Abstract
The molecular events regulating the elimination of cells to create a hollow lumen during tissue development are poorly understood. By using an in vitro morphogenesis model in which MCF-10A human mammary epithelial cells form hollow acini-like structures, we have observed both caspase-mediated apoptosis and autophagy associated with cells that are lost during lumen formation. Here, we show that the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) mediates induction of autophagic processes associated with lumen formation. TRAIL is up-regulated during morphogenesis of MCF-10A mammary epithelial cells in 3D basement-membrane cultures and inhibition of TRAIL signaling during morphogenesis blocks the formation of autophagic vacuoles. In addition, treatment with exogenous TRAIL induces extensive autophagy in monolayer and 3D cultures. When combined with inhibition of caspase 3 activity (by Bcl-XL overexpression), inhibition of TRAIL-induced autophagy results in luminal filling. Thus, TRAIL regulates an autophagic program during acinar morphogenesis, which together with caspase-mediated apoptotic events, results in lumen formation during MCF-10A morphogenesis.
Programmed cell death is important for the formation and maintenance of tissue architecture in multicellular organisms. Programmed cell death regulates the sculpting of structures, deletion of vestigial organs, and maintenance of proper cell number (reviewed in refs. 1 and 2). Cell death is involved in cavitation during the development of the inner-cell mass (3) and some glandular organs (4, 5). The specific molecular events that characterize and drive programmed cell-death processes associated with cavitation remain poorly understood. We have used an in vitro 3D cell-culture model employing the immortalized human mammary epithelial cell line MCF-10A to examine the molecular events involved in lumen formation. When seeded in a matrix-rich environment, MCF-10A cells form acinar-like structures that are notable for the presence of a hollow lumen (6, 7).
Lumen formation in MCF-10A spheroids is associated with the death of centrally located cells (6); furthermore, caspase 3 activation is observed in these dying cells, indicating that classical apoptosis is involved. In addition, overexpression of Bcl-2 or Bcl-XL, which abrogates caspase 3 activation, delays lumen formation significantly (6). Recently, we have found that the proapoptotic BH3-only protein Bim is an important regulator of apoptotic processes and lumen formation during MCF-10A morphogenesis (M.R., K.R.M., J.D., D. Lynch, and J.S.B., unpublished data). Apoptosis has been implicated also in the clearance of central cells in several other 3D spheroid-culture systems (8–12). Similarly, studies of mouse mammary-gland development indicate that apoptosis is involved in terminal end bud morphogenesis (13).
The evidence that inhibition of apoptotic events by overexpression of Bcl-2 or Bcl-XL delays, but does not inhibit, lumen formation in MCF-10A structures indicated that additional processes contribute to formation of the luminal cavity (6). Indeed, ultrastructural studies revealed abundant autophagic vacuoles (AVs) within dying cells in the lumen of in MCF-10A acini, suggesting that autophagic processes also may contribute to cell clearance in the lumen when classical apoptosis is blocked (6). Autophagy is a bulk-protein-degradation process characterized by the formation of double-membrane vacuoles, or AVs, which deliver cytoplasmic contents and organelles to the lysosome for destruction. Increasing evidence suggests that autophagy contributes to programmed cell death (14).
In this article, we identify tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) as a regulator of lumen formation during MCF-10A acini formation. Inhibition of TRAIL signaling alone did not prevent lumen formation; however, this event was blocked when TRAIL inhibition was combined with Bcl-XL overexpression. This cooperativity implicated TRAIL as a regulator of complementary processes involved in lumen formation. Indeed, inhibition of TRAIL signaling prevented formation of AVs, which was previously demonstrated to accompany lumen formation. Furthermore, treatment of MCF-10A monolayers and acini with exogenous TRAIL induced extensive AVs. Thus, our results implicate TRAIL in the induction of autophagy and cavitation during 3D morphogenesis of MCF-10A cells. Furthermore, these results suggest that autophagy and apoptosis represent two distinct but complementary processes involved in cell elimination during morphogenesis.
Materials and Methods
Materials. MCF-10A cells were obtained from the American Type Culture Collection and cultured as described (15). Growth factor-reduced Matrigel was purchased from BD Biosciences, and the protein concentrations of the lots that we used were 9–11 mg/ml. zVAD·f luoromethyl ketone (fmk) (Calbiochem), 3-methyladenine (Sigma), and TRAIL (Genentech, Vacaville, CA) were used at the specified concentrations. The following Abs were used: anti-Ki-67 (Zymed), antiactivated caspase 3 (Cell Signaling Technology, Beverly, MA), anti-TRAIL mAb (Pharmingen), anti-actin (C-11, Santa Cruz Biotechnology), and anti-laminin 5 (Chemicon).
Cell-Viability Assay. Assay media was removed from wells, and acini were washed once with PBS. Structures were then incubated for 15–30 min at 37°C with 1 μM ethidium bromide (EtBr). Cell death was quantified by counting the percentage of acini with at least two EtBr-positive cells by using an Eclipse TE300 microscope (Nikon).
Protein Extraction and Western Blot Analysis from MCF-10A Acini. Acinar structures were washed briefly with 4°C PBS with protease inhibitors (10 μg/ml PMSF/1 μg/ml leupeptin/1 μg/ml aprotinin/1 μg/ml pepstatin) and then treated for 15 min at 4°C with radioimmunoprecipitation assay lysis buffer (150 mM NaCl/20 mM Tris, pH 7.5/0.1% SDS/1.0% sodium deoxycholate/1.0% Triton X-100). Matrigel and acini were collected into a 1.5-ml centrifuge tube and pulled through a 27-gauge needle three to five times before being placed on ice for 15 min. We loaded 50 μl of lysate onto 12% polyacrylamide gels, transferred them to immunoblot poly(vinylidene difluoride) membrane (BioRad), and processed them as described (16).
Morphogenesis Assays and Generation of MCF-10A Cell Lines. MCF-10A morphogenesis assays were performed as described (15) (see Supporting Methods, which is published as supporting information on the PNAS web site).
pBABEpuro Bcl-2 and pBABEhygro Bcl-XL vectors were a gift from Or Gozani and Junying Yuan (Harvard Medical School, Boston). Truncated TRAIL-R1 and TRAIL-R2 cDNAs were generated by PCR using pCMV1-Flag-DR4 and pCMV1-Flag-DR5 (kindly provided by Avi Ashkenazi, Genentech) as templates. For cloning and retroviral-infection details, see Supporting Methods.
Microscopy and Image Analysis. Immunofluorescence analysis of MCF-10A acini was performed as described (39) (see Supporting Methods). Indirect immunofluorescence was performed on a TE300 microscope (Nikon), equipped with a mercury lamp and CCD camera. Images were acquired by using ip lab spectrum software (Version 3.1; Scanalytics, Fairfax, VA). Confocal analyses were performed by using an E800 microscope (Nikon) with the Radiance 2000 confocal system (Bio-Rad). All images were converted to TIFF format and formatted by using metamorph 4.0 software (Universal Imaging, Downingtown, PA).
Transmission Electron Microscopy (TEM). MCF-10A cells were plated onto plastic cover slips for 24 h before treatment. Media were replaced with growth media containing 10 mM 3-methyladenine (Sigma), 20 μM rapamycin, 50 nM zVAD·fmk, and/or 50 ng/ml TRAIL. After treatment, media was replaced with electron-microscopy fixative (1.2% paraformaldehyde/2.5% glutaraldehyde/0.03% picric acid) in 100 mM cacodylate buffer for 1 h at room temperature and then left overnight at 4°C. For TEM on acini, structures were fixed at specific time points in electron-microscopy fixative, scraped from the coverslip with a razor blade, pelleted, and processed. Detailed protocols are available at the Harvard Medical School Department of Cell Biology Research Facilities web site (http://cellbio.med.harvard.edu/research/facilities/electron_microscopy/conventional.html).
Results
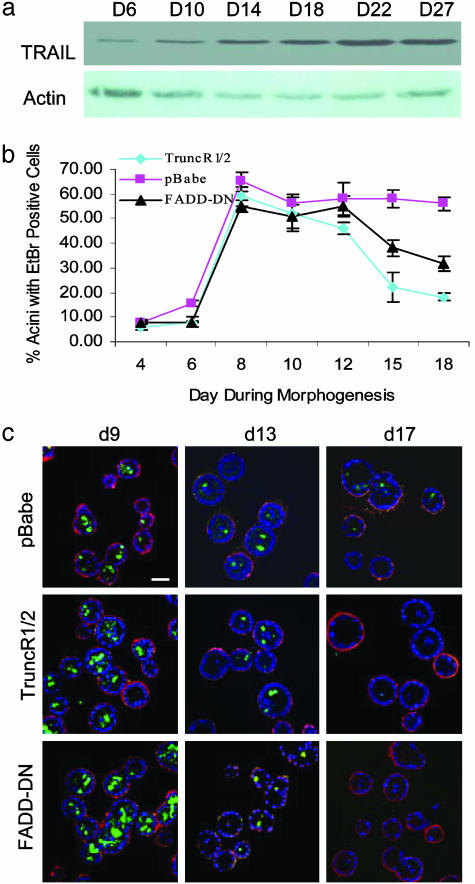
TRAIL Expression Is Regulated During MCF-10A Morphogenesis. To identify putative molecular regulators of lumen formation during morphogenesis of MCF-10A acini, we examined changes in expression of multiple genes regulating apoptosis. By using RNase protection assay apoptotic probe sets, we found that multiple apoptotic regulators are constitutively expressed throughout MCF-10A acini development; in contrast, TRAIL mRNA was up-regulated progressively during morphogenesis (see Fig. 5, which is published as supporting information on the PNAS web site). Consistent with RNA expression, TRAIL protein also increased during morphogenesis (Fig. 1a).
Fig. 1.
Effects of TRAIL inhibition on cell death during morphogenesis. (a) Cell lysates were prepared from acini on the indicated days and probed with an Ab to TRAIL or actin. (b) Indicated cell lines were seeded in Matrigel, and the percentage of acini with two or more EtBr-positive cells were quantified at the indicated time points. (c) Acini from b were immunostained with Abs against active caspase 3 (green) and laminin 5 (red). Nuclei were counterstained with TO-PRO III. Error bars represent SEM from four independent experiments in which at least 100 acini were counted. d, day. (Scale bars, 25 μm.)
Inhibition of TRAIL Signaling Decreases Late-Stage Apoptosis but Does Not Affect Lumen Formation. TRAIL up-regulation during morphogenesis intimated that it contributes to cell death and clearance during lumen formation. To examine whether TRAIL signaling is involved in formation of the luminal space, we inhibited TRAIL signaling using two dominant-inhibitory strategies. First, we generated dominant-inhibitory variants of the two known TRAIL death receptors, TRAIL-R1 and TRAIL-R2 (17), by truncation of their cytoplasmic tails. These truncated death receptors, TruncR1 and TruncR2, lack critical protein-interaction domains, known to recruit downstream apoptotic-signaling proteins. Second, we overexpressed a dominant-inhibitory variant of the Fas-associated death domain protein, FADD-DN, to disrupt the association between TRAIL receptors and caspase 8, impeding downstream signaling (18). Accordingly, on treatment with exogenous TRAIL, MCF-10A cells expressing TruncR1/2 or FADD-DN exhibited 8-fold and 3.5-fold reduced levels of caspase 3 activation, respectively, compared with control cells transduced with empty vector (pBabe) (data not shown). Therefore, TruncR1, TruncR2, and FADD-DN dominantly inhibit TRAIL-induced caspase 3 activation.
To examine the effects of blocking endogenous TRAIL-signaling during morphogenesis, MCF-10A cells expressing TruncR1/2 or FADD-DN were cultured in Matrigel and acinar cell death was quantified by staining with EtBr (Fig. 1b) or antiserum to activated caspase 3 (Fig. 1c). Cell death occurred normally during the development of the FADD-DN- and TruncR1/2-expressing acini. At day 8, 60% of FADD-DN and TruncR1/2-acini contained centrally localized EtBr positive cells, similar to controls. However, at day 15 the amount of cell death in FADD-DN and TruncR1/2-expressing acini decreased sharply, with only 20% of acini exhibiting cell death (Fig. 1 b and c). Similar results were observed with acini that developed from cells overexpressing either TruncR1 or TruncR2 alone (data not shown). Interestingly, the decrease in apoptosis in FADD-DN and TruncR1/2-expressing acini after day 15 did not affect lumen maintenance because the luminal space of acini did not fill, suggesting that protection from caspase activity was not sufficient to maintain cell viability in the lumen. The loss of TRAIL signaling did not result in a detectable change in proliferation or the development of an axis of polarity compared with pBabe control structures (data not shown). These results suggest that apoptotic events that take place after luminal clearance may depend on TRAIL; however, inhibition of the TRAIL-dependent apoptosis is not sufficient to cause luminal filling.
TRAIL Cooperates with Bcl-2 Family Members to Regulate Lumen Formation in MCF-10A Acini. Our recent studies indicate that upregulation of the BH3-only protein Bim is critically involved in cavitation of MCF-10A acini and that either Bcl-XL overexpression (which blocks the activity of Bim and other BH3-only proapoptotic proteins) or down-regulation of Bim by short-inhibitory RNA is sufficient to delay cavitation (M.R., K.R.M., J.D., D. Lynch, and J.S.B., unpublished results). Because Bim is still induced in TruncR1/2- and FADD-DN-expressing acini (data not shown), apoptosis induction by this proapoptotic protein could be sufficient to mediate lumen formation despite lack of TRAIL signaling. To determine whether TRAIL functions in concert with cell-death processes induced by BH3-only family members to elicit lumen formation, we overexpressed Bcl-XL along with the truncated TRAIL death receptors in MCF-10A cells (Bcl-XL plus TruncR1/2) and examined morphogenetic cell death and lumen formation. Luminal filling was quantified by counting the number of centrally localized cells without fragmented nuclei. Acini with two or more intact nuclei in the lumen were considered “filled” for this analysis.
At day 10, Bcl-XL- and Bcl-XL plus TruncR1/2-overexpressing acini retained more viable cells in the lumen (67% and 70%, respectively), than pBabe control acini (55%) or TruncR1/2 expressing cells (56%) (Fig. 2 a Left and b). By day 20, there was a dramatic reduction in acini containing filled lumen in Bcl-XL structures (33%). In contrast, the majority of XL plus TruncR1/2-overexpressing acini (62%) remained filled and some were structurally distorted (larger, with atypical morphology), suggesting that the combined expression of these proteins prevented luminal clearing (day 20, P = 0.0005). Because the defect in luminal clearance could reflect a failure of the XL plus TruncR1/2 acinar cells to arrest proliferation, we monitored cycling cells by using Ki67 staining. All cell lines examined exhibited a significant decrease in Ki67 positivity by day 15 (Figs. 2a and 6, which is published as supporting information on the PNAS web site). Therefore, the complementary effects of Bcl-XL-overexpression and inhibition of TRAIL-mediated signaling on luminal clearing suggest that these proteins may regulate distinct clearance pathways. Bcl-XL overexpression is sufficient to protect from TRAIL-induced apoptosis in MCF-10A acini (data not shown), suggesting that TRAIL must regulate a nonapoptotic process during cavitation. Previously, we demonstrated the involvement of autophagy-like processes during normal MCF-10A morphogenesis by using TEM. Based on these results, we speculated that both classical apoptosis and Bcl-XL-independent, autophagy-like process contribute to cavitation of MCF-10A acini. Because TruncR1/2 can complement Bcl-XL in blocking cavitation, we investigated if TRAIL regulated autophagy during cavitation.
Fig. 2.
Cooverexpression of Bcl-XL and dominant-inhibitory TRAIL receptors delays luminal clearance in MCF-10A acini. (a) Indicated cell lines were cultured in Matrigel for the indicated number of days (d). Images are representative confocal crossections through the center of acini immunostained with laminin 5 (red) and Ki67 (green). Nuclei were counterstained with TO-PRO III (blue). (Scale bars, 25 μm.) (b) The percentage of acini with two or more intact nuclei located within the lumen was quantified. Numbers are means of three independent experiments performed with a minimum of 100 acini scored for each cell line at all time points. *, P = 0.0005, by Fisher's exact test with Monte Carlo analysis.
TRAIL Treatment Induces Autophagy in MCF-10A Cells. To determine whether TRAIL is capable of inducing autophagy, we examined the ultrastructure of TRAIL-treated monolayer cells by using TEM. Although many cells (≈50%) detached from the coverslips during this 24-h treatment period, the remaining cells appeared to be viable. In the cells that remained viable, we observed characteristic features of autophagy, but not apoptosis. Specifically, cells did not have condensed cytoplasms or fragmented nuclei. Instead, 45% of pBabe-expressing control cells treated with 50 ng/ml TRAIL for 24 h, had evidence of extensive cytoplasmic vacuolization, whereas <5% of untreated cells exhibited such vacuoles (Fig. 3; see also Fig. 7, which is published as supporting information on the PNAS web site). At high magnifications (≥×35,000), a double membrane was clearly detectable around the majority of vacuoles (Fig. 3b). Moreover, most of the vacuoles contained electron dense material and some had engulfed entire organelles. These morphological features are characteristic of vacuoles associated with autophagy (14). Interestingly, overexpression of Bcl-XL did not inhibit the autophagic response to TRAIL treatment – 58% of cells displayed evidence of autophagy (Fig. 3 c–e). However, TruncR1/2 and FADD-DN overexpression significantly abrogated TRAIL-induced AV formation [6% (Fig. 3) or 11% (Fig. 7) of cells displayed evidence of autophagy].
Fig. 3.
TRAIL treatment induces AV formation in monolayer cultures. (a and b) MCF-10A cells infected with empty vector (pBabe) were treated with vehicle (a)or50ng/ml recombinant human TRAIL (b) for 48 h and analyzed by using TEM. b Inset is a representative high-magnification image of the outer membrane of an AV from a TRAIL-treated monolayer. (c–e) TEM images of Bcl-XL-expressing (c), TruncR1/2-expressing (d), or FADD-DN-expressing (e) structures treated with TRAIL as in b. AVs were observed in Bcl-XL cells (arrows) but not in TruncR1/2 or FADD-DN cells treated with TRAIL. (Scale bars, 200 nm.)
To investigate the processes involved in the formation of these autophagosome-like vacuoles in MCF-10A monolayers we examined the effects of two specific inhibitors on TRAIL-induced vacuoles: z-VAD·fmk, a relatively nonspecific caspase inhibitor that can block TRAIL-mediated caspase activation (data not shown); and 3-methyladenine (3-MA), which inhibits AV formation in many cell types (19). TRAIL-induced vacuole formation was inhibited in the presence of 10 mM 3-MA, but not 50 μM zVAD·fmk (see Fig. 8, which is published as supporting information on the PNAS web site). These data suggest that the vacuoles induced by TRAIL in MCF-10A cells are autophagic in origin and do not depend on caspase activity for their formation.
Also, we examined whether TRAIL is able to induce a similar phenotype in MCF-10A cells cultured in Matrigel by treating acinar structures for 24 h with TRAIL before induction of autophagy or caspase 3 activation (day 5). The central cells from TRAIL-treated structures contained many double-membrane, organelle-rich vacuoles (see Fig. 9, which is published as supporting information on the PNAS web site), similar to vacuoles found in treated monolayer cultures.
Inhibition of TRAIL Signaling Blocks Autophagic-Like Processes During Normal Morphogenesis. To determine whether TRAIL signaling is responsible for the induction of the AVs detected during morphogenesis of MCF-10A cells, we compared the ultrastructure of acini generated from cells overexpressing Bcl-XL, TruncR1/2, FADD-DN, and XL plus TruncR1/2. We did not observe significant numbers of AVs in acini from any cell line at day 5. At day 9, however, abnormal vacuoles, similar to those observed in TRAIL-treated monolayers, were apparent in many of the central cells in pBabe control and Bcl-XL-expressing acini (data not shown). At this time, the vacuoles were relatively small, and many contained entire mitochondria. By day 13, the number of vacuoles per cell, the overall number of cells with AVs, and the vacuole size increased (Figs. 4 and 10, which is published as supporting information on the PNAS web site). Contents included organelles; dark, unidentifiable electron-dense material; or highly degraded cellular contents. AVs were highly enriched in centrally localized cells and strongly inhibited in acini overexpressing either TruncR1/2 or FADD-DN (Fig. 4, bottom two rows). Together, these data indicate that TRAIL-mediated signaling is required for the induction of autophagy during mammary acinar morphogenesis and that it contributes to clearing the luminal space.
Fig. 4.
Inhibition of TRAIL signaling disrupts the formation of AVs in MCF-10A acini. Indicated cell lines were grown in Matrigel culture, fixed on day 13, and analyzed by TEM for evidence of autophagy. Images are representative TEMs of cells found within the luminal space. (Scale bars in Left, 1 μm.) (Right) Higher magnifications of inner cells (scale bars, 200 nm). Insets (2 μm) demonstrate the clear difference between the inner and outer cell populations.
Discussion
The results presented in this article indicate that TRAIL is a critical determinant of cavitation during MCF-10A morphogenesis. Although TRAIL is not required for early induction of apoptosis, the expression of this death ligand during morphogenesis mediates the induction of an autophagic process that occurs in parallel with apoptosis. Suppression of either apoptosis or TRAIL signaling alone does not prevent lumen formation, whereas simultaneous inhibition of both processes prevents cell clearance. These studies, together with our description of apoptosis regulation by the BH3-only protein Bim (M.R., K.R.M., J.D., D. Lynch, and J.S.B., unpublished data), suggest that two distinct death processes act in concert to elicit lumen formation in vitro.
TRAIL Induces Autophagy in MCF-10A Acini. Lumen formation occurs at days 8–15 of culture in Matrigel. During that period, inhibition of TRAIL by overexpression of dominant-inhibitory proteins did not inhibit caspase activation, indicating that although TRAIL is capable of inducing caspase-mediated apoptosis in MCF-10A cells, endogenous TRAIL is not functionally required for apoptosis during lumen formation. However, the evidence presented here strongly implicates TRAIL in the autophagic process that accompanies lumen formation. First, exogenous TRAIL is capable of inducing AVs that resemble the vacuoles detected in the presumptive lumen of acini. Secondly, overexpression of either dominant-inhibitory TRAIL death receptors or FADD-DN was sufficient to impede endogenous TRAIL-induced AV formation. Although our results strongly suggest that TRAIL regulates lumen formation by means of the induction of autophagic processes, further studies are required to prove that autophagic processes are exclusively responsible for the contribution of TRAIL to lumen formation.
TRAIL has not been implicated in autophagic processes; however, TNF-α, another member of the death ligand protein family, has been shown to induce autophagy after exogenous treatment of T lymphocyte and 3T3L1 cell lines (20, 21). TNF-α treatment of two T-lymphoblastic leukemic cell lines causes phenotypic effects that are characteristic of both autophagy and apoptosis (20). Together with our data, these results indicate that autophagy can occur coincident with apoptosis after treatment with death ligands, and suggest that TRAIL-induced autophagy might have been overlooked in studies that focused on markers of classical apoptosis. Although TNF treatment has been shown to induce autophagy in cultured cells, the finding that endogenous TRAIL mediates a spatially and temporally regulated event in the morphogenesis of acini in vitro provides the first evidence implicating an endogenous death ligand in a morphogenetic process. It will be of interest to identify the death ligand-induced signals that mediate activation of autophagic programs in vivo. The evidence that FADD-DN blocked autophagy in the MCF-10A cells indicates that autophagic processes depend on the factors that interact with the death domain of TRAIL receptors.
Cooperation Between Distinct Death Processes During Lumen Formation. Our data suggest that two distinct death processes are involved in cavitation during MCF-10A morphogenesis. Cooperation between death processes has been seen in both in vivo (22) and in vitro (23) models. Xue et al. (23) demonstrate that NGF-withdrawal in sympathetic neurons results in both a caspase-dependent and autophagic cell-death response. Inhibition of caspases by using Boc-Asp(Ome)·fmk prevented apoptosis, but not the subsequent death of neurons, by autophagy. Often, the secondary form of death is revealed only if caspase activity is inhibited.
Other studies have demonstrated that classical apoptotic events, like caspase activation, might be coupled functionally with autophagy under certain conditions. For example, during salivary-gland morphogenesis in Drosophila, several apoptosis-associated molecules are transcriptionally induced (24); however, the morphology of the dying cells more closely resembles autophagy (25). Indeed, when caspase activity is inhibited by ectopic overexpression of the caspase-inhibitory protein p35 in the salivary gland, autophagic cell death is inhibited (26). Although we have demonstrated that caspase activity is not required for the autophagic events induced by TRAIL, the precise relationship between TRAIL-induced apoptosis and autophagy requires further study.
Differential Sensitivity to TRAIL in Acinar Cells. TRAIL expression peaks after 3 weeks of morphogenesis, when acini are completely hollow. These data indicate that, although TRAIL is expressed in the outer cells, they are insensitive to the death signal. Furthermore, TRAIL-R1, TRAIL-R2, and the adaptor molecule FADD are expressed constitutively (data not shown), suggesting that TRAIL-signaling effectors are intact in both the inner and outer cell populations of developing acini. Nevertheless, treatment of acini with exogenous TRAIL induced autophagy exclusively in the central cells of MCF-10A acini. The hypersensitivity of centrally localized cells within solid masses to death signals has been observed in other in vitro epithelial-morphogenesis models (3, 8–10, 12). Data from several of these models suggest that the basement membrane provides survival signals that protect from death ligands. In addition, we found that cells deprived of matrix by incubation in suspension are differentially sensitive to TRAIL-induced apoptosis (data not shown). Thus, our data and previous reports implicate basement-membrane survival signals in establishment of the survival dichotomy that results in the specific removal of central cells.
The basis for the differential sensitivity to TRAIL is not understood. Several factors that modulate the sensitivity of cells to TRAIL signaling have been described. TRAIL signals can be blocked by expression of decoy receptors (reviewed in ref. 27) or the FLICE-inhibitory protein, FLIP (28). In addition, activation of Akt (29, 30) or Mek (31) also has been implicated in protection from TRAIL-induced cell death. We have described (6, 32) differential phosphorylation of Akt and mTOR in the inner and outer cells of developing acini; thus, the decreased signaling to Akt and mTOR could sensitize the cells to TRAIL because TOR has been shown to regulate autophagy in other cell systems (33). Other signals may influence protection also; notably, cell polarity may be observed to regulate mammary epithelial-cell survival in the presence of proapoptotic stimuli, such as TRAIL (34).
Although it is well established that transformed cells exhibit decreased apoptosis, recent evidence supports a role for reduced autophagy in mammary-tumor development. Overexpression of beclin, the mammalian homologue of the yeast autophagy gene apg6, in a tumorigenic mammary cell line (MCF-7) inhibits the formation of tumors in nude mice (35). Furthermore, beclin is monoallelically deleted in 40–75% of ovarian and breast cancers, strongly implicating it as a tumor suppressor. Last, studies from two laboratories have demonstrated that loss of one copy of beclin results in decreased autophagy and a high incidence of spontaneous tumor formation (36, 37).
One of the hallmarks of early breast cancer lesions is filling of the luminal space, and our results suggest that oncogenes that elicit luminal filling may do so by protecting excess cells in the lumen from both apoptosis and autophagy. These findings, thus, raise the question of whether misregulation of TRAIL or its death-inducing effectors may be involved in the pathogenesis of breast or other epithelial cancers, or whether it potentially plays a role in other diseases that arise at least in part because of inappropriate autophagic cell death, such as Parkinson's disease and Alzheimer's disease (38, 39).
Supplementary Material
Acknowledgments
We thank S. Muthuswamy for discussions during the early stages of this work; J. Paulus for assistance with cloning; E. Bennechi for help with TEM experiments; Michael Overholtzer for the critical reading of the manuscript; R. Gelman for statistical analyses; the Nikon Imaging Facility at Harvard Medical School for confocal equipment; V. Dixit, A. Ashkenazi, and Genentech for providing TRAIL and the TRAIL-R1 and TRAIL-R2 cDNA constructs; and J. Yuan for providing the FADD-DN vector. This work was supported by National Cancer Institute Grants CA80111, CA105134, and CA89393 and grants from the American Cancer Society. K.R.M. was funded by a National Science Foundation Predoctoral Fellowship; M.R. was funded by a Susan Komen Breast Cancer Postdoctoral Fellowship; and J.D. was funded by a Howard Hughes Medical Institute Postdoctoral Fellowship for Physicians and National Cancer Institute Grant 1KO8CA098419.
Abbreviations: AV, autophagic vacuole; EtBr, ethidium bromide; fmk, fluoromethyl ketone; TEM, transmission electron microscopy; TNF, tumor necrosis factor; TRAIL, TNF-related apoptosis-inducing ligand.
References
- 1.Baehrecke, E. H. (2002) Nat. Rev. Mol. Cell Biol. 3, 779-787. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson, M. D., Weil, M. & Raff, M. C. (1997) Cell 88, 347-354. [DOI] [PubMed] [Google Scholar]
- 3.Coucouvanis, E. & Martin, G. R. (1995) Cell 83, 279-287. [DOI] [PubMed] [Google Scholar]
- 4.Jaskoll, T. & Melnick, M. (1999) Anat. Rec. 256, 252-268. [DOI] [PubMed] [Google Scholar]
- 5.Hogan, B. L. & Kolodziej, P. A. (2002) Nat. Rev. Genet. 3, 513-523. [DOI] [PubMed] [Google Scholar]
- 6.Debnath, J., Mills, K. R., Collins, N. L., Reginato, M. J., Muthuswamy, S. K. & Brugge, J. S. (2002) Cell 111, 29-40. [DOI] [PubMed] [Google Scholar]
- 7.Muthuswamy, S. K., Li, D., Lelievre, S., Bissell, M. J. & Brugge, J. S. (2001) Nat. Cell Biol. 3, 785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, J., Hardy, J. D., Sun, Y. & Shively, J. E. (1999) J. Cell Sci. 112, 4193-4205. [DOI] [PubMed] [Google Scholar]
- 9.Blatchford, D. R., Quarrie, L. H., Tonner, E., McCarthy, C., Flint, D. J. & Wilde, C. J. (1999) J. Cell. Physiol. 181, 304-311. [DOI] [PubMed] [Google Scholar]
- 10.Coucouvanis, E. & Martin, G. R. (1999) Development (Cambridge, U.K.) 126, 535-546. [DOI] [PubMed] [Google Scholar]
- 11.Lin, H. H., Yang, T. P., Jiang, S. T., Yang, H. Y. & Tang, M. J. (1999) Kidney Int. 55, 168-178. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, M. P., Kibbey, M. C., Letterio, J. J. & Kleinman, H. K. (1996) J. Cell Sci. 109, 2013-2021. [DOI] [PubMed] [Google Scholar]
- 13.Humphreys, R. C., Krajewska, M., Krnacik, S., Jaeger, R., Weiher, H., Krajewski, S., Reed, J. C. & Rosen, J. M. (1996) Development (Cambridge, U.K.) 122, 4013-4022. [DOI] [PubMed] [Google Scholar]
- 14.Reggiori, F. & Klionsky, D. J. (2002) Eukaryot. Cell 1, 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debnath, J., Muthuswamy, S. K. & Brugge, J. S. (2003) Methods (San Diego, Calif.) 30, 256-268. [DOI] [PubMed] [Google Scholar]
- 16.Muthuswamy, S. K., Gilman, M. & Brugge, J. S. (1999) Mol. Cell. Biol. 19, 6845-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashkenazi, A. & Dixit, V. M. (1999) Curr. Opin. Cell Biol. 11, 255-260. [DOI] [PubMed] [Google Scholar]
- 18.Chinnaiyan, A. M., Tepper, C. G., Seldin, M. F., O'Rourke, K., Kischkel, F. C., Hellbardt, S., Krammer, P. H., Peter, M. E. & Dixit, V. M. (1996) J. Biol. Chem. 271, 4961-4965. [DOI] [PubMed] [Google Scholar]
- 19.Seglen, P. O. & Gordon, P. B. (1982) Proc. Natl. Acad. Sci. USA 79, 1889-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia, L., Dourmashkin, R. R., Allen, P. D., Gray, A. B., Newland, A. C. & Kelsey, S. M. (1997) Br. J. Haematol. 98, 673-685. [DOI] [PubMed] [Google Scholar]
- 21.Prins, J. B., Ledgerwood, E. C., Ameloot, P., Vandenabeele, P., Faraco, P. R., Bright, N. A., O'Rahilly, S. & Bradley, J. R. (1998) Biosci. Rep. 18, 329-340. [DOI] [PubMed] [Google Scholar]
- 22.Chautan, M., Chazal, G., Cecconi, F., Gruss, P. & Golstein, P. (1999) Curr. Biol. 9, 967-970. [DOI] [PubMed] [Google Scholar]
- 23.Xue, L., Fletcher, G. C. & Tolkovsky, A. M. (1999) Mol. Cell. Neurosci. 14, 180-198. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C. Y., Wendel, D. P., Reid, P., Lam, G., Thummel, C. S. & Baehrecke, E. H. (2000) Mol. Cell 6, 433-443. [DOI] [PubMed] [Google Scholar]
- 25.von Gaudecker, B. & Schmale, E. M. (1974) Cell Tissue Res. 155, 75-89. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C. Y. & Baehrecke, E. H. (2001) Development (Cambridge, U.K.) 128, 1443-1455. [DOI] [PubMed] [Google Scholar]
- 27.Sheridan, J. P., Marsters, S. A., Pitti, R. M., Gurney, A., Skubatch, M., Baldwin, D., Ramakrishnan, L., Gray, C. L., Baker, K., Wood, W. I., et al. (1997) Science 277, 818-821. [DOI] [PubMed] [Google Scholar]
- 28.Irmler, M., Thome, M., Hahne, M., Schneider, P., Hofmann, K., Steiner, V., Bodmer, J. L., Schroter, M., Burns, K., Mattmann, C., et al. (1997) Nature 388, 190-195. [DOI] [PubMed] [Google Scholar]
- 29.Thakkar, H., Chen, X., Tyan, F., Gim, S., Robinson, H., Lee, C., Pandey, S. K., Nwokorie, C., Onwudiwe, N. & Srivastava, R. K. (2001) J. Biol. Chem. 276, 38361-38369. [DOI] [PubMed] [Google Scholar]
- 30.Nesterov, A., Lu, X., Johnson, M., Miller, G. J., Ivashchenko, Y. & Kraft, A. S. (2001) J. Biol. Chem. 276, 10767-10774. [DOI] [PubMed] [Google Scholar]
- 31.Tran, S. E., Holmstrom, T. H., Ahonen, M., Kahari, V. M. & Eriksson, J. E. (2001) J. Biol. Chem. 276, 16484-16490. [DOI] [PubMed] [Google Scholar]
- 32.Debnath, J., Walker, S. J. & Brugge, J. S. (2003) J. Cell Biol. 163, 315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raught, B., Gingras, A. C. & Sonenberg, N. (2001) Proc. Natl. Acad. Sci. USA 98, 7037-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver, V. M., Lelievre, S., Lakins, J. N., Chrenek, M. A., Jones, J. C., Giancotti, F., Werb, Z. & Bissell, M. J. (2002) Cancer Cells 2, 205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H. & Levine, B. (1999) Nature 402, 672-676. [DOI] [PubMed] [Google Scholar]
- 36.Qu, X., Yu, J., Bhagat, G., Furuya, N., Hibshoosh, H., Troxel, A., Rosen, J., Eskelinen, E. L., Mizushima, N., Ohsumi, Y., et al. (2003) J. Clin. Invest. 112, 1809-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue, Z., Jin, S., Yang, C., Levine, A. J. & Heintz, N. (2003) Proc. Natl. Acad. Sci. USA 100, 15077-15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anglade, P., Vyas, S., Javoy-Agid, F., Herrero, M. T., Michel, P. P., Marquez, J., Mouatt-Prigent, A., Ruberg, M., Hirsch, E. C. & Agid, Y. (1997) Histol. Histopathol. 12, 25-31. [PubMed] [Google Scholar]
- 39.Jellinger, K. A. & Stadelmann, C. (2000) J. Neural Transm. Suppl. 59, 95-114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.