identification of yeast Mediator, a pol II holoenzyme that comprised both Mediator subunits [a subset of general transcription factors and additional proteins (i.e., Srb8-11)] was purified (11). Nine SRB genes [SRB2, -4, -5, -6, -7, -8, -9, and -10 (encoding the cyclin C-dependent kinase), and -11 (encoding cyclin C)] were identified in a genetic screen for suppressors of a cold-sensitive phenotype caused by a truncation of the C-terminal domain. Although Srb2, -4, -5, -6, and -7 are well established as Mediator subunits, the Srb8, -9, -10, and -11 proteins have not been identified in Mediator described by Kornberg and coworkers (1–4). In contrast, homologues of Srb10 and Srb11 are present in some Mediator preparations from human cells (12–16). Recent studies suggest that Mediator exists in two specific forms. Mediator containing the Srb8 to -11 subunits is always present in free form whereas Mediator lacking this submodule may associate with pol II. The relative levels of Srb8–11 vary because this submodule is degraded when yeast is grown under conditions of nutrient limitation (17, 18), and the relative levels are also sensitive to the extraction procedure (19).
Activators contact individual Mediator subunits and recruit the Mediator complex to promoters during transcriptional activation. However, repressors have also been shown to interact directly with Mediator subunits (20, 21). If repressors also recruit holo-pol II to specific promoters, it is difficult to understand why recruitment in this case would lead to down-regulation of transcription. Recruitment might therefore be required for regulation of transcription, both activation and repression, but yet unknown mechanisms may determine the effect of the recruitment. Recent reports suggest that recruitment of holoenzyme is not needed for each round of transcription. Rather, the mediator-pol II interaction is dynamic, and both Mediator and several general transcription factors remain at the promoter after release of pol II where they function as a scaffold for reinitiation by pol II. It is therefore of interest to study how interactions between Mediator subunits, and interactions between Mediator, general transcription factors, and pol II can be regulated.
We here present evidence that posttranslational modifications may significantly modify the ability of Mediator to support transcription of specific genes. We believe that this observation suggests a mechanism by which Mediator may switch between being a positive or a negative coregulator of transcription.
Materials and Methods
Yeast Strains. All yeast strains except Rgr1-TAP (22) and its derivative MHY2 were congenic to W303-1A and thus carry the ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 markers. Strains H707 (MATa med1-Δ2::HIS3), H844 (MATa srb10-Δ1::LEU2) and H906 (MATαmed2-Δ1::HIS3) have been described (8, 23). To create the site-directed serine-208 to alanine mutation we used overlap extension PCR. The resulting PCR fragment was cloned into a derivative of the pFL34 vector (24) in which the HindIII site had been destroyed. The resulting plasmid was digested with HindIII before transformation of the Rgr1 TAP-tagged strain and the W303-1A strain. Transformants were selected on plates lacking uracil. Mutated revertants selected on plates containing 5-fluoro-orotic acid were analyzed by sequencing.
Growth Media. Low-phosphate medium (LPM) contains 30 mg of KH2PO4, 25 mg of MgSO4 × 7 H2O, 100 mg of NaCl, 100 mg of CaCl2 × 2 H2O, 2 g of asparagine, 1 g of KCl, amino acids (SC media), and 20 g of glucose per liter. No-phosphate medium is identical to LPM without KH2PO4.
Expression and Purification of Proteins. Cloning, expression, and purification of a 6× histidine-tagged Med1 has been described (8). The Med1 used in this study was further purified by chromatography on a UnoQ column (Bio-Rad). The coding sequences for Srb10 and Srb11 were amplified from genomic DNA by PCR and then cloned into the pGEX-6p-2 vector. We expressed the GST-tagged Srb10 and Srb11 in Escherichia coli BL21 cells and purified the recombinant proteins by using glutathione-Sepharose 4B beads according to the manufactures recommendations (Amersham Pharmacia Biosciences). For analysis of cyclin-dependent kinase (CDK)-phosphorylated proteins, Mediator was purified from a strain expressing a tandem affinity purification (TAP)-tagged Rgr1 as described previously (25). For preparation of whole-cell extracts (WCE), S. cerevisiae strains were grown to an OD600 of 1.2 in 100 ml of YNB (W303-1A) or YNB lacking uracil and tryptophane [H905 (med2) and H844 (srb10)]. Isolation of WCE was performed as described (23). The Med2/Pgd1 dimer purification has been described (J.B., and C.M.G., unpublished results).
In Vivo Phosphorylation Assays. Yeast strains W303-1A, H707 (med1), and H844 (srb10) were grown at 30°C in low-phosphate medium to a concentration of 1 × 107 cells per ml. The cells were harvested by centrifugation and resuspended at 5 × 107 cells per ml in no-phosphate medium. To these cells, 0.5 ml of 32PO4 (10 mCi/ml)(1 Ci = 37 GBq) was added, and the cultures were incubated at 30°C with shaking for 20 min. The cultures were then diluted 10 times with low-phosphate medium and incubated with shaking at 30°C for 4–6 h. Mediator was immunoprecipitated from the whole-cell extract by using anti-Med1 antibodies (8) coupled to Protein A agarose beads.
In Vitro Phosphorylation Assays. Samples containing 0.5 μg of substrate (Med1 or Med2/Pgd1) were incubated at 30°C for 20 min with GST-Srb10 (0.25 μg) and GST-Srb11 (0.25 μg) in CDK-buffer (20 mM Hepes-KOH/10 mM MgCl2/1 mM EGTA/1 mM DTT/2 mg/ml BSA and protease inhibitors), 100 μM ATP, and 1 μl [γ-32P]ATP (3,000 Ci/mmol) in a final volume of 25 μl. Labeled proteins were visualized by separation on 10% SDS/PAGE followed by autoradiography.
Immunoprecipitation Assays. For studies of the in vivo substrate for Srb10-phosphorylation, whole-cell extracts (350 μg) from the W303-1A, med2, and srb10 strains expressing a lexA-Srb7 fusion protein were isolated, and Mediator was immunopurified by using monoclonal lexA antibodies coupled to protein A-agarose beads as described (23). For studies of interaction between Med2 and Srb10/Srb11, we used the following: (i) 20 μg of GST-protein, (ii) 25 μg of GST-Srb10, (iii) 25 μg of GST-Srb11, and (iv) 25 μg of GST-Srb10 plus 25 μg of GST-Srb11. Proteins were then immunoprecipitated by using anti-GST monoclonal antibodies (Amersham Pharmacia Biotech) coupled to protein G-agarose with 25 μg of Med2/Pgd1 protein being added to each tube. After 2 h incubation by rotation at 4°C, the beads were washed three times with buffer W (1× PBS/0.8 M potassium acetate/0.5% Nonidet P-40) and once with 1× PBS. The precipitated proteins were analyzed by Western blotting by using antibodies specific for Med2.
Affymetrix GeneChip Probe Array Analyses and RT-PCR. Total yeast RNA was isolated by using a hot acid phenol extraction protocol (26). Poly(A)+ RNA was prepared from total RNA by using a Qiagen (Valencia, CA) Oligotex Midi Kit. The cDNA synthesis, cRNA synthesis, and labeling, as well as array hybridization to Affymetrix yeast S98 arrays, were performed at the Karolinska Institute Affymetrix core facility as described in the Affymetrix users' manual [Affymetrix GeneChip Expression Analysis Technical Manual (2000), Affymetrix, Santa Clara, CA]. Arrays were washed and stained by using the GeneChip Fluidics Station 400 and scanned in an Affymetrix GeneArray Scanner. Acquisition and quantification of array images as well as primary data analysis were performed by using the Affymetrix software package microarray suite 5.0. Data were used for all probe sets representing transcripts, called “present” by microarray suite 5.0, in at least two arrays. The transcription analysis was performed with RNA prepared from two WT and two med2-S208A cell cultures. Each of the two WT RNA preparations was compared with both the med2-S208A RNA preparations, generating in total four comparisons (Exps. 1–4). The fold changes were calculated by using the average of the four comparisons.
Expression of REP1, REP2, FLP1, RAF1, and MED1 mRNAs was quantified by using the Titanium one-step RT-PCR Kit (CLONTECH). In brief, 2 ng of total RNA from either the WT or the med2-S208A strain were reverse transcribed at 50°Cfor1h and then amplified by using 30 PCR cycles. RT-PCR products were subsequently analyzed by agarose gel electrophoresis.
Results
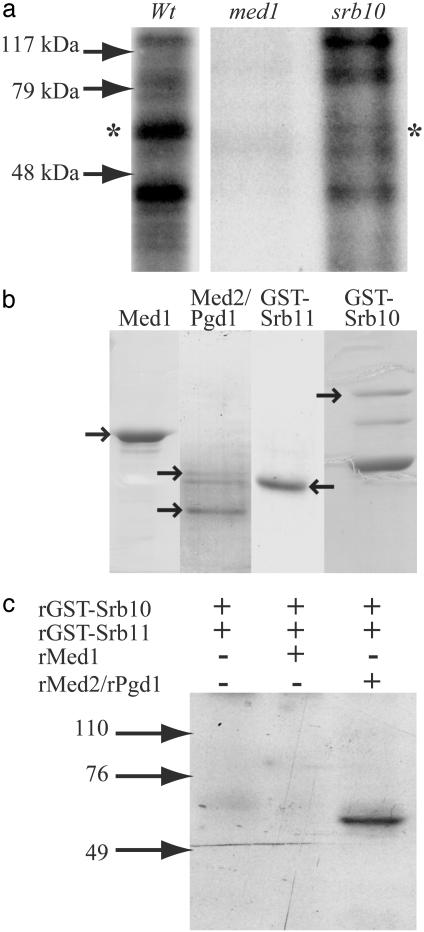
Phosphorylation of Mediator Subunits in Vivo. We have previously reported that a LexA fusion to the Med1 protein may strongly activate transcription from promoters containing LexA binding sites and that this LexA-Med1 dependent activation is regulated by the Srb10/Srb11 cdk/cyclin complex (23). We therefore wanted to test whether Med1 was a target for the Srb10 kinase activity. To this end, we made in vivo phosphorylation experiments where Mediator was immunopurified by using Med1 antibodies, both from WT cells and from cells lacking either Srb10 or Med1 (negative control for immunoprecipitation) in the presence of 32PO4. We identified two labeled polypeptides of ≈40 and 60 kDa that were present in Mediator purified from WT cells but absent in Mediator purified from cells lacking Srb10 (Fig. 1a). We have previously reported that Med4 is phosphorylated in vivo (23). The labeled 40-kDa polypeptide could therefore correspond to Med4 because it has a predicted molecular mass of 32 kDa but migrates as an ≈38-kDa protein. However, we found no indication that Srb10 phosphorylates Med4 in any of the other experiments presented in this article. As for the labeled 60-kDa polypeptide, the only Mediator subunits in that molecular mass range are Med1 and Med2. We therefore proceeded to investigate whether Srb10 can phosphorylate either of these two proteins.
Fig. 1.
Phosphorylation of a Mediator subunit by Srb10 and Srb11. (a) WT, med1Δ, and srb10Δ yeast strains were labeled in vivo with 32PO4. Mediator was immunoprecipitated from each strain by using Med1 antibodies coupled to Protein A agarose beads, and bound proteins were separated on a 10% SDS/PAGE gel. Immunoprecipitation from extracts isolated from cells lacking Med1 was made as a control for nonspecific immunoprecipitation. The asterisks indicate the position of the 60-kDa protein, which is specifically labeled only in the WT strain. (b) Coomassie-stained protein gels of the peak fractions from the final purification steps of Srb10, Srb11, Med1, and Med2/Pgd1. Arrows indicate the position of each protein as indicated above each lane. The identity of Med1, Srb10, and Srb11 was confirmed by N-terminal amino acid sequencing. (c) Purified, recombinant Srb10/Srb11 was incubated with [32P]ATP alone, together with purified recombinant Med1, or with purified recombinant Med2/Pgd1 as indicated in the figure. Labeled proteins were separated on a 10% SDS/PAGE gel and subjected to autoradiography. Arrows to the left (in a and c) indicate the positions of prestained molecular mass markers.
Phosphorylation of Med2 by Srb10 in Vitro. We next investigated whether the Srb10 kinase could phosphorylate Med1 or Med2 in recombinant form (Fig. 1b). Expression and purification of 6 × histidine-tagged Med1 has been described (8). Med2 is insoluble when expressed alone, but soluble untagged Med2 protein was obtained by coexpression with the Mediator subunit Pgd1 in a baculovirus expression system. The Med2/Pgd1 dimer was then purified to near homogeneity. Finally, the Srb10 and Srb11 proteins were expressed as GST-fusion proteins in E. coli and purified as described in Materials and Methods.
Initially, we performed in vitro phosphorylation experiments where Srb10/Srb11 was incubated with [32P]ATP alone or with either Med1 or Med2/Pgd1. We could not observe any phosphorylation of Med1 or Srb10. Thus, the phosphorylated 60-kDa polypeptide identified in Fig. 1a was not due to autophosphorylation of Srb10/Srb11 or to phosphorylation of Med1. We did, however, observe a phosphorylated polypeptide of ≈60 kDa when Srb10/Srb11 was incubated with Med2/Pgd1 (Fig. 1c). This observation suggested to us that Med2 was phosphorylated, but we could not rule out Pgd1 because a phosphorylation could change the protein's behavior during SDS/PAGE.
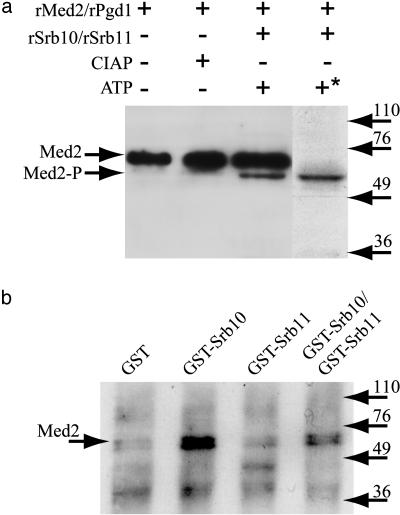
To identify which protein of Med2 and Pgd1 that is phosphorylated by Srb10, we performed in vitro phosphorylation experiments with purified, recombinant Srb10, Srb11, Med2/Pgd1, and unlabeled ATP. After incubation, the proteins were separated by SDS/PAGE, transferred to poly(vinylidene difluoride) (PVDF) membranes, and blotted with Med2 antibodies. Fig. 2a shows that a fraction of Med2 migrated faster when incubated with Srb10, Srb11, and ATP. A similar increase in the migration rate after phosphorylation has been reported previously, i.e., when Cdk2 is phosphorylated by CDK-activating kinase (CAK) 1, another cyclin-dependent kinase (27). Alignment of the filter from this Western blot with the autoradiograph from the previous in vitro phosphorylation experiment showed that the faster migrating Med2 species migrated at the same position as the labeled 60-kDa protein (the rightmost lane in Fig. 1c is aligned with lane 3 in Fig. 2a). We conclude that the phosphorylated protein was Med2 and that the phosphorylated form of Med2 migrated slightly faster than the unphosphorylated form. Finally, it should be noted that only a fraction of Med2 was phosphorylated under the conditions used here.
Fig. 2.
Med2 is phosphorylated by and interacts with the Srb10 kinase in vitro. (a) Purified, recombinant Med2/Pgd1 was incubated alone, with calf intestine alkaline phosphatase (CIAP), or with Srb10/Srb11 and unlabeled ATP. Proteins were separated on a 10% SDS/PAGE gel, transferred to PVDF membranes, and blotted with antibodies specific for the Med2 protein. The same batch of prestained molecular mass markers was used to align this Western blot with the autoradiograph presented in Fig. 1c. The asterisk next to the plus sign above the rightmost lane in the figure indicates that [32P]ATP was added in this experiment. Arrows to the left indicate the positions of Med2 and phosphorylated Med2 (Med2-P). (b) Copurified, recombinant untagged Med2 and Pgd1 were subjected to coimmunoprecipitation with recombinant GST protein (lane 1), GST-tagged Srb10 alone (lane 2), GST-tagged Srb11 alone (lane 3), or with GST-Srb10 and GST-Srb11 together (lane 4), by using monoclonal anti-GST antibodies. Precipitated proteins from each reaction were separated on a 10% SDS/PAGE gel and immunoblotted with antibodies specific for the Med2 protein. Arrows to the right indicate the positions of prestained molecular mass markers.
Med2 Interacts with Srb10 in Vitro. Phosphorylation of Med2 by the Srb10/Srb11 cyclin-kinase suggested a direct physical interaction between Med2 and Srb10 or Srb11. To test whether such an interaction could be detected in vitro, we analyzed whether Med2 could be coimmunoprecipitated with either GST-tagged Srb10 alone, GST-Srb11 alone, or with GST-Srb10 and GST-Srb11 together. Fig. 2b shows that Med2 is specifically precipitated only when GST-Srb10 is prebound to the beads, i.e., lane 2 (GST-Srb10 alone) and lane 4 (GST-Srb10 and GST-Srb11). In contrast, Med2 did not interact with GST-Srb11 alone (lane 3) or with recombinant GST (lane 1).
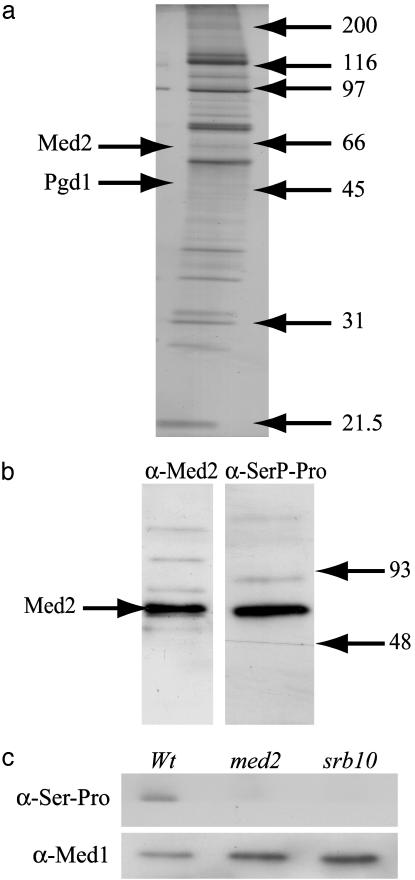
Phosphorylation of Med2 by Srb10 in Vivo. We conclude from our experiments that Srb10 can phosphorylate Med2 in vitro, but it was still unclear whether the 60-kDa phosphoprotein identified in the in vivo phosphorylation experiments (Fig. 1a) was Med2. We therefore isolated Mediator from a yeast strain expressing a TAP-tagged Rgr1 Mediator subunit (Fig. 3a). The Med2 protein appeared as a weak, fuzzy band in the purified Mediator, and the Pgd1 subunit was nearly undetectable. The observed pattern is consistent with earlier reports of purified Mediator where both Med2 and Pgd1 usually are present in substoichiometric amounts (4). The purified fraction was then analyzed by Western blotting by using either Med2 antibodies (Fig. 3b Left), or anti-SerP-Pro/Lys antibodies (monoclonal antibody PSER-16B4, nano-Tools Antikörpertechnik, Teningen, Germany; Fig. 3b Right) specific for proteins containing a phosphorylated CDK phosphorylation site (a phosphorylated serine or threonine followed by proline). In agreement with the in vivo labeling result (Fig. 1a), the Med2 antibodies detected one protein of ≈60 kDa in the purified Mediator, which migrated at the same position as the protein detected with the anti-SerP-Pro/Lys antibody. This result corroborates the in vitro phosphorylation results presented in Fig. 2a. The absence of the slower migrating species seen in Fig. 2a further suggests that only the phosphorylated, fast migrating form of Med2 is present in Mediator.
Fig. 3.
Med2 is phosphorylated by Srb10 in vivo.(a) A silver-stained gel from the peak-fraction of TAP-purified Mediator. Arrows to the right indicate the molecular mass of protein standards. (b) The Mediator fraction shown in a was transferred to PVDF membranes and blotted with anti-Med2 antibodies (left filter) and anti-SerP-Pro/Lys antibodies (right filter). The two filters were aligned in silico by using the prestained protein marker proteins as reference. (c) Mediator was immunoprecipitated with anti-lexA antibodies from wholecell extracts isolated from WT, med2Δ, and srb10Δ strains. Each strain also expressed a lexA-Srb7 fusion protein from a low copy number CEN plasmid (23). The same amount of protein from each strain was separated on a 10% SDS/PAGE gel, transferred to PVDF membranes, and blotted with either anti-Med1 antibodies (Lower) or anti-SerP-Pro/Lys antibodies (Upper).
To further establish that Med2 is an in vivo substrate for the Srb10 kinase, Mediator was immunoprecipitated with anti-lexA antibodies from yeast strains expressing the lexA-Srb7 fusion protein in different genetic backgrounds. Mediator isolated from WT, med2, and srb10 strains was first blotted with anti-Med1 antibodies to confirm that equal amounts of Mediator was loaded to each lane (Fig. 3c Lower). The same filter was then blotted with the anti-SerP-Pro/Lys antibodies (Fig. 3c Upper). A phosphorylated protein of ≈60 kDa was detected only in Mediator isolated from the WT strain but absent in Mediator isolated from strains lacking either Med2 or Srb10. We conclude that the 60-kDa polypeptide reflects Srb10-dependent phosphorylation of Med2 in vivo.
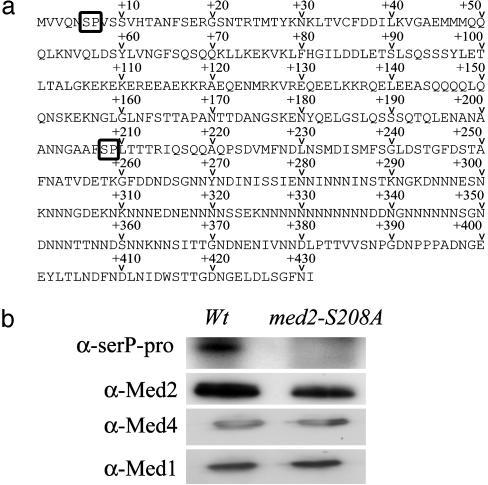
Identification of the Phosphorylation Site in Med2. The predicted amino acid sequence of Med2 contained only two potential CDK-phosphorylation sites, at positions 6 and 208 (Fig. 4a). For technical reasons, we first chose to mutate serine-208 to alanine (see Materials and Methods). Mediator was then affinity purified from a med2-S208A strain expressing a TAP-tagged Rgr1, and proteins were transferred to PVDF membranes. Immunoblotting with anti-Med2, anti-Med4, and anti-Med1 antibodies (Fig. 4b) showed that Med2 was precipitated equally efficiently from extracts isolated from both the WT strain and the med2-S208A mutant. We also blotted the same filter with Pgd1 antibodies to study whether the Med2-S208A mutation had any effect on the interaction between Med2 and Pgd1. However, the anti-Pgd1 antibodies we had available showed crossreactivity to several proteins, which made it impossible to interpret the results (data not shown). Reblotting of the same membrane using the anti-SerP-Pro/Lys antibodies showed that a phosphorylated polypeptide with the same molecular mass as Med2 is present in the WT, but absent in the med2-S208A mutant. This finding demonstrated that the serine-208 to alanine mutation abolished the Srb10-dependent phosphorylation of Med2. Finally, the equally efficient precipitation of Med1, Med2, and Med4 from both extracts showed that phosphorylation of Med2 has no major effect on the subunit composition of Mediator.
Fig. 4.
Mutation of the Med2 serine-208 to alanine abolishes the Srb10-dependent phosphorylation of Med2. (a) The predicted amino acid sequence of Med2. The positions of the two potential CDK-dependent phosphorylation sites are indicated. (b) Mediator was purified from a WT (Wt) strain and from the med2-S208A strain. Both strains also expressed TAP-tagged Rgr1, which was used for affinity purification. Purified Mediator from each strain was immunoblotted with anti-SerP-Pro/Lys, anti-Med2, anti-Med4, and anti-Med1 antibodies as indicated.
The med2-S208A Mutation Strongly Reduces Expression of Genes Encoded on the Endogenous 2-μm Plasmid. To investigate whether the Med2-S208A mutation has a detectable phenotype, we compared its growth to that of the W303-1A parental strain under a number of different growth conditions, for example, growth on galactose or at 37°C. However, the med2-S208A mutant behaved similarly to the WT strain under all conditions tested.
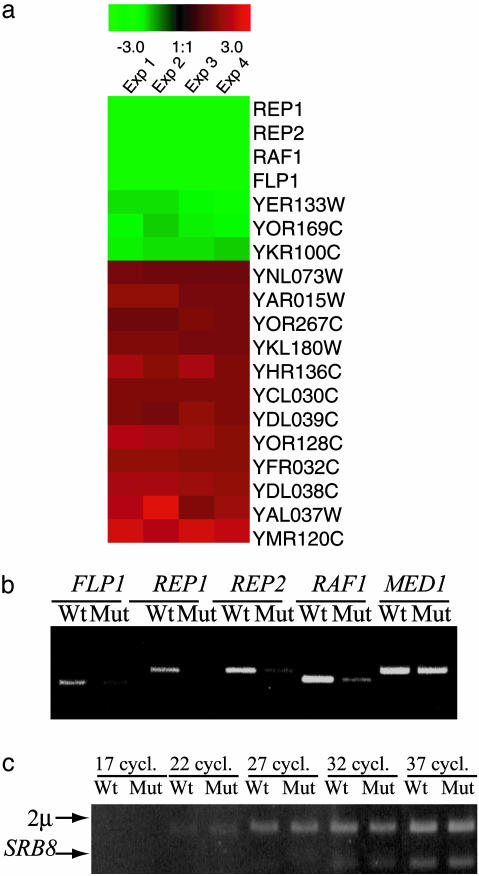
To search for more specific differences between the WT strain and the med2-S208A mutant, whole-genome expression profiles were determined with DNA chip technology (Affymetrix). Differences in specific transcript levels were determined under rich conditions [yeast extract/peptone/dextrose (YPD)]. In agreement with the mild phenotypes observed, the transcription of only a subset of 19 genes was changed >1.4-fold in the med2-S208A mutant (Fig. 5a). Most of these genes were only mildly affected, but the transcription of four genes was dramatically decreased in the med2-S208A mutant strain. The REP1, REP2, FLP1, and RAF1 genes are all encoded for on the endogenous 2-μm plasmid, and the transcription of these genes was high in the WT strain, but essentially quenched in the med2-S208A mutant strain. These results were confirmed by quantitative RT-PCR assays (Fig. 5b). As a control, we also determined the relative levels of the 2-μm plasmid in both strains by using quantitative PCR. Fig. 5c shows that the levels of the 2-μm plasmid are comparable in the two strains. Apparently, a single amino acid substitution at position 208 of Med2 inhibits gene expression from the 2-μm plasmid.
Fig. 5.
Mutation of the Med2 serine-208 to alanine strongly reduces expression of REP1, REP2, FLP1, and RAF1.(a) List of genes whose expression is most affected by the Med2 serine-208 to alanine mutation. Only genes that are down-regulated at least 1.4-fold (green) or up-regulated at least 1.4-fold (red) have been included. The expression analysis was made with the Affymetrix yeast S98 arrays as described in Materials and Methods.(b) Quantification of the FLP1, REP1, REP2, and RAF1 mRNAs in the WT and med2-S208A strains by using RT-PCR. (c) Quantification of the 2-μm plasmid in the WT and med2-S208A strains. An equal amount of genomic DNA isolated from the two strains was used as templates for PCRs with primers specific for the 2-μm plasmid and the SRB8 cDNA. Samples were removed from the PCRs after 17 (lanes 1 and 2), 22 (lanes 3 and 4), 27 (lanes 5 and 6), 32 (lanes 7 and 8), and 37 (lanes 9 and 10) cycles as indicated.
Discussion
Srb10 and Srb11 have been shown to have a negative effect on transcription of a limited set of genes, and it was suggested that this effect operates through phosphorylation of the C-terminal domain before formation of the initiation complex on promoter DNA (17, 28). The Srb10/Srb11 complex has also been shown to operate through mechanisms acting directly on DNA-bound transcriptional activators. Thus, phosphorylation of Gal4, Ste12, Sip4, Gcn4, and Msn2 by Srb10 suggests that modulation of specific transactivators in response to physiological signals is involved in coordinating inducible transcription with the cellular environment (18, 29–31).
In previous experiments, we used fusion proteins where the lexA DNA-binding domain was fused to different Mediator subunits (8, 23). We found that lexA-Med1 was unable to activate transcription in WT cells although the fusion protein was functionally incorporated into the Mediator complex (23). However, lexA-Med1 was a potent activator (400-fold) in cells lacking components of the Srb8-11 complex or in cells overexpressing lexA-Med1 (8). In both cases, the ability of lexA-Med1 to activate transcription depended on Med2. Furthermore, Mediator purified from yeast mutants lacking Med1 also lacks Med2. Taken together, our data demonstrated that the Srb10/Srb11 complex functionally interacts with Med1 and Med2 within the Mediator complex.
We here provide yet another example of a functional interaction between the Srb10/Srb11 complex and Med2. Our results show that Med2 is a target for the Srb10 kinase activity both in vivo and in vitro. It is interesting to note that Med2 physically interacts with Srb10, and not with Srb11, because target specificity in CDK phosphorylation usually is mediated through interaction between the cyclin and the substrate (32, 33). However, it has recently been shown that the activator protein Gal4, which is phosphorylated by Srb10, also physically interacts with the catalytic subunit of the kinase, i.e. Srb10 itself (34).
We performed several unsuccessful experiments searching for a specific growth phenotype of the med2-S208A mutant strain under different conditions. We proceeded with a gene expression study where we examined the expression profiles of the WT and med2-S208A strains by using Affymetrix microarrays. In total, we found 19 genes that were affected >1.4-fold. Most of these genes encoded proteins with diverse functions, and we found no correlation to previous microarray results obtained with cells lacking Med2 (7). However, those experiments described the effects of a deletion of Med2 only when cells were grown in galactose medium and after heat shock but not under normal conditions in rich glucose medium (YPD) as we use here. Surprisingly, the four most strongly affected genes (FLP1, RAF1, REP1, and REP2) are all located on the endogenous 2-μm plasmid. These four genes are highly expressed in the WT strain, but their expression is strongly reduced in the mutant strain. Increased amounts of Flp1 protein have previously been shown to induce 2-μm recombination and thus increase the 2-μm copy number (35). However, PCR analysis showed that the copy number of the 2-μm plasmid in med2-S208A mutant yeast strains was indistinguishable from that of congenic WT strains. Thus, although the phosphorylation status of Med2 regulates FLP1 expression, it does not affect the 2-μm plasmid copy number. The reduced FLP1 expression in the med2-S208A mutant was unexpected because Srb10 plays a negative role in regulating the expression of many genes (36). One might therefore expect that loss of an Srb10-dependent phosphorylation site should have positive effects on gene expression. However, it is not clear whether any of the known negative effects of Srb10 on gene expression is mediated by its phosphorylation of Med2. Interestingly, it was recently reported that deletion of Hst3, a Sir2 histone deacetylase homolog, leads to an up-regulation of FLP1 gene expression (37). Similar to our results presented here, they found that the altered expression of FLP1 was not a result of differences in copy number of the 2-μm plasmid between the WT and mutant strains.
Based on the data presented here, we suggest that posttranslational modifications of Mediator subunits represent a so far uncharacterized mechanism for regulation of gene expression that can function as a molecular switch that controls the ability of Mediator to act as either a corepressor or a coactivator in transcription.
Acknowledgments
We thank Roger Kornberg and Tilman Borggrefe for generous gifts of strains and antibodies. This work was supported by grants from the Swedish Research Council (to S.B., H.R., and C.M.G.), the Swedish Cancer Society, the Swedish Foundation for Strategic Research, and the Human Frontier Science Program (to S.B and C.M.G.), and by the Erik and Mai Pehrsson Foundation (to H.R.), and the Kempe Foundation (to M.H.).
Abbreviations: pol II, RNA polymerase II; PVDF, poly(vinylidene difluoride); CDK, cyclin-dependent kinase; TAP, tandem affinity purification.
References
- 1.Kelleher, R. J., Flanagan, P. M. & Kornberg, R. D. (1990) Cell 61, 1209-1215. [DOI] [PubMed] [Google Scholar]
- 2.Flanagan, P. M., Kelleher, R. J., Sayre, M. H., Tschochner, H. & Kornberg, R. D. (1991) Nature 350, 436-438. [DOI] [PubMed] [Google Scholar]
- 3.Kim, Y. J., Björklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. (1994) Cell, 77, 599-608. [DOI] [PubMed] [Google Scholar]
- 4.Myers, L. C., Gustafsson, C. M., Bushnell, D. A., Lui, M., Erdjument-Bromage, H., Tempst, P. & Kornberg, R. D. (1998) Genes Dev. 12, 45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, Y., Björklund, S., Jiang, Y. W., Kim, Y. J., Lane, W. S., Stillman, D. J. & Kornberg, R. D. (1995) Proc. Natl. Acad. Sci. USA 92, 10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, Y. C. & Kim Y. J. (1998) Mol. Cell. Biol. 18, 5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers, L. C., Gustafsson, C. M., Hayashibara, K. C., Brown, P. O. & Kornberg, R. D. (1999) Proc. Natl. Acad. Sci. USA 96, 67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balciunas, D., Gälman, C., Ronne, H. & Björklund, S. (1999) Proc. Natl. Acad. Sci. USA 96, 376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asturias, F. J., Jiang, Y. W., Myers, L. C., Gustafsson, C. M. & Kornberg, R. D. (1999) Science 283, 985-987. [DOI] [PubMed] [Google Scholar]
- 10.Dotson, M. R., Yuan, C. X., Roeder, R. G., Myers, L. C., Gustafsson, C. M., Jiang, Y. W., Li, Y., Kornberg, R. D. & Asturias, F. J. (2000) Proc. Natl. Acad. Sci. USA 97, 14307-14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koleske, A. J. & Young, R. A. (1994) Nature 368, 466-469. [DOI] [PubMed] [Google Scholar]
- 12.Chao, D. M., Gadbois, E. L., Murray, P. J., Anderson, S. F., Sonu, M. S., Parvin, J. D. & Young, R. A. (1996) Nature 380, 82-85. [DOI] [PubMed] [Google Scholar]
- 13.Boyer, T. G., Martin, M. E., Lees, E., Ricciardi, R. P. & Berk, A. J. (1999) Nature 399, 276-279. [DOI] [PubMed] [Google Scholar]
- 14.Gu, W., Malik, S., Ito, M., Yuan, C., Fondell, J., Zhang, X., Martinez, E., Qin, J. & Roeder, R. G. (1999) Mol. Cell 3, 97-108. [DOI] [PubMed] [Google Scholar]
- 15.Cho, H., Maldonado, E. & Reinberg, D. (1997) J. Biol. Chem. 272, 11495-11502. [DOI] [PubMed] [Google Scholar]
- 16.Rachez, C., Suldan, Z., Ward, J., Chang, C. P., Burakov, D., Erdjument-Bromage, H., Tempst, P. & Freedman, L. P. (1998) Genes Dev. 12, 1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengartner, C. J., Myer, V. E., Liao, S. M., Wilson, C. J., Koh, S. S. & Young, R. A. (1998) Mol. Cell 2, 43-53. [DOI] [PubMed] [Google Scholar]
- 18.Nelson, C., Goto, S., Lund, K., Hung, W. & Sadowski, I. (2003) Nature 421, 187-190. [DOI] [PubMed] [Google Scholar]
- 19.Samuelsen, C. O., Baraznenok, V., Khorosjutina, O., Spahr, H., Kieselbach, T., Holmberg, S. & Gustafsson, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 6422-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, S. J., Lee, J. S., Kang, J. S. & Kim, Y. J. (2001) J. Biol. Chem. 276, 37020-37026. [DOI] [PubMed] [Google Scholar]
- 21.Papamichos-Chronakis, M., Conlan, R. S., Gounalaki, N., Copf, T. & Tzamarias, D. (2000) J. Biol. Chem., 275, 8397-8403. [DOI] [PubMed] [Google Scholar]
- 22.Borggrefe, T., Davis, R., Bareket-Samish, A. & Kornberg, R. D. (2001) J. Biol. Chem. 276, 47150-47153. [DOI] [PubMed] [Google Scholar]
- 23.Balciunas, D., Hallberg, M., Björklund, S. & Ronne, H. (2003) J. Biol. Chem. 278, 3831-3839. [DOI] [PubMed] [Google Scholar]
- 24.Bonneaud, N., Ozier-Kalogeropoulos, O., Li, G. Y., Labouesse, M., Minvielle-Sebastia, L. & Lacroute, F. (1991) Yeast 7, 609-615. [DOI] [PubMed] [Google Scholar]
- 25.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M. & Seraphin, B. (1999) Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt, M. E., Brown, T. A. & Trumpower, B. L. (1990) Nucleic Acids Res. 18, 3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown, N. R., Noble, M. E., Lawrie, A. M., Morris, M. C., Tunnah, P., Divita, G., Johnson, L. N. & Endicott, J. A. (1999) J. Biol. Chem. 274, 8746-8756. [DOI] [PubMed] [Google Scholar]
- 28.Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717-728. [DOI] [PubMed] [Google Scholar]
- 29.Hirst, M., Kobor, M. S., Kuriakose, N., Greenblatt, J. & Sadowski, I. (1999) Mol. Cell 3, 673-678. [DOI] [PubMed] [Google Scholar]
- 30.Chi, Y., Huddleston, M. J., Zhang, X., Young, R. A., Annan, R. S., Carr, S. A. & Deshaies, R. J. (2001) Genes Dev. 15, 1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vincent, O., Kuchin, S., Hong, S. P., Townley, R., Vyas, V. K. & Carlson, M. (2001) Mol. Cell. Biol. 21, 5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulman, B. A., Lindstrom, D. L. & Harlow, E. (1998) Proc. Natl. Acad. Sci. USA 95, 10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown, N. R., Noble, M. E. M., Endicott, J. A. & Johnson, L. N. (1999) Nat. Cell Biol. 1, 438-443. [DOI] [PubMed] [Google Scholar]
- 34.Ansari, A. Z., Koh, S. S., Zaman, Z., Bongards, C., Lehming, N., Young, R. A. & Ptashne, M. (2002) Proc. Natl. Acad. Sci. USA 99, 14706-14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Som, T., Armstrong, K. A., Volkert F. C. & Broach, J. R. (1988) Cell 52, 27-37. [DOI] [PubMed] [Google Scholar]
- 36.Carlson, M. (1997) Annu. Rev. Cell. Dev. Biol. 13, 1-23. [DOI] [PubMed] [Google Scholar]
- 37.Grünweller, A & Ehrenhofer-Murray, A. E. (2002) Genetics 162, 59-71. [DOI] [PMC free article] [PubMed] [Google Scholar]