Abstract
The biosynthesis in vitro of phosphorylcholine oligosaccharides in Caenorhabditis elegans has been investigated. Here we show that extracts of C. elegans' microsomes transfer phosphorylcholine from L-α-dipalmitoyl phosphatidylcholine to hybrid and complex type N-linked oligosaccharides containing mannose residues disubstituted with N-acetylglucosamine. The reaction products are consistent with structures reported for C. elegans as well those found in the filarial nematodes Acanthocheilonema viteae, Onchocerca volvulus, and Brugia malayi, strongly supporting the concept that the phosphorylcholine oligosaccharide biosynthetic enzymes are conserved in this group of organisms. Because it is thought that phosphorylcholine substitution of oligosaccharides modulates host immune response in filarial infections, this in vitro system may help in gaining an understanding of the basis for this response.
An important strategy used by filarial nematodes to avoid host immune response is to esterify their secreted N-glycoproteins and glycosphingolipids with phosphorylcholine. Humans are infected by eight species of filarial nematodes; of these, Wuchereria bancrofti, Brugia malayi, and Onchocerca volvulus infect 150 million humans with one billion more at risk (1). Phosphorylcholine-substituted oligosaccharide N-glycans (PCOs) have been detected in many of these species (2, 3) and their structures are all very similar. Caenorhabditis elegans, a well characterized free-living nematode, also contains PCOs (4–6). Because its genome has been sequenced and many glycosyltransferases (7–9) and some nucleotide sugar transporters have been described (10, 11), C. elegans offers an attractive system to study PCO biosynthesis. Previous studies on Acanthocheilonema vitae PCO biosynthesis in vivo by Harnett and colleagues (12, 13) suggest that either phosphatidylcholine (PTC) or CDP-choline is the phosphorylcholine donor of the PCO-modified secreted N-glycoprotein ES-62 and that the site of phosphorylcholine addition may be the Golgi lumen.
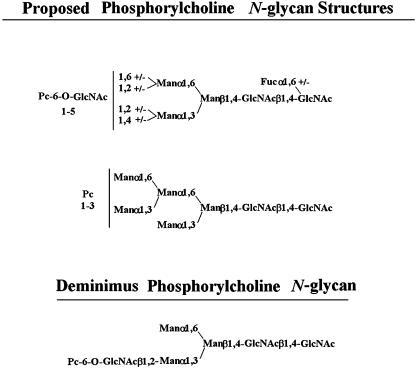
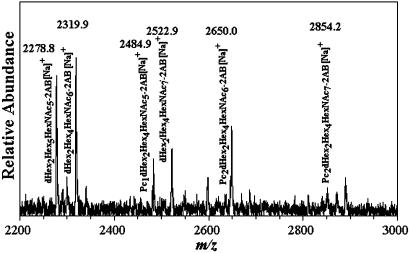
Three studies have reported the occurrence of PCOs in C. elegans (4–6) as shown in Scheme 1. By studying individual life stages of this nematode, we observed in Dauer larvae a series of PCOs from PC1Hex3HexNAc3 to PC2 deoxyHex2Hex4HexNAc7. Fig. 1 shows a matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) MS spectrum of 2-aminobenzamide-labeled C. elegans Dauer larvae N-glycans. The sodium-substituted molecular ions observed at m/z 2484.9, 2650.0, and 2854.2 are consistent with Pc1deoxyHex2Hex4HexNAc6-2AB, Pc2deoxyHex2Hex4HexNAc6-2AB, and Pc2deoxyHex2-Hex4HexNAc7-2AB, respectively, and demonstrate that relatively high molecular weight PCOs exist in C. elegans. In another study, we found that phosphorylcholine can substitute not only terminal N-acetylglucosamine as previously thought (2, 4, 6, 14, 15) but also core N-acetylglucosamine residues (data not shown). Based on the above structural and metabolic results, we have developed a system in vitro to study PCO biosynthesis in C. elegans. We show that phosphorylcholine is derived from PTC and that specific oligosaccharides similar to the PCOs found in vivo are optimal acceptors.
Scheme 1.
C. elegans phosphorylcholine oligosaccharides. The top structure is that proposed by Haslam et al. (4). Phosphorylcholine-6-O-substitution is present on up to five N-acetylglucosamine residues. The center structure is the PCO proposed by Cipollo et al. (5). The position of the phosphorylcholine substitutions are unknown, but core N-acetylglucosamine and mannose substitutions are both possible. The bottom structure is the diminimus PCO reported by Haslam et al. (4).
Fig. 1.
MALDI-TOF MS of phosphorylcholine oligosaccharides from C. elegans Dauer larvae obtained as described in Materials and Methods. The composition and m/z of the molecular ions detected are shown.
Materials and Methods
Materials. All chemicals were of reagent grade or better. PTC, l-α-dipalmitoyl[choline-methyl-3H] [60 Ci/mmol (1 Ci = 37 GBq)], CDP-choline[choline-methyl-3H] (80 Ci/mmol), and sphingomyelin [choline-methyl-3H] (60 Ci/mmol) were purchased from American Radiolabeled Chemicals (St. Louis). Acceptor structures are shown in Table 2. Bovine IgG and chicken ovalbumin were purchased from Sigma–Aldrich. Compound 1 (see Table 2 for compound structures) was from a previous study (5). Compound 2 was purchased from Sigma. Compounds 3 and 4 were released from ovalbumin with endoglycosidase H (endo H; New England Biolabs), and compounds 6–8 and 10 were released with peptide N-glycosidase F (New England Biolabs). Ovalbumin-derived oligosaccharides were isolated by sequential chromatographic separation. First, glycans were separated on a 1.0 cm × 120 cm Bio-Gel P-4 column calibrated with authentic oligosaccharide standards from S. cerevisiae. Individual glycan species were enriched by chromatographic separation on porous graphite (Hypersil) (16). The HPLC system included a Waters 1525 binary pump and a 2407 dual-wavelength absorbance detector. Compound 5 was obtained by peptide N-glycosidase F release from bovine IgG after digestion with bovine testes β-galactosidase (Glyko, San Leandro, CA), passed through a Sep Pak C-18 (Waters) cartridge, and was finally isolated with the porous graphite system described above. Compound 9 was obtained by digestion of Gal4GlcNAc4Man3GlcNAc2 (Sigma–Aldrich) with bovine testes β-galactosidase.
Table 2. Acceptor specificity of oligosaccharide phosphorylcholine transferase activity of C. elegans microsomes.
| Acceptor | Product, cpm | |
|---|---|---|
| 1. | 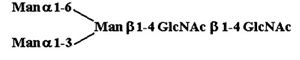 |
12,010 ± 2,050 |
| 2. | 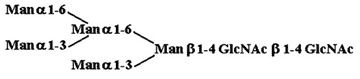 |
300 ± 2,020 |
| 3. | 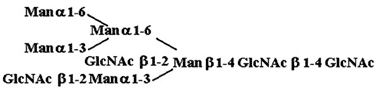 |
200 ± 1,830 |
| 4. | 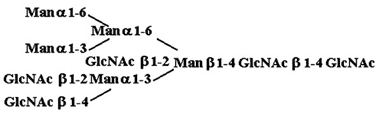 |
80,630 ± 2,620* |
| 5. | 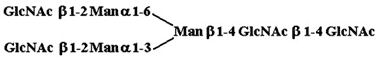 |
1,190 ± 410 |
| 6. | 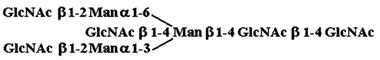 |
240 ± 2,410 |
| 7. | 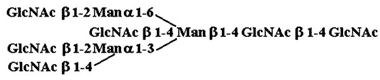 |
6,520 ± 3,260 |
| 8. | 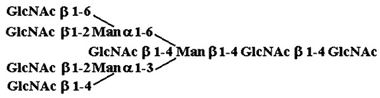 |
52,760 ± 1,440 |
| 9. | 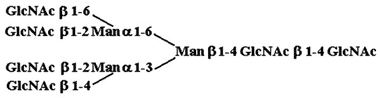 |
55,830 ± 1,910 |
| 10. | 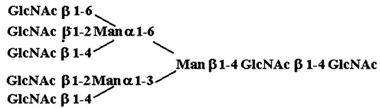 |
32,860 ± 1,930 |
Concentration of phosphatidylcholine donor was 100 μM, whereas the acceptor concentration was 60 μM. Reactions, including without acceptors, were performed for 30 minutes at 30°C in duplicate. All compounds were 95% pure or greater, except compound 4, which could not be purified in sufficient quantities for testing in pure form. The activity reported for this compound is that obtained with a mixture of substrates used in Table 1.
Only compound 4 was a substrate based on purification and testing of each of the other substrates individually.
Phosphorylcholine: Oligosaccharide Transferase Assay. The incubation mixture contained 2.5 mM 5′AMP, 20 mM MnCl2, 1 mg/ml BSA, 0.1% Triton X-100, 10 mM DTT, 20 mM GlcNAc, 100 mM 2[N-morpholino]ethanesulfonic acid (pH 6.8), and 11 mg/ml C. elegans Bristol N2 strain-derived microsomes, prepared as described in ref. 18. The concentration of choline donors and acceptors was 100 μM and 50 μM, respectively. Reactions were carried out for 30 min at 30°C, unless indicated otherwise, and were stopped by flash freezing in a methanol bath. Products were isolated by application of the reaction mixture to a prewashed Sep Pak C-18 cartridge and were eluted with distilled water. Aliquots were subjected to liquid scintillation spectrometry or used for structural analyses.
MS of Reaction Products. Reactions products were lyophilized and then reductively aminated with 2-aminobenzamide (17). After paper chromatography in acetonitrile, the aminated products were visualized by using a BLAK-RAY UVL-21 lamp. Products were eluted from the paper with 1% 1-butanol and evaporated. The particulate material was removed with a Costar 0.22-μm nylon filter. The oligosaccharides were redissolved in 5 μl of acetonitrile, and 0.5-μl aliquots were spotted on a MALDI target with an equal volume of 20 mg/ml 2,5-dihydroxybenzoic acid containing 20 mM sodium acetate. Samples were analyzed with a Bruker Reflex IV MALDI-TOF MS instrument operated in the positive reflectron mode with a nitrogen laser (337 nm, 3-nsec pulse width). Spectra shown are the sum of 50–100 shots.
Results
Strategy for Identification of Substrates for Phosphorylcholine Oligosaccharides of C. elegans. We postulated that PTC, CDP-choline, and sphingomyelin could be possible phosphorylcholine donors, the first two selections being based on previous studies on the biosynthesis of sphingomyelin and PTC (18). Our general strategy was first to test the transfer in vitro of the above donors to a mixture of endo H-released, ovalbumin-derived oligosaccharides that would be expected to contain possible acceptor substrates, based on the above described structural studies. Once a donor substrate was identified, purified oligosaccharides derived from endo H or N-glycosidase F-treated α-acid glycoprotein ovalbumin and bovine IgG were tested as specific acceptors. The glycan structures of these glycoproteins are well known and occur as part of PCOs.
Identification of PTC As Phosphorylcholine Donor. The major phospholipids of C. elegans are phosphatidylethanolamine (54%), PTC (32%), and sphingomyelin (8%) (19). Thin-layer chromatographic analyses of microsomal membranes essentially confirmed the expected phospholipid composition (data not shown).
In separate incubations, either choline radiolabeled CDP-choline, PTC, or sphingomyelin was used as a substrate, and a mixture of endo H-released oligosaccharides from ovalbumin were introduced as acceptors. As shown in Table 1 (lines 1–3), both CDP-choline and PTC appeared to be effective donors of phosphorylcholine.
Table 1. Evaluation of donor substrates for C. elegans PCO synthesis in vitro.
| Donor substrates | Product, cpm* | Product, pmol |
|---|---|---|
| PTC | 200,650 ± 170 | 1,000 |
| Sphingomyelin | 18,640 ± 2,090 | 92 |
| CDP-choline | 132,430 ± 4,550 | 2,600 |
| [3H]PTC alone | 18,960 ± 1,410 | 95 |
| [3H]PTC + 3-fold PTC | 5,350 ± 440 | 112 |
| [3H]PTC + 3-fold CDP-choline | 25,090 ± 1,830 | 126 |
Shown is donor specificity of phosphorylcholine oligosaccharide transferase activity of C. elegans microsomes. Concentration of donor substrates was 100 μM. Specific activities of the donors were as follows: [3H]PTC, 2 Ci/mmol; [3H]sphingomyelin, 2 Ci/mmol; and [3H]CDP-choline, 500 mCi/mmol. The acceptor substrate mixture contained Man5GlcNAc1 (20%), Man6GlcNAc1 (25%), Man5GlcNAc3 (25%), Man4GlcNAc3 (10%), and Man5GlcNAc4 (20%). Total acceptor concentration was 60 μM. Reactions, including without acceptors, were performed for 30 min at 30°C in duplicate. Incubations for dilution studies (lines 4–6) were performed as others, except that in one case either nonradioactive PTC (15 nmol) or CDP-choline (15 nmol) was added to a final concentration of 0.4 mM and 0.3 mM, respectively. Dilution reactions were performed in duplicate for 7 min at 30°C.
Data are cpm ± SEM
We next determined whether PTC or CDP-choline was the direct donor of phosphorylcholine. We reasoned that the phosphorylcholine moiety derived from CDP-choline would first be converted to PTC and that, after a time lag, phosphorylcholine would be incorporated into oligosaccharides. This hypothesis was based on the fact that most of the oligosaccharide acceptors are synthesized in the lumen of the Golgi apparatus and that no transport system across the endoplasmic reticulum or Golgi apparatus membrane has been described for CDP-choline (which is synthesized in the cytosol). PTC, on the other hand, is synthesized on the cytosolic side of the endoplasmic reticulum membrane and its head group is translocated to the luminal side of the endoplasmic reticulum membrane bilayer (18).
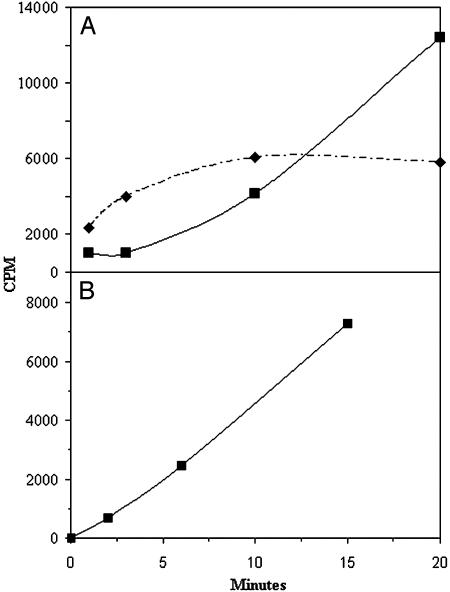
Two experimental approaches were used to demonstrate that PTC was the direct phosphorylcholine donor in the above reaction. We reasoned that, during incubations with choline radiolabeled PTC as described in Table 1, the addition of excess nonradiolabeled PTC (line 5) would result in lower amounts of radioactive PCOs (as a result of the lower specific activity of the donor PTC), whereas addition of CDP-choline (line 6) in amounts similar to the nonradioactive PTC added to the above incubation would have no effect. As shown in Table 1 (lines 4–6), this was indeed the case. The reason for the slight stimulation by CDP-choline of radiolabeled phosphorylcholine incorporation in the latter reaction is not clear but may be related either to increased stability of PTC, decreased degradation of products, or perhaps a direct stimulation of the transferase. A second approach demonstrating that PTC is the direct donor of phosphorylcholine in the reactions shown in Table 1 is shown in Fig. 2. Fig. 2 shows that phosphorylcholine derived from CDP-choline was first incorporated into PTC and thereafter into the oligosaccharide acceptors. No such time lag was observed when choline-radiolabeled PTC was used as substrate, as shown in Fig. 2. From these studies, we conclude that PTC is most likely the in vivo donor of phosphorylcholine to the oligosaccharides.
Fig. 2.
Phosphorylcholine transfer to oligosaccharides. (A) Microsomes were incubated for 1, 3, 10, and 20 min at 30°C with CDP-[methyl-3H]choline and an endo H-released ovalbumin oligosaccharide mixture as described in Materials and Methods. The dashed line shows radioactive lipid, and the solid line shows radioactive oligosaccharides. (B) Microsomes were incubated for 2, 7, and 15 min at 30°C with PTC l-α-dipalmitoyl[choline-methyl-3H] and the same oligosaccharide mixture as A. Appearance of radioactivity in oligosaccharides is shown.
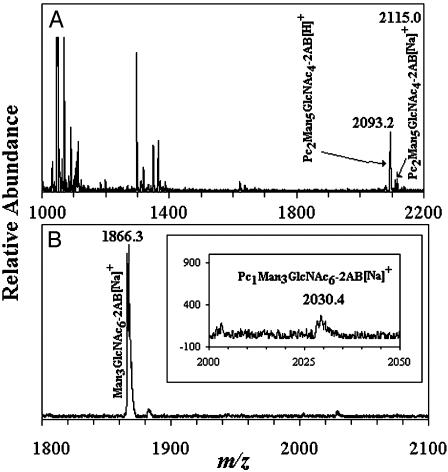
Identification of Oligosaccharide Acceptors for Phosphorylcholine Oligosaccharide Biosynthesis. Our first indication of possible oligosaccharide acceptors for phosphorylcholine came from incubations performed with oligosaccharides from ovalbumin that had been released by endo H treatment (Table 1 and Fig. 2 A and B). The substrate mixture contained Man5GlcNAc1 (20%), Man6GlcNAc1 (25%), Man5GlcNAc3 (25%), Man4GlcNAc3 (10%), and Man5GlcNAc4 (20%). To identify oligosaccharides that served as acceptors in the reaction described in Table 1, in which PTC was the phosphorylcholine donor, the reaction products were reductively aminated with 2-aminobenzamide and analyzed by MALDI-TOF MS. As shown in Fig. 3A, molecular ions of m/z 2093.2 and 2115.0, consistent with protonated and sodium-substituted PC2Man5GlcNAc4, respectively, were observed. Other molecular ions, such as those observed with m/z 1053.7 Man3GlcNAc2 and 1297.8 Man2GlcNAc4, are likely remnant oligosaccharides destroyed during the incubation.
Fig. 3.
MALDI-TOF MS of 2-aminobenzamide-labeled oligosaccharides. (A) The reaction contained PTC l-α-dipalmitoyl[choline-methyl-3H] and an endo H-released ovalbumin oligosaccharide mixture that consisted of Man5GlcNAc1 (20%), Man6GlcNAc1 (25%), Man5GlcNAc3 (25%), Man4GlcNAc3 (10%), and Man5GlcNAc4 (20%). (B) The reaction contained PTC l-α-dipalmitoyl[cholinemethyl-3H] and compound 9. Reaction products are shown in Inset. The composition and m/z of the molecular ions detected are shown.
To further test possible oligosaccharide acceptor substrates, we purified N-glycans prepared by N-glycosidase F treatment of bovine IgG and ovalbumin. We also obtained Gal4GlcNAc4Man3GlcNAc2 that had been derived from α-acid glycoprotein and was then treated with bovine β-galactosidase. The set of oligosaccharides tested as individual substrates, with PTC as the phosphorylcholine donor, are shown in Table 2. Compound 4 could not be obtained in pure form and was incubated as part of the oligosaccharide mixture described in Table 1.
Certain trends of acceptor structural requirements can be inferred from the results. All acceptors that incorporated >30,000 cpm contained GlcNAcβ1,2[GlcNAcβ1,4]Man-lower arm. Compounds 8 and 9, which yielded relatively high amounts of product, contained both GlcNAcβ1,2[GlcNAcβ1,6]Man-upper and GlcNAcβ1,2[GlcNAcβ1,4]Man-lower arms. Fig. 3B shows a MALDI-TOF MS spectrum from in vitro reactions with compound 9 as the acceptor. A molecular ion consistent with the product PC1Man3GlcNAc6 is observed at m/z 2030.4, whereas the unreacted acceptor appears at a m/z 1866.3. Given that both compounds 8 and 9 yield similar amounts of product, the presence of core bisecting N-acetylglucosamine in compound 8 apparently does not affect its substrate recognitions suitability. Indeed, PC2deoxyHex2Man3GlcNAc7 purified from C. elegans Dauer larvae (Fig. 1) probably has a similar branching pattern and contains core β1,4GlcNAc.
Compound 10, which has a GlcNAcβ1,2[GlcNAcβ 1,6][GlcNAcβ1,4]Man-upper arm is a significantly poorer acceptor than compounds 8 and 9 (Table 2), suggesting that in the presence of a GlcNAcβ1,2[GlcNAcβ1,4]Man-lower arm, the upper arm structure may affect substrate suitability. Compound 7, which contains a [GlcNAcβ1, 2]Man-upper arm was not a good substrate. Although compound 4 was not tested in pure form, it yielded the highest amount of product (Table 2). Thus, substrates with a Manα1,2[Manα1,6]Man-upper arm appear to be optimal acceptor substrates, although we cannot totally rule out a synergistic effect by other oligosaccharide components of the reaction mixture.
No high mannose or hybrid acceptors, other than compound 4, which contained the GlcNAcβ1,2[GlcNAcβ1,4]Man-lower arm, yielded significant amounts of product under the conditions tested. These include the hybrid compound 3, with a [GlcNAcβ1,2]Man-lower arm and GlcNAcβ1,4 core bisection and the high mannose compound 2. Compounds with GlcNAcβ1,2 mono substitutions of both the upper and lower arms and compound 7, with GlcNAcβ1,2 mono substitution in only the upper arm, were not good acceptors. Of all substrates tested containing GlcNAcβ1,2[GlcNAcβ1,4]Man-lower arm, compound 7, which has GlcNAcβ1,2Man-upper arm, is the worst acceptor, suggesting that the GlcNAcβ1,2 substitution of the upper or lower arms is detrimental to substrate recognition under the conditions tested.
Surprisingly, compound 1 yielded low but significant product, although it contains no Golgi-type monosaccharide additions. Compound 1 resembles the deminimus phosphorylcholine oligosaccharides reported (see Scheme 1).
Discussion
Here we report the in vitro biosynthesis of a nematode PCO. The reaction products are either consistent with the reported structures or represent reasonable processing intermediates that can be predicted from PCOs of several nematodes (2, 3). Our results strongly suggest that PTC is the phosphorylcholine donor, whereas CDP-choline can serve as donor only after being converted to PTC. For oligosaccharides to serve as acceptor substrates for phosphor ylcholine, the presence of GlcNAcβ1,2[GlcNAcβ1,4]Man α1,3 lower arm was sufficient as shown by product formation with compounds 4 and 8–10 (Table 2). We have not determined whether the phosphorylcholine substitution is at a mannose and/or N-acetylglucosamine residue. Core GlcNAcβ1,4Man bisection has no apparent effect on acceptor ability (compare compounds 8 and 9 in Table 2).
The presence of a high mannose upper arm (Man1, 3[Manα1,6]Man) is compatible as an acceptor when in the presence of GlcNAcβ1,2[GlcNAcβ1,4]Man (compound 4) but is not sufficient per se (compound 2) or in context of a GlcNAcβ1,2 lower arm (compound 3). This latter result is surprising, because some PCOs from C. elegans and other nematodes contain what appears to be a PC-6-O-GlcNAcβ1,2-mono-substituted lower arm (4–6). This structure, however, may be the result of oligosaccharide processing. For instance, it is possible that the GlcNAcβ1,4 of the GlcNAcβ1,2[GlcNAcβ1,4]Manα1,3 arm is removed by hexosaminidase(s) subsequent to phosphorylcholine addition, but such processing reactions have not yet been described in nematodes. Compound 1 yielded significant amounts of product, suggesting that core N-acetylglucosamine or mannose were acceptors for phosphorylcholine.
We cannot rule out that conditions used in our assay may influence substrate specificity. Whether or not the protein structure influences the transfer of phosphorylcholine to glycoprotein oligosaccharide acceptors is at present unknown and should be the subject of further investigations.
It is possible that several phosphorylcholine transferases with different substrate specificities occur in C. elegans. To date, no protein that catalyzes the phosphorylcholine transferase reaction has been identified in a eukaryotic system although similar enzymatic activities have been described in bacteria. For example, LIC2 is thought to catalyze phosphorylcholine addition during LPS biosynthesis in Haemophilus influenzae (20). The use of bioinformatics should help to search the C. elegans genome for possible candidate genes.
Acknowledgments
This work was supported by National Research Service Award F32 GM66486 to J.F.C. and National Institutes of Health Grants R01 GM30365 (to C.B.H.), R01 GM31318 (to P.W.R.), and P41 RR10888 and S10 RR15942 (to C.E.C).
Abbreviations: PCO, phosphorylcholine oligosaccharide; MALDI-TOF, matrix-assisted laser desorption ionization/time-of-flight; PTC, phosphatidylcholine; endo H, endoglycosidase H.
References
- 1.U.S. Congress, Office of Technology Assessment (1985) Status of Biomedical Research and Related Technology for Tropical Diseases, OTA-H-258 (U.S. Government Printing Office, Washington, D.C.), NTIS order no. PB87-139614.
- 2.Haslam, S. M., Houston, K. M., Harnett, W., Reason, A. J., Morris, H. R. & Dell, A. (1999) J. Biol. Chem. 274, 20953-20960. [DOI] [PubMed] [Google Scholar]
- 3.Lal, R. B. & Ottesen, E. A. (1989) Trans. R. Soc. Trop. Med. Hyg. 83, 652-655. [DOI] [PubMed] [Google Scholar]
- 4.Haslam, S. M. & Dell, A. (2003) Biochimie 85, 25-32. [DOI] [PubMed] [Google Scholar]
- 5.Cipollo, J. F., Costello, C. E. & Hirschberg, C. B. (2002) J. Biol. Chem. 277, 49143-49157. [DOI] [PubMed] [Google Scholar]
- 6.Haslam, S. M., Gems, D., Morris, H. R. & Dell, A. (2002) Biochem. Soc. Symp. 117-134. [PubMed]
- 7.Chen, S., Zhou, S., Sarkar, M., Spence, A. M. & Schachter, H. (1999) J. Biol. Chem. 274, 288-297. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, W., Cao, P., Chen, S., Spence, A. M., Zhu, S., Staudacher, E. & Schachter, H. (2003) Biochem. J. 372, 53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBose-Boyd, R. A., Nyame, A. K. & Cummings, R. D. (1998) Glycobiology 8, 905-917. [DOI] [PubMed] [Google Scholar]
- 10.Berninsone, P., Hwang, H.-Y., Zemtseva, I., Horvitz, H. R. & Hirschberg, C. B. (2001) Proc. Natl. Acad. Sci. USA 98, 3738-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luhn, K., Wild, M. K., Eckhardt, M., Gerardy-Schahn, R. & Vestweber, D. (2001) Nat. Genet. 28, 69-72. [DOI] [PubMed] [Google Scholar]
- 12.Houston, K. M., Lochnit, G., Geyer, R. & Harnett, W. (2002) Mol. Biochem. Parasitol. 123, 55-66. [DOI] [PubMed] [Google Scholar]
- 13.Houston, K. M., Cushley, W. & Harnett, W. (1997) J. Biol. Chem. 272, 1527-1533. [DOI] [PubMed] [Google Scholar]
- 14.Haslam, S. M., Khoo, K. H., Houston, K. M., Harnett, W., Morris, H. R. & Dell, A. (1997) Mol. Biochem. Parasitol. 85, 53-66. [DOI] [PubMed] [Google Scholar]
- 15.Morelle, W., Haslam, S. M., Morris, H. R. & Dell, A. (2000) Mol. Biochem. Parasitol. 109, 171-177. [DOI] [PubMed] [Google Scholar]
- 16.Lipniunas, P. H., Neville, D. C., Trimble, R. B. & Townsend, R. R. (1996) Anal. Biochem. 243, 203-209. [DOI] [PubMed] [Google Scholar]
- 17.Bigge, J. C., Patel, T. P., Bruce, J. A., Goulding, P. N., Charles, S. M. & Parekh, R. B. (1995) Anal. Biochem. 230, 229-238. [DOI] [PubMed] [Google Scholar]
- 18.Carman, G. M. & Henry, S. A. (1999) Prog. Lipid Res 38, 361-399. [DOI] [PubMed] [Google Scholar]
- 19.Satouchi, K., Hirano, K., Sakaguchi, M., Takehara, H. & Matsuura, F. (1993) Lipids 28, 837-840. [DOI] [PubMed] [Google Scholar]
- 20.Schweda, E. K., Brisson, J. R., Alvelius, G., Martin, A., Weiser, J. N., Hood, D. W., Moxon, E. R. & Richards, J. C. (2000) Eur. J. Biochem. 267, 3902-3913. [DOI] [PubMed] [Google Scholar]