Abstract
Circular dichroism is commonly used to investigate the conformations of nucleic acids. However, many biochemical processes implicate conformational changes of particular nucleotide residues within DNA or RNA that cannot be studied by this method, because the CD of these residues is buried in the total signal of the polynucleotide. Here, we report a method to study local conformations of DNA or RNA that is based on the use of the CD of 2-aminopurine (AP) residues as a probe. AP is readily incorporated into DNA in place of adenine and does not significantly alter DNA structure. Unlike adenine, AP is fluorescent and this property has been used for many years to investigate local nucleic acid structure. We show here that the CD spectrum of AP dinucleotide, (AP)2, exhibits a positive CD band at 326 nm, a spectral region in which nucleic acids (and proteins) do not absorb. Our results show that the bases of (AP)2 are stacked in a right-handed helical conformation. A low-energy CD band is also observed when this nucleotide dimer is incorporated into double-stranded DNA. Control experiments show that this signal comes from the stacking of adjacent AP residues. We have used this CD signal to provide information about the conformation of the AP dinucleotide at a defined position within single- and double-stranded nucleic acids.
Keywords: dinucleotide conformations, nucleic acid conformations, spectroscopy, exciton coupling, base stacking
CD is widely used to obtain information about the conformations of nucleic acids. Exciton coupling provides a theoretical framework for understanding the dependence of the CD signal on the conformation of dinucleotides and polynucleotides (1). In this formalism, the sign and amplitude of the CD spectrum depend on a geometric relationship between coupled electronic transition dipoles on adjacent nucleotides and the distance between the bases. The relation between CD signal and dinucleotide structure is degenerate, in that different conformations can give rise to identical spectra. Nevertheless, much useful information about the extent and dynamics of base stacking of dinucleotides and polynucleotides has been determined by using this method (1–5).
The CD of a polynucleotide effectively comes from the sum of all base–base interactions. As a consequence, CD measurements are not useful for the study of many interesting questions that focus on the conformation of particular nucleotides within a larger polymer. One might like, for example, to measure local structural changes that occur during RNA folding or when a protein binds at a specific nucleic acid site. A possible method of extending CD techniques to address these questions is to use nucleic acid analogs that absorb above 300 nm, a region of the spectrum where DNA and RNA (and the proteins that manipulate them) are transparent. Exciton coupling of these low-energy transitions with those of adjacent base analogs should give a CD signal that is separated from the polynucleotide spectrum and sensitive to base stacking of the analog. Here, we report an application of this approach using 2-aminopurine (AP).
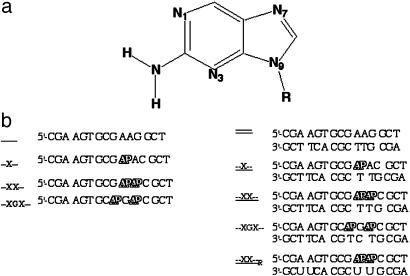
AP is a structural isomer of adenine in which the amino group has been moved from position 6 to position 2 of the purine ring (Fig. 1a). This molecule has a low-energy electronic transition at 305 nm that is fluorescent (6, 7). AP behaves in other ways like adenine. It forms a base pair with thymine and can be incorporated into B-form DNA without significantly changing the DNA structure (8, 9). The AP deoxyribonucleoside triphosphate is efficiently used during template-directed DNA synthesis by DNA polymerases in vitro and in vivo (10–17). AP is a mutagen causing A·T → G·C and G·C → A·T transitions. This mutagenicty occurs because AP·C mispairs form at a higher frequency than do A·C mispairs (18, 19). AP has been extensively used as a fluorescent probe of nucleic acid structure because of its functional equivalence to adenine.
Fig. 1.
Structures and abbreviations. (a) Structure of AP. (b) DNA and RNA oligonucleotides used in this study and their abbreviations.
Here, we report the spectroscopic properties of AP dinucleoside monophosphate (AP)2 in isolation and incorporated into DNA. As expected, (AP)2 exhibits a CD spectrum characteristic of exciton coupling between the adjacent aminopurine bases in a stacked, right-handed helical conformation. The CD of the low-energy AP transition can be used to monitor the conformation of the aminopurine dinucleotide embedded in DNA. Monitoring the CD of AP offers a useful new approach to the determination and analysis of local conformations and conformational changes in nucleic acid polymers.
Materials and Methods
AP was purchased from Aldrich. (AP)2 and DNA oligonucleotides (Fig. 1b) were purchased from TriLink BioTechnologies (San Diego). RNA oligonucleotide was from Dharmacon (Lafayette, CO). Unless otherwise stated, experiments were carried out in 20 mM phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA (pH 7.5) and made with deionized water (Barnstead). AP and (AP)2 concentrations were determined from absorbance at 305 nm by using an extinction coefficient of 6,000 M-1·cm-1 (6). Concentrations of single-stranded (ss) oligonucleotides were determined from the maximum intensity of the UV spectrum near 260 nm, using extinction coefficients of 8,650 M-1·cm-1 for the canonical nucleotide residues and 1,700 M-1·cm-1 for AP (6).
Solutions of ss oligonucleotides were divided into two portions. One portion was used to characterize the ss oligonucleotides and the other portion was mixed with the complementary ss oligonucleotide to form double-stranded (ds) complexes. The ds nature of the polynucleotide was confirmed by thermal denaturation, which was monitored by UV absorbance changes. Fluorescence and CD measurements were then carried out on the same samples.
Fluorescence spectra were taken by using a Jobin-Yvon (Longjumeau, France) Fluorolog spectrometer. Samples in a 10 × 4-mm cuvette, with the long pathlength oriented in the direction of the incident light, were excited at 305 nm, and emission spectra were recorded at 20°C. Fluorescence lifetimes were determined with the same machine by using frequency domain measurements (20); plots of phase and modulation vs. frequency, over the 1–200 MHz range, were fit with software provided with the Jobin-Yvon apparatus. CD spectra were taken between 450 and 195 nm on a Jasco model J-720 CD spectrometer equipped with a thermostated cell holder. Results are the average of four to eight spectra and are presented as Δε, the difference in molar extinction coefficient of left and right circularly polarized light per mol residue (1). CD per mol nucleotide below 300 nm was compared with CD per mol AP above 300 nm; to compare the CD intensities above 300 nm of modified and unmodified oligonucleotides, Δε of unmodified oligonucleotides (per mol nt) were multiplied by the appropriate ratio of nt/AP.
Results
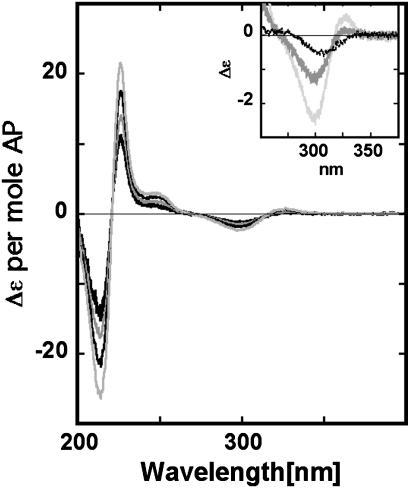
AP Dinucleotide. We wished to use the CD signal of adjacent AP residues to measure local conformation in DNA. Because the CD spectrum of this system had not previously been characterized, we first studied the simpler dinucleoside monophosphate (AP)2 molecule, in which two AP deoxyribonucleoside residues are linked 3′–5′ by a phosphate. The CD spectrum of (AP)2 exhibits exciton coupling at 220 and 317 nm (Fig. 2). The transition at 317 nm has a low-energy peak and a high-energy trough that are indicative of a right-handed helical conformation for the (AP)2 molecule (2). High temperatures and added ethyl alcohol (EtOH) decreased the CD intensity, as expected for perturbations that unstack adjacent bases.
Fig. 2.
CD spectrum of (AP)2. The CD of (AP)2 per mol of AP in 20 mM phosphate buffer, pH 7.5, at various temperatures. The spectra, in order from greater to lesser intensity, were obtained at 20, 40, 60, and 80°C. (Inset) The molar CD of (AP)2 in phosphate buffer at 20°C (highest intensity) and 80°C (intermediate intensity), and in absolute EtOH (20°C) (lowest intensity).
The absorption spectrum of (AP)2 in 20 mM phosphate buffer (pH 7.5) has peaks at 305 and 216 nm and a shoulder near 245 nm (data not shown). In dilute aqueous buffer, the UV spectrum of (AP)2 increased by 2% when the temperature was raised from 15°C to 75°C. Similar results were observed in control experiments with adenine dinucleoside monophosphate and interpreted to reflect thermal unstacking of the bases (1). When the dimer was dissolved in absolute EtOH, the UV maximum shifted 4–5 nm toward the red, and the peak intensity increased by 20% relative to the intensity in dilute aqueous buffer. The extinction coefficient of AP is the same in water and in EtOH (6), suggesting that this hyperchromicity reflects base unstacking.
Addition of a ribose moiety reduces the fluorescence quantum yield of AP from 0.86 to 0.68; further addition of a phosphate alters the fluorescence of the nucleoside only slightly (6, 7). In contrast, the fluorescence intensity of (AP)2 was 20-fold less than that obtained for the equivalent concentration of AP (Table 1). If this quenching was due to interactions between the aminopurine residues, fluorescence should increase as the bases unstacked. Ethanol increased the fluorescence intensity of the dinucleotide (Table 1), consistent with the solvent-induced unstacking suggested by the CD results (Fig. 2). Solvent quenching of AP, which increases with temperature (7), was much larger than the dequenching effect of unstacking; as a result, the fluorescence intensity of (AP)2 decreased with temperature.
Table 1. Relative fluorescence intensities of AP and (AP)2.
| Solvent | AP | (AP)2 | ssDNA (--XX--) |
|---|---|---|---|
| H2O | 100 ± 5 | 5 ± 0.1 | — |
| Buffer | 100 ± 5 | 4 ± 0.1 | 6 ± 1 |
| Ethanol | 67 ± 2 | 22 ± 1 | — |
Fluorescence intensity relative to AP in water. Values are relative fluorescence intensities per AP residue.
The effect of ethanol was further investigated by fluorescence lifetime experiments (Table 2). AP exhibits a single (≈10 ns) lifetime in water and buffer, and this lifetime decreases to ≈5 ns in ethanol. In contrast, lifetime measurements showed that the excited state of (AP)2 decays by a process that can be fit by two exponentials. The slow component corresponds to the lifetime of the aminopurine base, both in magnitude and in the effects of solvent. Hence, this component can be used as a spectroscopic signal for the unstacked fraction of the AP bases. An additional rapid decay (1.1–1.4 ns) was observed only with the AP dinucleotide and thus can perhaps be considered to correspond to a “stacked bases” signal for (AP)2. If this interpretation is correct, these data show that ethanol decreases the stacked population of (AP)2 from 35% to 17% (Table 2) and concomitantly increases the quantum yield of this species (Table 1). Observation of a short-lived fluorescent species in the dimer, with amplitude inversely correlated with fluorescence intensity, is also good evidence for dynamic quenching due to collisions between the purine bases in (AP)2 (20).
Table 2. Excited state lifetimes of AP and (AP)2.
| AP τ, ns | (AP)2 τ, ns (fraction population) | ||
|---|---|---|---|
| H2O | 11.6 ± 0.3 | 10.4 ± 0.05 | 1.1 ± 0.05 |
| (0.68 ± 0.02) | (0.32 ± 0.02) | ||
| Buffer | 9.0 ± 0.01 | 8.1 ± 0.02 | 1.14 ± 0.1 |
| (0.64 ± 0.02) | (0.36 ± 0.02) | ||
| Ethanol | 5.0 ± 0.05 | 4.8 ± 0.2 | 1.35 ± 0.25 |
| (0.83 ± 0.05) | (0.17 ± 0.05) | ||
Taken together, these results show that the bases of an (AP)2 dinucleotide are stacked in a right-handed helical conformation that is disrupted by temperature and solvent in the same way as previously observed for adenosine dinucleotide. Furthermore, the CD of (AP)2 centered at 317 nm appears to be a result of band splitting due to exciton coupling between adjacent AP bases, and its magnitude reflects the extent of base stacking in the dinucleotide. Because the low-energy CD band of this transition is in a transparent region of the nucleic acid UV spectrum, it should be possible to measure base stacking of pairs of AP bases incorporated into DNA or RNA.
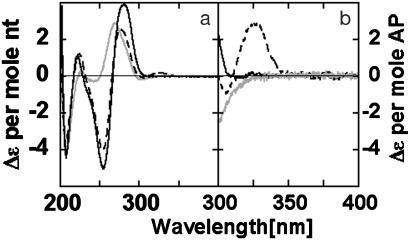
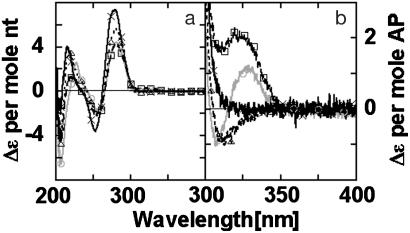
AP Dinucleotide in DNA. Fig. 3 shows the CD spectrum of a ds 15-mer oligonucleotide that contains (--XX--), or does not contain (-------), an (AP)2 dimer. (These symbolic designations of oligonucleotides are defined in Fig. 1b.) Fig. 3 compares the CD per mol nucleotide residue below 300 nm with the CD per mol AP residue above 300 nm. The CD spectrum of (AP)2 dimer-containing DNA above 300 nm resembles the spectrum of the dinucleotide (AP)2 (Fig. 2). In both spectra, a peak is observed at 326–328 nm. This peak has an intensity of Δε = 3.2 ± 0.4 (M AP)-1·cm-1 for --XX--; this intensity is 6-fold larger than in the dinucleotide, consistent with increased base-stacking expected between the neighboring AP bases that have been incorporated into dsDNA.
Fig. 3.
CD Spectra of ds oligonucleotides containing AP dimer. (a) CD per mol nucleotide residues. (b) CD per mol AP residues. Oligonucleotides: ----- (dark solid line), --XX-- (dashed line), and --XX--R (light solid line).
A trough at 305 nm was also observed in the CD spectrum of --XX--. The position and intensity of this trough was sensitive to small changes in the 280-nm peak. Fig. 3b shows the “spillover” from the 280-nm CD peak. The magnitude of the spillover will, of course, increase with oligonucleotide length. The molar CD per AP residue above 300 nm is similar to that obtained at 280 nm with canonical nucleotides in dsDNA [i.e., Δε = 2–4 (M residue)-1·cm-1]. The CD band at 226 nm in the dinucleotide (A P)2 spectrum has a much larger intensity [20 (M residue)-1·cm-1; Fig. 2] and appears to be present in the CD spectrum of --XX-- (Fig. 3).
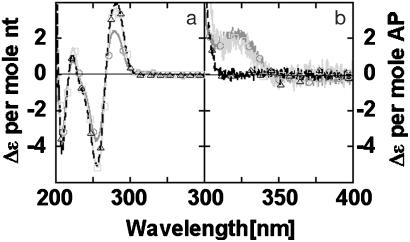
A number of experiments were carried out to further characterize the 328-nm peak of the CD spectra. Double-stranded oligonucleotides containing a single AP residue (--X--) or two AP residues separated by an intervening base (--XGX--), showed similar CD spectra above 300 nm (Fig. 4). These spectra had a single peak with maximum intensity at 320 nm, differing markedly from the CD spectrum of the --XX-- species (Fig. 3).
Fig. 4.
CD Spectra of ds oligonucleotides containing AP. (a) CD per mol nucleotide residues. (b) CD per mol AP residues. Oligonucleotides: --------- (triangles), --X-- (squares), and --XGX-- (circles).
These results indicate that the low-energy CD bands in Figs. 3 and 4 have different origins. Exciton coupling between electronic transitions on two chromophores generally gives rise to a conservative CD spectrum with two bands of equal intensity and opposite sign centered around the wavelength of maximum UV absorbance (1, 2, 4). In the case of the --XX-- species (Fig. 3), the low-energy CD signal appears to come from two bands of opposite sign, as would be expected for exciton coupling of transitions near 320 nm on adjacent bases. This is a significantly higher wavelength than the corresponding low-energy transition of AP that occurs at 305 nm. This wavelength change can be explained by the large red shift in the low-energy electronic transition of AP that is known to occur on incorporation of isolated AP residues into dsDNA (7, 21). Unlike --XX--, the CD spectra of the --X-- and --XGX-- species have maximum intensities at 320 nm (Fig. 4), consistent with the idea that these bands may have the shape of the UV absorbance of AP in dsDNA. These considerations suggest that the chirality of --X-- and --XGX-- is not a result of exciton coupling but rather reflects another mechanism such as interband coupling with the high-energy transitions of adjacent nucleotides in the DNA.
CD spectra of ss oligonucleotides ------, --X--, --XGX--, and --XX-- are shown in Fig. 5. The addition of a single AP residue reduced the intensity of the peak at 280 nm from 7 to 5.5 (M nt)-1·cm-1. The presence of a second AP residue, whether --XGX-- or --XX--, further decreased this peak to 4.4 (M nt)-1·cm-1. The low-energy CD spectrum of --XX-- shows two bands of opposite sign, with a crossover at 315 nm. This CD spectrum resembles the exciton coupling observed for the --XX-- species (Fig. 3). However, the peak at 328 nm is 2.5-fold less intense in the ss oligonucleotide, probably because of weaker base stacking. Similarly, the intensity of this band in the spectrum of the dinucleotide (AP)2 was 2 times smaller than that seen with the --XX-- species, indicating very weak stacking in the dinucleotide (Fig. 2). These results show that the CD signal characteristic of stacked nearest-neighbor AP residues is also apparent in ssDNA. In contrast, the spectrum of ssDNA with non-nearest-neighbor AP residues does not have this appearance. The spectrum of --XGX-- had a positive band at 320 nm, whereas the spectrum of --X-- had a negative band at 310 nm.
Fig. 5.
CD Spectra of ss oligonucleotides containing AP. (a) CD per mol nucleotide residues. (b) CD per mol AP residues. Oligonucleotides: ------ (X), --X-- (triangles), --XGX-- (squares), and --XX-- (circles).
We also examined the fluorescent properties of these oligonucleotides. Samples excited at 305 nm gave an emission spectrum with a maximum intensity at 370 nm. Fluorescence intensities of --X--, --XX--, and (AP)2 were the same (Tables 1 and 3), suggesting that the mobility of the fluorophore in the dinucleotide and in ssDNA are similar. The addition of complementary ssDNA to --XX-- and --X-- further decreased the fluorescence (by 90–95%; Table 3). In contrast, the fluorescence of the ssDNA species --XGX-- was 60% higher than that of the --XX-- species, and less quenching was observed when the dsDNA species --XGX-- was formed. The cause of this fluorescent increase is unknown, but it may indicate that the environment of the AP base is less structured in this sequence (9).
Table 3. Spectroscopic properties of ss and ds oligonucleotides.
| H, %* | Tm, °C | % F1 ss oligos† | % F1 ds oligos‡ | |
|---|---|---|---|---|
| --XX-- | 100 | |||
| --X-- | 100 | |||
| --XGX-- | 160 | |||
| ------- | 19 | 63 | ||
| --X-- | 18 | 62 | 8 | |
| --XX-- | 15 | 63 | 5 | |
| --XGX-- | 16 | 63 | 15 | |
| --XX--R | 15 | 52 | 35 |
Percent increase in intensity of UV absorbance of dsDNA relative to ssDNA
Fluorescence intensity relative to --XX-- (per mole AP)
Fluorescence intensity relative to the fluorescent of the ss oligonucleotide in the ds complex
Some Applications. Biochemical processes involving DNA and RNA (such as replication and transcription) often involve local conformational changes from ss to ds structures, or the reverse. Our results show that the CD of AP can be used to monitor this phenomenon at the nucleotide residue level. The CD intensity at 328 nm of an embedded AP dimer increased by a factor of 2–3 on formation of dsDNA from ssDNA (Figs. 3 and 5). As shown above, this comes from increased stacking of the adjacent APs and should therefore be independent of the flanking DNA bases. The insertion of a single AP residue into an oligonucleotide resulted in a negative low-energy CD signal for the ss molecule --X-- and a positive signal for duplex DNA --X-- (Figs. 4 and 5). Here, the origin of these CD spectroscopic changes is less clear, and the signal may depend on the sequence context of AP.
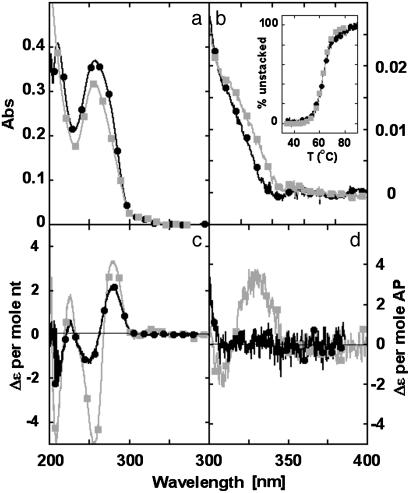
Thermal denaturation of ds oligonucleotide --XX-- provides a simple demonstration of the validity of this method for the study of local “strandedness” in polynucleotides. The temperature-dependent intensity of the UV spectrum reflects overall base stacking in the polynucleotide, while CD changes at 328 nm come from interactions of AP residues that can be used to monitor local base stacking of the AP dimer. The temperature-dependent changes in the CD spectrum at 330 nm and in the UV absorbance spectrum at 256 nm are superimposable (Fig. 6b Inset). Hence, global and local DNA conformations showed the same dependence on temperature, as expected for a two-state cooperative conformational change involving ds oligonucleotides of this size and sequence.
Fig. 6.
Temperature-dependent UV absorbance and CD spectra of (AP)2-containing oligonucleotide --XX--. (a and b) Absorption spectra at 20°C (squares) and 80°C (circles). (c and d) CD spectra at 0°C (squares) and 80°C (circles). (b Inset) Melting curves (% unstacked conformation vs. T) from absorbance at 256.5 nm (squares) and CD at 330 nm (circles).
The UV spectrum above 310 nm decreased during thermal denaturation of --XX-- (Fig. 6b). This hypochromism was also observed for the ds oligonucleotides --X-- and --XGX--, as reported (21). The UV absorbance of ssDNA containing an isolated AP is known to exhibit a large red shift on annealing with complementary DNA (7, 21). We also observed a 1- to 2-nm blue shift in the low-energy UV spectrum of the dinucleotide (AP)2 at 75°C that is consistent with such wavelength changes. Hence, hypochromism above 300 nm at high temperatures can be explained by a blue shift in the absorption spectrum of AP as dsDNA unwinds.
Finally, we investigated the spectroscopic properties of ds polynucleotides with different conformations. Single-stranded DNA (--XX--) was mixed with stoichiometric concentrations of complementary ssDNA (------) or ssRNA (—R) oligonucleotides. These mixtures were heated to 80°C and slowly cooled. The ds character of the polynucleotide species that resulted was confirmed by UV absorbance. The Tm of the RNA/DNA hybrid duplex was 52°C, which is 10° lower than the Tm of the equivalent DNA/DNA duplex (--XX--) (Table 3). Fluorescence and CD measurements were then performed on these samples.
The fluorescence intensity of the (AP)2 dimer in the DNA/DNA duplex (--XX--) decreased by 95%, compared to the signal for the equivalent ssDNA oligomer (--XX--), whereas the fluorescence intensity of the RNA/DNA hybrid was significantly greater (Table 3). Furthermore, the exciton splitting that characterized the low-energy CD spectrum of the dsDNA molecule was not observed in the RNA/DNA hybrid (Fig. 3). Double-stranded DNA is expected to be in the B conformation, with 10 bp per helical turn and 3.4 Å between adjacent base pairs, whereas the RNA/DNA hybrid is expected to have a conformation similar to A-form, with 11 bp per turn, 2.6 Å between base pairs, and a tilt in the bases with respect to the helix axis (5, 22). Consequently, these results show that the CD spectrum of AP residue pairs incorporated into polynucleotides is sensitive to small changes in nucleic acid conformation and/or mobility.
Discussion
The main conclusion of this paper is that the low-energy CD spectrum of an AP dimer can, in principle, be used to investigate the local conformation of specific sequences within DNA and RNA at various stages of interesting biological processes. Thus, we have shown that an AP dimer embedded in dsDNA exhibits a CD spectrum above 300 nm, i.e., at wavelengths where the canonical nucleic acid residues do not absorb (Fig. 3). Furthermore, we have shown that adjacent AP bases in DNA are stacked in right-handed helical conformations and that the CD spectrum of these AP pairs is sensitive to local and global conformational changes in the nucleic acids within which they reside.
The optical activity of AP can also be used to study fundamental spectroscopic questions. For example, we can experimentally investigate the relative contributions of nearest-neighbor and non-nearest-neighbor bases to the CD of polynucleotides. A conservative signal due to exciton coupling between AP residues was observed in the low-energy CD spectrum of the --XX-- species (Fig. 3) but not the --XGX-- species (Fig. 4). The absence of exciton coupling in the CD of --XGX-- could be the result of geometric factors that are predicted to reduce the CD intensity when bases are in this conformation (23). However, the intensity of the CD signal of the oligonucleotide --XGX-- indicates that significant nonconservative interactions contribute to this spectrum. Furthermore, the CD of --XGX-- resembles the spectrum of --X-- (Fig. 4), suggesting that similar interactions with adjacent nucleotides may be responsible for the chirality of the two oligonucleotides. Taken together, these results indicate that effects of base stacking between nearest-neighbor bases dominates the CD spectra of polynucleotides, as previously suggested on the basis of theoretical calculations (2, 4).
The CD of the AP dimer also gives an important insight into the mechanism of the fluorescent quenching of AP in DNA. Loss of fluorescent intensity generally occurs by two mechanisms (20). Dynamic quenching is caused by collisions that increase the nonradiative decay from the excited state. Static quenching, on the other hand, is caused by formation of a dark complex between the fluorophore and a quenching molecule. Both static and dynamic quenching contribute to the reduced fluorescence of an isolated AP residue in dsDNA. It is thought that static quenching is the result of base stacking, and that dynamic quenching reflects the conformational mobility of the fluorophore (24). Quantum mechanical calculations by Jean and Hall (25) indicate that static quenching by adenine or guanine involves mixing of the ground state orbitals of AP and the adjacent purine.
For most fluorescent molecules, static quenching would not be associated with increased optical activity. However, in nucleic acids the contiguity of the fluorophore and the quencher imposes an asymmetric conformation on the dark complex and the statically quenched complex should be chiral. In our experience, a strong conservative CD signal for the AP dimer is associated with a weak fluorescence intensity, and vice versa. This correlation between optical activity and fluorescence quenching could be explained if stacking of the AP bases produces an optically active dark complex. Hence, these results lend experimental support to the mechanism of static quenching proposed by Jean and Hall (25). They also suggest that the fluorescence quenching and optical activity of AP in DNA share a common physical origin.
The optical activity of AP should be a useful complement to its fluorescence. The reciprocal relation between CD and fluorescence intensities means that it is now possible, using CD, to investigate the environment of AP residues in polynucleotides under conditions in which AP fluorescence has been quenched. Furthermore, optical activity measurements may give geometric information that is not available from fluorescence. The ss oligonucleotide species --XX-- and --X-- illustrate this point. Although these oligonucleotides show the same fluorescence quenching per AP residue (Table 3), there are dramatic differences in their CD spectra (Fig. 5) that reflect the stacked conformation of adjacent AP bases in --XX--.
Fluorescence methods such as fluorescence resonance energy transfer (FRET) can also give information about the relative positions of two fluorophores. FRET is the result of dipole–dipole interactions that depend on the distance between fluorophores (r) as 1/r6 (1). Exciton coupling, on the other hand, depends on distance between chromophores as 1/r2 (4). Hence, CD has the potential to measure through-space interactions over a greater distance than FRET, although the apparent sensitivity may be offset by weak splitting, geometric effects, or conformational averaging over orientations with opposite chirality.
In conclusion, we present a method to monitor local conformations within DNA and RNA molecules. CD of adjacent AP bases can be used to measure base stacking in nucleic acid polymers at the dinucleotide level. Isolated aminopurine residues also display significant CD signals because of nearest-neighbor interactions with adjacent bases. Further work will be required to investigate the relation between nucleic acid structure and the details of the CD spectra of embedded AP. To date, we have used this method to observe local conformational changes in polynucleotides in the following experimental situations: (i) transformation from ssDNA to dsDNA; (ii) conformational changes in dsDNA, such as B-form to A-form conversions; (iii) effects of temperature and denaturing solvents; (iv) protein–nucleic acid interactions; and (v) RNA conformational changes (N.P.J., unpublished data). This list illustrates the types of structural questions that can be addressed by means of the low-energy CD spectrum of AP.
Acknowledgments
We thank John Schellman and members of our laboratory for many helpful discussions. This work was supported in part by National Institutes of Health Grants GM-15792 and GM-29158 (to P.H.v.H.) and by partial sabbatical salary support from Centre National de la Recherche Scientifique (to N.P.J.). P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Abbreviations: AP, 2-aminopurine; ds, double-stranded; ss, single-stranded.
References
- 1.Cantor, C. R. & Schimmel, P. R. (1980) Biophysical Chemistry (Freeman, San Francisco).
- 2.Johnson, W. C., Jr., & Tinoco, I., Jr. (1969) Biopolymers 8, 715-731. [Google Scholar]
- 3.Johnson, W. C., Jr. (1996) in Circular Dichroism and the Conformational Analysis of Biomolecules, ed. Fasman, G. D. (Plenum, New York), pp. 433-468.
- 4.Johnson, W. C., Jr., & Tinoco, I., Jr. (1969) Biopolymers 7, 727-749. [Google Scholar]
- 5.Bloomfield, V. A., Crothers, D. M. & Tinoco, I., Jr. (1974) Physical Chemistry of Nucleic Acids (Harper & Row, New York).
- 6.Smagowicz, J. & Wierzchowski, K. L. (1974) J. Luminescence 8, 210-232. [Google Scholar]
- 7.Ward, D. C., Reich, E. & Stryer, L. (1969) J. Biol. Chem. 244, 1228-1237. [PubMed] [Google Scholar]
- 8.Sowers, L. C., Fazakerley, G. V., Eritja, R., Kaplan, B. E. & Goodman, M. F. (1986) Proc. Natl. Acad. Sci. USA 83, 5434-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordlund, T. M., Andersson, S., Nilsson, L., Rigler, R., Graslund, A. & McLaughlin, L. W. (1989) Biochemistry 28, 9095-9103. [DOI] [PubMed] [Google Scholar]
- 10.Fidalgo da Silva, E., Mandal, S. S. & Reha-Krantz, L. J. (2002) J. Biol. Chem. 277, 40640-40649. [DOI] [PubMed] [Google Scholar]
- 11.Frey, M. W., Sowers, L. C., Millar, D. P. & Benkovic, S. J. (1995) Biochemistry 34, 9185-9192. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, M. F., Hopkins, R. & Gore, W. C. (1977) Proc. Natl. Acad. Sci. USA 74, 4806-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandal, S. S., Fidalgo da Silva, E. & Reha-Krantz, L. J. (2002) Biochemistry 41, 4399-4406. [DOI] [PubMed] [Google Scholar]
- 14.Wang, M. L., Stellwagen, R. H. & Goodman, M. F. (1981) J. Biol. Chem. 256, 7097-7100. [PubMed] [Google Scholar]
- 15.Purohit, V., Grindley, N. D. F. & Joyce, C. M. (2003) Biochemistry 42, 10200-10211. [DOI] [PubMed] [Google Scholar]
- 16.Petruska, J. & Goodman, M. F. (1985) J. Biol. Chem. 260, 7533-7539. [PubMed] [Google Scholar]
- 17.Zhong, X., Patel, S. S., Werneburg, B. G. & Tsai, M. D. (1997) Biochemistry 36, 11891-11900. [DOI] [PubMed] [Google Scholar]
- 18.Goodman, M. F. & Fygenson, D. K. (1998) Genetics 148, 1475-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe, S. M. & Goodman, M. F. (1982) Proc. Natl. Acad. Sci. USA 79, 6429-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakowicz, J. R. (1999) Principles of Fluorescence Spectroscopy (Kluwer/Plenum, New York), 2nd Ed.
- 21.Eritja, R., Kaplan, B. E., Mhaskar, D., Sowers, L. C., Petruska, J. & Goodman, M. F. (1986) Nucleic Acids Res. 14, 5869-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, S. H., Yu, Q., Gray, D. M. & Ratliff, R. L. (1994) Nucleic Acids Res. 22, 4326-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush, C. A. & Tinoco, I., Jr. (1967) J. Mol. Biol. 23, 601-614. [DOI] [PubMed] [Google Scholar]
- 24.Rachofsky, E. L., Osman, R. & Ross, J. B. A. (2001) Biochemistry 40, 946-956. [DOI] [PubMed] [Google Scholar]
- 25.Jean, J. M. & Hall, K. B. (2001) Proc. Natl. Acad. Sci. USA 98, 37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]