Abstract
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins constitute the core of the fusion machinery, and isolated SNAREs fuse membranes with exquisite specificity by cognate pairing. Most SNAREs have a membrane-spanning region, an N-terminal domain, and a membrane proximal SNARE motif domain. Although the SNARE motif is critical for SNARE complex formation, is it the sole determinant of the specificity of SNARE-dependent fusion? To test this, we make use of a SNARE complex functioning in the late endosomal compartment in yeast. Studying this complex and the previously identified early endosomal SNARE complex, we find that the specificity of fusion resides in the SNARE motifs.
One fundamental difference between prokaryotes and eukaryotes is the presence of compartments. Prokaryotes have their synthesis machinery, from DNA transcription to protein synthesis, packed within a single membrane, the plasma membrane. Eukaryotes, on the other hand, segregate each step into specific compartments, separated from the others by membranes, with proteins trafficking among them by vesicle budding and fusion. Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are not only responsible for mediating fusion, but they also contribute centrally to its specificity (1–6). In vitro studies of different yeast target SNARE (t-SNARE) complexes [Sso1p/Sec9p (Golgi–plasma membrane), Sed5p/Bos1p,Sec22p (endoplasmic reticulum-Golgi), Sed5p/Gos1p,Ykt6p (intra-Golgi), Vam3p/Vam7p,Vti1p (vacuole fusion), and Tlg2p/Tlg1p,Vti1p (early endosome/trans-Golgi network)] have validated that each membrane t-SNARE is specific for a limited number of vesicle SNAREs (v-SNAREs) (6–8) and therefore allows only the vesicles carrying the matching v-SNARE to fuse. This specificity is independent of any other proteins, demonstrating that specificity is inherent to the SNAREs themselves (6). However, it is not known how the various domains within each SNARE protein establish this specificity.
The cytoplasmic portion of a SNARE has two domains: an N-terminal domain and a SNARE motif. The SNARE motif is a coiled-coil domain of ≈70 amino acids critical for SNARE complex formation. When a t-SNARE and a v-SNARE interact, four SNARE motifs, three from the t-SNARE (one syntaxin and two light-chains, coming from one or two proteins) and one from the v-SNARE, rearrange in a parallel four-helix bundle (9–12). This motif is highly conserved through all isoforms and among species (13, 14). The N-terminal domains, on the other hand, are much more diverse (15). Many t-SNARE N-terminal domains form a three-helix bundle or a β-strand/α-helix structure, which seems to regulate the SNARE assembly when they fold back onto the SNARE motif (15–21).
Here, we ask whether the fusion specificity is encoded in the SNARE motifs or in the N-terminal domains or both. To address this question, we have investigated the SNARE endocytic complexes in a simplified in vitro fusion assay.
Materials and Methods
Plasmid Constructs. The coding sequence of Pep12p was amplified by PCR from Saccharomyces cerevisiae genomic DNA (Novagen) with primers FO5 (GGGCATATCCATATGTCGGAAGACGAATTTTTTGGTGG) and FO6 (CGCGGATCCTTACAATTTCATAATGAG). The PCR product was digested with NdeI and BamHI and ligated either in pET28a vector (Novagen), resulting in FD03, or in pGEX-2T (Amersham Pharmacia), resulting in FD15. The coding sequences of Vti1p, Ykt6p, and Nyv1p were obtained as described (22); Snc1p, Snc2p, and the cytosolic domain of Snc2p were obtained as described (6); Bos1p, Gos1p, Sft1p, Bet1p, and Sec22p were obtained as described (23); and Tlg2p and Tlg1p were obtained as described (8). All plasmids are propagated in Escherichia coli strain DH5α (GIBCO). Truncated forms of each sequence were obtained by PCR amplification with primers FO36 (GGGCATATCATATGACGTTGCAGAGACAGCAACAG) and FO11 (CGGGATCCTCATTTCAACATAACAAAGAA) for ΔTlg2p; FO38 (GGGAATTCCATATGGCTGAGAACAATGATGGT) and FO8 (CGCGGATCCTCAAGCAATGAATGCCAAAAC) for ΔTlg1p; FO44 (GCGGATCCATGAATATTGACGATGACCAAAGGCAA) and FO17 (GCGAATTCCCGGGTTATTTAAACTTTGAGAACAAAAC) for ΔVti1p; and finally FO48 (GGGCATATCATATGAGAGACCCGATAAAACAATAT) and FO6 (CGCGGATCCTTACAATTTCATAATGAG) for ΔPep12p. The PCR products were digested with NdeI/BamHI for ΔTlg2p, ΔTlg1p, and ΔPep12p or with EcoRI/BamHI for ΔVti1p and ligated in pET28a or pGEX-2T, resulting in FD109 (ΔTlg1p), FD113 (ΔPep12p), FD116 (ΔVti1p), and FD117 (ΔTlg2p).
Protein Expression and Purification. Snc2p-his6, Snc1p-his6, GST-Snc2p cytosolic domain, GST-Gos1p, GST-Ykt6p, GST-Sft1p, his6-Vti1p, GST-Vti1p, GST-Nyv1p, his6-Bos1p, his6-Sec22p, his6-Bet1p, his6-Tlg2p, his6-Tlg1p, and his6-Vti1p were prepared as described (6, 8, 22, 24). For GST-Pep12p, GST-ΔTlg2p, GST-ΔTlg1p, GST-ΔPep12p, and GST-ΔVti1p, plasmids used for protein expression were transformed into the E. coli strain BL21star (DE3) (Invitrogen). Transformed cells were grown at 37°C to an absorbance at 600 nm of 0.8, and protein expression was induced with 0.2 mM isopropyl β-d-thiogalactoside (Boehringer Mannheim) for 2 h at 37°C. Cells were collected by centrifugation and lysed in buffer A (8). Lysates were clarified by centrifugation. Lysates containing His-tagged proteins were bound to nickel nitrilotriacetic (Ni-NTA) agarose (Qiagen, Valencia, CA) and washed with buffer B containing 50 mM imidazole; proteins were then eluted with a 50 mM to 1 M imidazole gradient (in buffer B) (8). Lysates containing GST-tagged protein were bound to glutathione agarose beads (Sigma) and washed with buffer B, and proteins were eluted with 20 mM glutathione reduced (Boehringer Mannheim) in buffer B. GST-Sft1p, GST-Pep12p, GST-ΔTlg2p, GST-ΔTlg1p, GST-ΔPep12p, and GST-ΔVti1p were cleaved from GST on liposomes with 0.05 unit/μl thrombin (Sigma).
SNARE Reconstitution into Liposomes. All t-SNAREs were reconstituted from individual proteins as described (8, 25). Briefly, acceptor liposomes were prepared by adding ≈13 nmol of each protein in a volume of 500 μl of 400 mM KCl reconstitution buffer and incubated 20 h at 4°C to form the t-SNARE complex. For donor liposomes, ≈7.5 nmol of v-SNARE in 100 μl was used. All lipids were obtained from Avanti Polar Lipids, and the composition of acceptor and donor liposomes are described in ref. 25. The typical lipid recovery efficiency in the recovered Nycodenz fraction was ≈50% for acceptor liposomes containing Tlg2p and Pep12p complexes and ≈30% for donor liposomes containing Snc2p.
Fusion Assay. The lipid mixing assay was conducted as described (25, 26). Briefly, 45 μl of acceptor liposomes were mixed with 5 μl of donor liposomes in a 96-well FluoroNunc microtiter plate (Nunc). For some experiments, 3.5 nmol of Snc2-C-peptide (8) or 6 nmol of cytosolic domain of Snc2p were added as indicated in the figure legends. Microtiter plates were then placed in a Fluoroscan II Platereader (Labsystems, Chicago) equilibrated at 37°C, and 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) fluorescence was measured over 2 h at 2-min intervals (excitation 460 nm, emission 538 nm). After 2 h, 10 μl of 2.5% wt/vol n-dodecylmaltoside (Boehringer Mannheim) was added to dissolve the lipids and measure the maximum NBD fluorescence. The data were converted to rounds of fusion as described (26). Note that the small decrease observed during the first 10 min of each fusion reaction is caused by the temperature equilibration of the fluorophore.
Results and Discussion
The Late Endocytic Complex Is Composed of Pep12p/Tlg1p,Vti1p, and Snc2p. Along the endocytic pathway, several compartments can be distinguished by the presence of different SNAREs of the syntaxin subfamily: Tlg2p on the trans-Golgi network and on early endosomes (27, 28), Pep12p mainly on the prevacuolar compartment (also present on the trans-Golgi network) (29), and Vam3p on the vacuole (30). However, only the early endocytic and the vacuolar SNARE complexes have been identified in yeast (Tlg2p/Vti1p,Tlg1p fusing with Snc2p and Vam3p/Vti1p,Vam7p fusing with Nyv1p, respectively) and functionally confirmed in vitro (8, 22). We have now investigated the SNARE complex(es) controlling the trafficking to the prevacuolar compartment.
It has been shown that the syntaxin Pep12p interacts with the light chain Vti1p (31). By using this binary complex, we searched for the second light chain as well as the v-SNARE in the fusion assay. This assay consists of two sets of artificial liposomes in which purified SNARE proteins have been reconstituted. In one proteoliposome population, two fluorescent lipids (rhodamine and NBD) have been incorporated at a concentration such that the NBD fluorescence is quenched. When both populations fuse (the labeled and the nonlabeled), the NBD fluorescence increases due to the dequenching of the fluorophore (ref. 25; see ref. 32 for more technical details). By using recombinant proteins expressing GST or His-tag, we preformed different complexes composed of Pep12p/Vti1p and a combination of t-SNARE (X) and v-SNARE (Y) (where X and Y are Ykt6p, Vam7p, Tlg1p, Nyv1p, or Snc2p, the best SNARE candidates to fulfill a role in endocytosis). Each t-SNARE complex was reconstituted into the unlabeled liposome population (also called acceptor liposomes), whereas the v-SNAREs were reconstituted into the labeled liposome population (also called donor liposomes). We tested the fusion capacity of each combination by mixing the t and the v liposomes at 37°C and monitoring the NBD fluorescence. Knowing that some complexes can be locked in a nonfunctional state, we tested all of the possibilities in the presence or absence of an activator, the corresponding v-SNARE peptides (data not shown). These peptides correspond to the membrane proximal C-terminal half of the v-SNARE motif. They bind to the t-SNARE complex, likely prestructuring the SNARE motif into a coil ready to bind the v-SNARE. This conformational change induces an increase of the fusion rate (33).
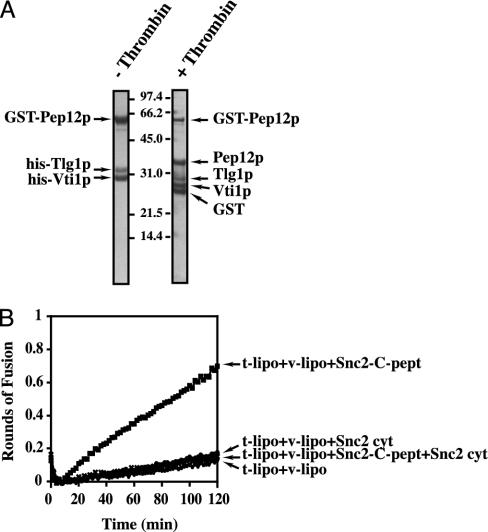
As a result, we have identified a functional complex composed of Pep12p/Tlg1p,Vti1p as the t-SNARE and Snc2p as the v-SNARE (Fig. 1). Note that the Pep12p/Tlg1p,Vti1p complex typically fuses with Snc2p to an average of ≈0.5 rounds of fusion. No fusion is observed after mixing the liposomes together unless a C-peptide corresponding to the v-SNARE is added to the fusion reaction (Fig. 1B). Such an absolute requirement for the complex to be activated to fuse has previously been observed for SNAREs marking early endosomes (8). As expected, this fusion is SNARE-dependent, because the cytosolic v-SNARE is able to compete with the reconstituted v-SNARE (Fig. 1B). Thus, an endocytic SNARE complex constituted by the t-SNARE Pep12p/Tlg1p,Vti1p and the v-SNARE Snc2p is functionally active and is likely involved in the transport of soluble enzymes from the trans-Golgi network to the prevacuolar compartment (29). During this investigation, the in vivo interaction of these same four proteins was reported, confirming the relevance of this complex in the cell (34).
Fig. 1.
Pep12p/Tlg1p, Vti1p, and Snc2p form a functional fusogenic complex. (A) Reconstitution of acceptor and donor liposomes. GST-Pep12p, his-Tlg1p, and his-Vti1p were preincubated to form a complex and then reconstituted into acceptor liposomes. After liposome flotation, the GST moiety was cleaved with thrombin (0.05 unit/μl) during 2 h at room temperature. Ten percent (4.5 μl) of the acceptor liposomes used in a typical fusion reaction were analyzed by SDS/PAGE and Coomassie blue staining (before and after thrombin cleavage). Only the thrombin-cleaved population was used in the fusion assay. Note that the full-length GST-Pep12p remaining after thrombin cleavage is facing the inside of the liposome and therefore is not accessible to the enzyme. (B) Membrane fusion. Donor and acceptor liposomes were mixed (5:45 μl) in a microtiter plate. Depending on the experiment, 5 μl of snc2-C-pept (3.5 nmol) plus 5 μl of buffer, 5 μl of snc2p cytosolic domain (6 nmol) plus 5 μl of buffer, or 5 μl snc2-C-pept plus 5 μl of snc2p cytosolic domain were added in the reaction as indicated on the figure. The plate was transferred to a 37°C fluorescent plate reader, and NBD fluorescence was monitored and converted to rounds of fusion as described (26).
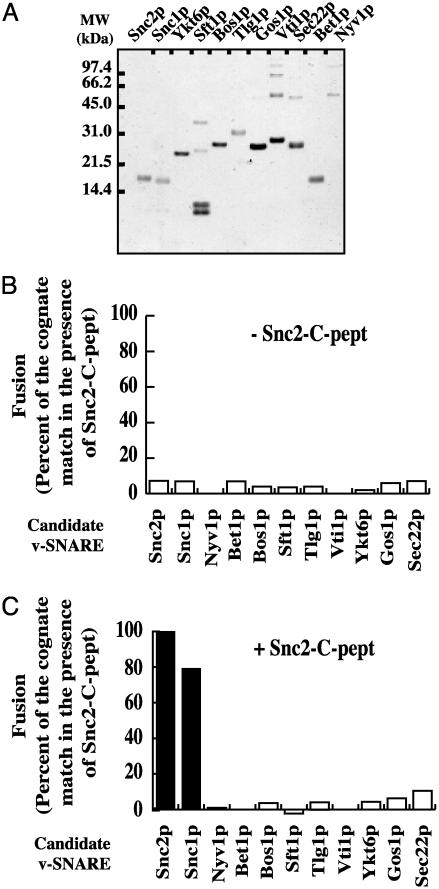
The Late Endocytic Complex Is Specific for Snc2p. Many vacuolar proteins such as carboxypeptidase Y go from the trans-Golgi network through the prevacuolar compartment before reaching the vacuole. The prevacuolar compartment also receives proteins from the early endosomes and the vacuole as well as from the plasma membrane. It has been shown that all these pathways require the function of the endosomal syntaxin Pep12p, suggesting that Pep12p is involved in more than one complex (35). In this study, we kept the t-SNARE composition constant (Pep12p/Tlg1p,Vti1p) and tested the fusion capacity of all of the potential v-SNAREs. Therefore, we reconstituted all of the v-SNAREs (non-syntaxin SNAREs) in donor liposomes (Fig. 2A) and tested their fusion capacity with Pep12p/Tlg1p,Vti1p t-SNARE acceptor liposomes. All of these experiments were done in the absence (Fig. 2B) and in the presence of Snc2-C-peptide (Fig. 2C) or of the corresponding peptide (data not shown). None of the v-SNAREs is able to mediate fusion with the Pep12 complex in the absence of peptide (Fig. 2B). On the other hand, only Snc2p and Snc1p (which are close homologues, 77% identical and genetically interchangeable) (36) are able to mediate fusion in the presence of peptide (Fig. 2C). Therefore, Pep12p must function as a common heavy chain in multiple distinct t-SNARE complexes in the endocytic pathway. Interestingly, additional interactions between Pep12p and two other SNAREs, Ykt6p and Syn8p, were recently found (34). This is reminiscent of the role of Sed5p as the common heavy chain in Golgi t-SNAREs (23, 26).
Fig. 2.
Pep12p/Tlg1p,Vti1p is specific for Snc2p/Snc1p v-SNAREs. (A) Characterization of v-SNARE liposomes. Donor liposomes (5 μl) were loaded and stained by Coomassie blue. GST was cleaved from Sft1p after flotation (resulting bands on the gel are GST-Sft1p, GST, Sft1p, and a degradation product). Based on their molecular mass, Vti1p and Sec22p high molecular bands are likely multimers (6). (B) Fusion between t-SNARE liposomes and all potential v-SNARE liposomes in absence of peptide. Pep12p/Tlg1p,Vti1p acceptor liposomes were incubated with each v-SNARE donor liposome (45:5), and the normalized fluorescence signal after 2 h at 37°C was plotted. (C) Fusion between t-SNARE liposomes and all potential v-SNARE liposomes in the presence of Snc2-C-pept. t-SNARE Pep12p/Tlg1p,Vti1p acceptor liposomes were incubated with each v-SNARE donor liposomes (45:5). Snc2-C-pept (3.5 nmol) was added to all reactions immediately before the fusion started. The normalized fluorescence signal obtained after2his represented. For B and C, the extent of fusion at 2 h was normalized to the amount of cognate v-SNARE fusion in the presence of peptide (Snc2p-black bars in C). Fusion with protein-free liposomes was subtracted from all of the fusion results. The maximal extent of fusion (100%) with Snc2p donor liposomes and snc2-C-pept was 0.512 rounds. Ykt6p was anchored to the membrane by the attachment of a synthetic geranylgeranyl lipid anchor as described (37). This experiment is representative of three different assays.
v-SNARE Specificity Is Governed by the SNARE Motif. All syntaxins have a predominant N-terminal domain. Interestingly, each of the light chains belonging to the late endocytic t-SNARE complex also possesses a relatively large N-terminal domain (≈10 kDa). It is possible that one or another of these three N-terminal domains creates a surface that could bind the v-SNARE or control its accessibility to the SNARE motifs of the t-SNARE and thus contribute to the specificity of fusion. To test this, we deleted all three N-terminal domains. We cloned and expressed the truncated forms of each t-SNARE protein Pep12p183–289 (ΔPep12p), Tlg1p118–224 (ΔTlg1p), and Vti1p111–216 (ΔVti1p) as GST-recombinant proteins. We reconstituted the truncated complex into the acceptor liposome and characterized its fusion capacity with Snc2p liposomes. Interestingly, the truncated t-SNARE complex ΔPep12p/ΔTlg1p,ΔVti1p still requires activation by the C-peptide (Fig. 3A), showing that the apparent off-state of this complex is not entirely due to autoinhibition by its N-terminal domains. As it has already been shown that the peptide acts by binding the membrane-proximal portion of the t-SNARE (8, 33), this result suggests that the critical peptide-dependent activation switch resides on the SNARE motif itself.
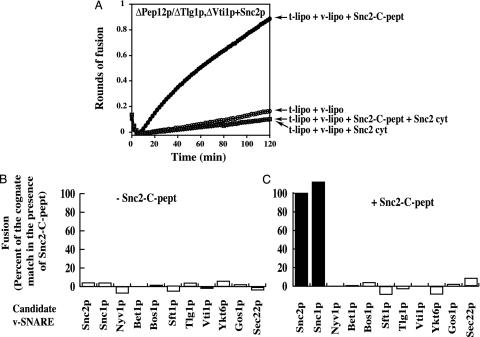
Fig. 3.
t-SNARE ΔPep12p/ΔTlg1p,ΔVti1p remains specific for Snc1p/Snc2p v-SNAREs. (A) Characterization of the t-SNARE ΔPep12p/ΔTlg1p,ΔVti1p fusion capacity. GST-ΔPep12p, GST-ΔTlg1p, and GST-ΔVti1p were preincubated to form a complex and then reconstituted into acceptor liposomes. After flotation, the GST moiety was thrombin-cleaved from the liposomes. Donor and cleaved acceptor liposomes were mixed (5:45 μl) in a microtiter plate with Snc2-C-pept plus buffer, Snc2p cytosolic domain plus buffer, or Snc2-C-pept plus Snc2p cytosolic domain as indicated on the figure. The plate was transferred to a 37°C fluorescent plate reader, and the converted fluorescence (rounds of fusion) was plotted. (B and C) Membrane fusion between ΔPep12p/ΔTlg1p,ΔVti1p t-SNARE liposomes and potential v-SNARE liposomes in the absence or presence of Snc2p-C-pept. t-SNARE ΔPep12p/ΔTlg1p,ΔVti1p acceptor liposomes were incubated with each v-SNARE donor liposome (45:5) in the absence (B) or presence (C) of peptide. The fluorescence signal was monitored over2hat37°C. As in Fig. 2, the extent of fusion at 2 h was normalized to the amount of cognate v-SNARE fusion in the presence of peptide (Snc2p, black bars in C). Fusion with protein-free liposomes was subtracted from all of the fusion results. The maximal extent of fusion (100%) with Snc2p donor liposomes and snc2-C-pept was 0.925 rounds. This experiment is representative of at least three different assays.
We also investigated whether the truncated complex, freed of N-terminal domains, retained the specificity of the full-length complex. For this purpose, we tested the fusion capacity of ΔPep12p/ΔTlg1p,ΔVti1p with a panel of all of the potential v-SNAREs in yeast in the presence or in the absence of the Snc2-C peptide. As demonstrated in Fig. 3, the specificity of fusion is fully conserved in the absence of any of the N-terminal domains. Once again, no fusion is observed in the absence of peptide.
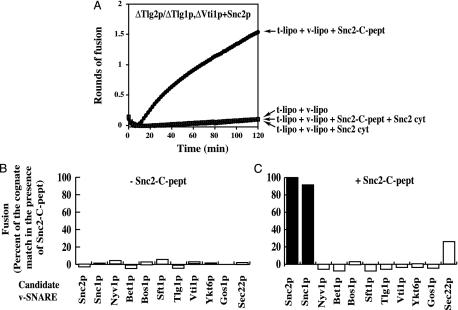
To confirm that this result is not unique to the late endocytic complex, we investigated a second t-SNARE complex Tlg2p/Tlg1p,Vti1p involved in the early endocytic pathway (8). We cloned and expressed Tlg2p231–334 (ΔTlg2p) and reconstituted the truncated t-SNARE complex ΔTlg2p/ΔTlg1p,ΔVti1p into acceptor liposomes. As for the truncated Pep12 complex, ΔTlg2 complex still requires activation to be fusogenic (Fig. 4A), suggesting that both complexes may have analogous regulatory mechanisms. Fig. 4C shows that the v-SNARE specificity is fully retained in the absence of the N-terminal domains (note that the small amount of fusion observed with Sec22p in the presence of Snc2-C-pept is also observed with the full-length complex; see figure 6a in ref. 8) but disappeared in the presence of the specific peptide Sec22-C-pept (see figure 6b in ref. 8).
Fig. 4.
t-SNARE ΔTlg2p/ΔTlg1p,ΔVti1p remains specific for Snc1p/Snc2p v-SNAREs. (A) Characterization of ΔTlg2p/ΔTlg1,ΔVti1p fusion capacity. GST-ΔTlg2p, GST-ΔTlg1p, and GST-ΔVti1p were preincubated to form a complex and reconstituted into acceptor liposomes, and the GST moiety was thrombin-cleaved from the liposomes after flotation. Donor and cleaved acceptor liposomes were mixed (5:45 μl) in a microtiter plate in different conditions as indicated. During 2 h at 37°C, the NBD fluorescence was monitored before being converted to rounds of fusion. (B and C) Membrane fusion between ΔTlg2p/ΔTlg1p,ΔVti1p t-SNARE liposomes and all potential v-SNARE liposomes in the absence or presence of Snc2p-C-pept. t-SNARE ΔTlg2p/ΔTlg1p,ΔVti1p acceptor liposomes were incubated with each v-SNARE donor liposome (45:5) in the absence (B) or presence (C) of Snc2-C peptide. The normalized fluorescence signal after2hat37°C was plotted. Fusion with protein-free liposomes was subtracted from all of the fusion results. The maximal extent of fusion (100%) with Snc2p donor liposomes and Snc2-C-pept was 1.61 rounds. This experiment is representative of at least three different assays.
Conclusion. With isolated SNAREs, t-SNARE N-terminal domains do not influence the specificity of fusion, suggesting that the SNARE motif is solely responsible for specificity at this stage of the process. However, the N-terminal domains are likely involved in controlling the rate of the complex formation and fusion (15–20, 26); in this respect, additional regulatory proteins may contribute to influence them.
Acknowledgments
We thank the members of the Rothman laboratory, especially Peter Antinozzi and Thalia Becker, for critical comments on the manuscript; Thomas Melia for all our stimulating discussions and for helpful critiques of the manuscript; Sonia Sequeira and James McNew for great input during the writing of this paper; and Thomas Söllner for the critical reading of the manuscript. We particularly acknowledge Hedy Druskin for her efficient management of the laboratory, which made this project so much easier to carry on. This research was supported by a National Institutes of Health grant (to J.E.R.).
Abbreviations: SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; t-SNARE, target SNARE; v-SNARE, vesicle SNARE; NBD, 7-nitrobenz-2-oxa-1,3-diazol-4-yl.
References
- 1.Ungar, D. & Hughson, F. M. (2003) Annu. Rev. Cell Dev. Biol. 19, 493-517. [DOI] [PubMed] [Google Scholar]
- 2.Duman, J. G. & Forte, J. G. (2003) Am. J. Physiol. 285, C237-C249. [DOI] [PubMed] [Google Scholar]
- 3.Lodish, H., Berk, A., Matsudaira, P., Kaiser, C. A., Krieger, M., Scott, M. P., Zipursky, L. & Darnell, J. (2003) Molecular Cell Biology (Freeman, New York).
- 4.Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K. & Walter, P. (2002) Molecular Biology of the Cell (Taylor & Francis, New York).
- 5.Scales, S. J., Chen, Y. A., Yoo, B. Y., Patel, S. M., Doung, Y. C. & Scheller, R. H. (2000) Neuron 26, 457-464. [DOI] [PubMed] [Google Scholar]
- 6.McNew, J. A., Parlati, F., Fukuda, R., Johnston, R. J., Paz, K., Paumet, F., Sollner, T. H. & Rothman, J. E. (2000) Nature 407, 153-159. [DOI] [PubMed] [Google Scholar]
- 7.Parlati, F., Varlamov, O., Paz, K., McNew, J. A., Hurtado, D., Sollner, T. & Rothman, J. E. (2002) Proc. Natl. Acad. Sci. USA 99, 5424-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paumet, F., Brugger, B., Parlati, F., McNew, J. A., Sollner, T. H. & Rothman, J. E. (2001) J. Cell Biol. 155, 961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canaves, J. & Montal, M. (1998) J. Biol. Chem. 273, 34214-34221. [DOI] [PubMed] [Google Scholar]
- 10.Poirier, M. A., Xiao, W., Macosko, J. C., Chan, C., Shin, Y. K. & Bennett, M. K. (1998) Nat. Struct. Biol. 5, 765-769. [DOI] [PubMed] [Google Scholar]
- 11.Sutton, R., Fasshauer, D., Jahn, R. & Brunger, A. (1998) Nature 395, 347-353. [DOI] [PubMed] [Google Scholar]
- 12.Antonin, W., Fasshauer, D., Becker, S., Jahn, R. & Schneider, T. R. (2002) Nat. Struct. Biol. 9, 107-111. [DOI] [PubMed] [Google Scholar]
- 13.Weimbs, T., Low, S. H., Chapin, S. J., Mostov, K. E., Bucher, P. & Hofmann, K. (1997) Proc. Natl. Acad. Sci. USA 94, 3046-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock, J. B., Matern, H. T., Peden, A. A. & Scheller, R. H. (2001) Nature 409, 839-841. [DOI] [PubMed] [Google Scholar]
- 15.Misura, K. M. S., Bock, J. B., Gonzalez, L. C., Jr., Scheller, R. H. & Weis, W. I. (2002) Proc. Natl. Acad. Sci. USA 99, 9184-9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tochio, H., Tsui, M. M. K., Banfield, D. K. & Zhang, M. (2001) Science 293, 698-702. [DOI] [PubMed] [Google Scholar]
- 17.Dulubova, I., Yamaguchi, T., Wang, Y., Sudhof, T. C. & Rizo, J. (2001) Nat. Struct. Biol. 8, 258-264. [DOI] [PubMed] [Google Scholar]
- 18.Munson, M., Chen, X., Cocina, A. E., Schultz, S. M. & Hughson, F. M. (2000) Nat. Struct. Biol. 7, 894-902. [DOI] [PubMed] [Google Scholar]
- 19.Antonin, W., Dulubova, I., Arac, D., Pabst, S., Plitzner, J., Rizo, J. & Jahn, R. (2002) J. Biol. Chem. 277, 36449-36456. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez, L. C., Jr., Weis, W. I. & Scheller, R. H. (2001) J. Biol. Chem. 276, 24203-24211. [DOI] [PubMed] [Google Scholar]
- 21.Dietrich, L. E. P., Boeddinghaus, C., LaGrassa, T. J. & Ungermann, C. (2003) Biochim. Biophys. Acta 1641, 111-119. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda, R., McNew, J. A., Weber, T., Parlati, F., Engel, T., Nickel, W., Rothman, J. E. & Sollner, T. H. (2000) Nature 407, 198-202. [DOI] [PubMed] [Google Scholar]
- 23.Parlati, F., McNew, J. A., Fukuda, R., Miller, R., Sollner, T. H. & Rothman, J. E. (2000) Nature 407, 194-198. [DOI] [PubMed] [Google Scholar]
- 24.McNew, J. A., Coe, J. G., Sogaard, M., Zemelman, B. V., Wimmer, C., Hong, W. & Sollner, T. H. (1998) FEBS Lett. 435, 89-95. [DOI] [PubMed] [Google Scholar]
- 25.Weber, T., Zemelman, B., McNew, J., Westermann, B., Gmachl, M., Parlati, F., Söllner, T. & Rothman, T. (1998) Cell 92, 759-772. [DOI] [PubMed] [Google Scholar]
- 26.Parlati, F., Weber, T., McNew, J. A., Westermann, B., Sollner, T. H. & Rothman, J. E. (1999) Proc. Natl. Acad. Sci. USA 96, 12565-12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abeliovich, H., Grote, E., Novick, P. & Ferro-Novick, S. (1998) J. Biol. Chem. 273, 11719-11727. [DOI] [PubMed] [Google Scholar]
- 28.Holthuis, J. C., Nichols, B. J., Dhruvakumar, S. & Pelham, H. R. (1998) EMBO J. 17, 113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becherer, K. A., Rieder, S. E., Emr, S. D. & Jones, E. W. (1996) Mol. Biol. Cell 7, 579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada, Y., Nakamura, N., Ohsumi, Y. & Hirata, A. (1997) J. Cell Sci. 110, 1299-1306. [DOI] [PubMed] [Google Scholar]
- 31.Fisher von Mollard, G. F., Nothwehr, S. F. & Stevens, T. H. (1997) J. Cell Biol. 137, 1511-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott, B. L., Van Komen, J. S., Liu, S., Weber, T., Melia, T. J. & McNew, J. A. (2003) Methods Enzymol. 372, 274-300. [DOI] [PubMed] [Google Scholar]
- 33.Melia, T. J., Weber, T., McNew, J. A., Fisher, L. E., Johnston, R. J., Parlati, F., Mahal, L. K., Sollner, T. & Rothman, J. E. (2002) J. Cell Biol. 158, 929-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, M. J. & Pelham, H. R. (2002) Traffic 3, 922-929. [DOI] [PubMed] [Google Scholar]
- 35.Gerrard, S. R., Levi, B. P. & Stevens, T. H. (2000) Traffic 1, 259-269. [DOI] [PubMed] [Google Scholar]
- 36.Protopopov, V., Govindan, B., Novick, P. & Gerst, J. E. (1993) Cell 74, 855-861. [DOI] [PubMed] [Google Scholar]
- 37.McNew, J. A., Weber, T., Parlati, F., Johnston, R. J., Melia, T. J., Sollner, T. H. & Rothman, J. E. (2000) J. Cell Biol. 150, 105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]