Abstract
Background
The live attenuated yellow fever vaccine 17D (YF-17D) is one of the most effective vaccines. Despite its excellent safety record, some cases of viscerotropic adverse events develop, which are sometimes fatal. The mechanisms underlying such events remain a mystery. Here, we present an analysis of the immunologic and genetic factors driving disease in a 64-year-old male who developed viscerotropic symptoms.
Methods
We obtained clinical, serologic, virologic, immunologic and genetic data on this case patient.
Results
Viral RNA was detected in the blood 33 days after vaccination, in contrast to the expected clearance of virus by day 7 after vaccination in healthy vaccinees. Vaccination induced robust antigen-specific T and B cell responses, which suggested that persistent virus was not due to adaptive immunity of suboptimal magnitude. The genes encoding OAS1, OAS2, TLR3, and DC-SIGN, which mediate antiviral innate immunity, were wild type. However, there were heterozygous genetic polymorphisms in chemokine receptor CCR5, and its ligand RANTES, which influence the migration of effector T cells and CD14+CD16bright monocytes to tissues. Consistent with this, there was a 200-fold increase in the number of CD14+CD16bright monocytes in the blood during viremia and even several months after virus clearance.
Conclusion;
In this patient, viscerotropic disease was not due to the impaired magnitude of adaptive immunity but instead to anomalies in the innate immune system and a possible disruption of the CCR5-RANTES axis.
Yellow fever is a mosquitoborne hemorrhagic disease that is endemic in sub-Saharan Africa and tropical South America. The etiologic agent, the yellow fever virus (YFV) is a single-stranded RNA virus in the family Flaviviridae, which also includes the dengue and West Nile viruses. After a natural YFV infection, viral replication initially occurs in tissues at the site of infection, but it rapidly spreads to the lymph nodes, blood, and liver [1]. The live attenuated yellow fever vaccine (YF-17D) was developed in the 1930s through experimental attenuation of the Asibi strain of YFV by serial passaging in cell culture [2]. The vaccine is considered safe and extremely effective, and it has been administered to >500 million people worldwide [3, 4]. Protection is achieved in >98% of recipients, with a duration of at least 10 years and probably much longer, given that significant neutralizing antibody titers may persist for ≥35 years after a single vaccination [3, 4].
Although YF-17D is usually a well-tolerated vaccine, in rare cases (approximately 1 in 250,000 vaccinees) individuals develop severe viscerotropic adverse reactions within 2 to 5 days after vaccination; these reactions are sometimes fatal [5–8]. Yellow fever vaccine–associated viscerotropic disease is characterized by the failure of multiple organ systems [5–8]. Within 2–5 days after vaccination, patients develop high fever, malaise, and myalgia, followed by jaundice, oliguria, cardiovascular instability, hemorrhage, and renal and respiratory failure. The case fatality rate is over 50%, and large amounts of YFV antigen may be found in the liver, heart, and other organs, primarily in tissue-associated macrophages [5–9].
The syndrome was first described in 2001, but cases in 1975 and the 1990s were identified retrospectively. To date, a total of 36 cases have been reported worldwide. Genetic mutations in the YFV do not seem to be the cause of the adverse reactions, because in several instances YFV isolated from subjects has had the same consensus nucleotide sequence as the original vaccine strain virus [8]. Furthermore, the YFV isolates recovered from the subjects showed no reversion to virulence in animal models, suggesting that host factors may be involved in disease [8].
METHODS
The research was approved by the Emory University institutional review board. The patient signed a written informed consent form. Specimens obtained for virus isolation and serological analysis were stored at −70°C until used. Control blood samples were obtained from unvaccinated individuals or healthy vaccinees, matched for age and sex, aged 21–45 years. Viral load data was obtained using a TaqMan real-time polymerase chain reaction (PCR) assay (Applied Biosystems), as described elsewhere [9]. YFV neutralization assays were performed as described elsewhere [10].
Flow cytometry
All antibodies were obtained from BD Pharmingen. Antibodies were added to 200 μL of whole blood and incubated at room temperature for 30 min, followed by a 10 min lysis of red blood cells by use of FACS Lysing Solution (BD Pharmingen). Samples were analyzed on a FACScalibur flow cytometer (BD Pharmingen) and data were analyzed with Flowjo (Treestar) software.
Peripheral blood mononuclear cell (PBMC) stimulation assays
PBMCs were stimulated by infection with recombinant vesicular stomatitis virus (VSV) expressing YFV proteins at an MOI of 1 at 37°C. After 5 h, brefeldin A was added and the cultures were incubated overnight. Cultures were then incubated with anti-CD3 and anti-CD8 monoclonal antibodies and fixed and permeabilized (Cytofix/Cytoperm; BD Pharmingen), and intracellular interferon (IFN)–γ and CD3 were detected using specific antibody. Samples were analyzed on a FACScalibur flow cytometer (BD Pharmingen), and data were analyzed using Flowjo (Treestar) software.
Serum cytokine detection using Multiplex assay
Cytokine assays were done with the Bio Rad human 18-plex multicytokine detection panel. The data were acquired using the Luminex 100 reader and analyzed with Masterplex Quantitation software (Miraibio).
Genotyping of genetic polymorphisms
PCR was used to genotype CCR5-Δ32 [11]. PCR restriction fragment length polymorphism was used to genotype RANTES-403 and RANTES-28 [12]. The sequences of a region of a CCR5 exon, the CCR5 promoter, and the RANTES promoter were determined by direct sequencing and from clone sequences [13]. For the Oas1, Oas2 [13–15], and Tlr3 [16] genes, the promoter regions and each of the exons were amplified by PCR.
RESULTS
Clinical Presentation
On October 27, 2004, a 64-year-old, previously healthy white male presented to his primary care provider with a 5-day history of fever, chills, urinary frequency, and 1 day of nausea, vomiting, and diarrhea (table 1). These symptoms began 2 days after vaccination on October 20, 2004, with the yellow fever vaccine licensed for use in the United States (YF-VAX; Sanofi Pasteur). On the same day, he was vaccinated against hepatitis A (Havrix; SmithKline Beecham) and typhoid fever (Typhim Vi; Sanofi Pasteur). Physical examination revealed a blood pressure of 90/70 mm Hg and temperature of 38.5°C. The rest of the physical examination was unremarkable. Laboratory findings were also unremarkable, with the exception of the patient’s low platelet count (table 1). He was diagnosed with viral gastroenteritis and treated symptomatically.
Table 1.
Clinical, viral, and immunological data for a 64-year-old man who developed viscerotropic symptoms after yellow fever vaccination, according to time after vaccination.
| Variable | Time after vaccination | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference or normal values | 7 daysa | 9 daysb | 11 days | 15 days | 17 days | 27 daysc | 33 days | 43 days | 55 days | 113 days | |
| Clinical signs and symptoms | … | Nausea, vomiting, diarrhea | Weakness, diarrhea, vomiting, confusion, erythematous rash on trunk and legs, petechial rash on face | NA | NA | NA | NA | NA | NA | NA | NA |
| Temperature, °C | 36.5–37.3 | 38.5 | 38.1 | 38.9 | 37 | NA | NA | 36.8 | 36.9 | 36.9 | 37.4 |
| Blood pressure, mm Hg | 120/80 | 90/70 | 70/60 | NA | 118/60 | NA | NA | 100/80 | 100/80 | 130/80 | 120/70 |
| Receipt of vasopressors | … | ND | Yes | ND | ND | ND | ND | ND | ND | ND | ND |
| WBC count, × 103 WBCs/mm3 | 3.4–10.8 | 6.3 | 8.2 | 7.9 | 10.9 | ND | 6.7 | ND | 11.3 | 7.7 | ND |
| Neutrophils, % | 24–75 | 97 | 93 | 90 | 99 | ND | 70 | ND | ND | 44 | ND |
| Lymphocytes, % | 14–65 | 1 | ND | 1 | 1 | ND | 17 | ND | ND | 32 | ND |
| Platelets, × 103 platelets/mm3 | 150–440 | 120 | 36 | 30 | 73 | ND | 321 | ND | 543 | 339 | ND |
| PBMCs, cells/mL | ND | ND | ND | ND | ND | ND | 107 | ND | ND | 106 | ND |
| Calcium level, mg/dL | 8.4–10.2 | 8.6 | 6.7 | 6.4 | 7.4 | ND | 7.7 | ND | 9.1 | ND | ND |
| Aspartate aminotransferase level, IU/L | 10–42 | 79 | 210 | 318 | 143 | ND | 50 | ND | 31 | ND | ND |
| Alanine aminotransferase level, IU/L | 8–50 | 37 | 65 | 82 | 105 | ND | 44 | ND | 24 | ND | ND |
| Alkaline phosphatase level, IU/L | 38–126 | 64 | 66 | 115 | 298 | ND | 168 | ND | 100 | ND | ND |
| Direct bilirubin level, mg/dL | 0.2–1.3 | 1.2 | 1.7 | 3.3 | 3.4 | ND | 1.4 | ND | 1.2 | ND | ND |
| Creatinine level, mg/dL | 0.8–1.5 | 1.5 | 3.6 | 2.6 | 3.1 | ND | 2.0 | ND | 1.4 | ND | ND |
| Blood urea nitrogen level, mg/dL | 9–20 | 19 | 80 | 63 | 46 | ND | 18 | ND | 11 | ND | ND |
| Creatine kinase level, IU/L | 55–170 | ND | ND | 3443 | 455 | ND | ND | ND | ND | ND | ND |
| Lactic acid level, mmol/L | 0.7–2.1 | ND | 3.6 | 1.3 | ND | ND | ND | ND | ND | ND | ND |
| Prothrombin time, s/INR | 11.7–15.0/0.84–1.15 | ND | ND | 16.2/1.21 | 22.2/2.0 | ND | ND | ND | ND | ND | ND |
| D-Dimer, ng/mL | 68–500 | ND | ND | 5136 | 1577 | ND | ND | ND | ND | ND | ND |
| Plasma viral load, YFV RNA copies/mL | … | ND | ND | ND | ND | ND | 145 | 75 | ND | ND | 0 |
| Immune response to YFV vaccination | |||||||||||
| Adaptive | |||||||||||
| HLA-DR+CD38+ CD8+ CD3+ T cells, % | >10 | ND | ND | ND | ND | ND | 20 | 52 | ND | ND | 40 |
| Neutralizing antibody titers against YFV | >1/600 | ND | ND | ND | ND | 1/2560 | >1/10,240 | 1/40,960 | ND | ND | ND |
| Innate | |||||||||||
| Cytokines and chemokines | |||||||||||
| IL-1α, pg/mL | >80 | ND | ND | ND | ND | ND | 600 | 400 | ND | ND | 200 |
| IL-6, pg/mL | >10 | ND | ND | ND | ND | 10 | 20 | 10 | ND | ND | 10 |
| IP-10, pg/mL | >200 | ND | ND | ND | ND | 2800 | 2500 | 1500 | ND | ND | 500 |
| MCP-1, pg/mL | >25 | ND | ND | ND | ND | 1300 | 300 | 100 | ND | ND | 100 |
| RANTES, ng/mL | >0.05 | ND | ND | ND | ND | >0.05 | 0.4 | 0.5 | ND | ND | 0.6 |
| CD14+CD16 bright monocytes, cells/mL | 500 | ND | ND | ND | ND | ND | 145,000 | ND | ND | ND | 5000 |
NOTE. IL, interleukin; INR, international normalized ratio; IP, interferon-inducible protein; MCP, macrophage chemoattractant protein; NA, not available; ND, not determined; PBMCs, peripheral blood mononuclear cells; WBC, white blood cells; YFV, yellow fever virus. Cells containing ellipses (…) indicate categories that are not applicable.
First outpatient visit.
Hospital admission.
Hospital discharge.
The patient returned on October 29 with increasing weakness, diarrhea, vomiting, and confusion. His blood pressure had decreased to 70/60 mm Hg and his temperature remained elevated at 38.1°C. He had developed a mild erythematous rash on his legs and trunk and a petechial rash on his face. Laboratory findings showed a white blood cell count of 8,200 cells/mm3 (93% neutrophils), a platelet count of 36,000 platelets/mm3, a blood urea nitrogen level of 80 mg/dL, a lactic acid level of 2.6 mg/dL, an alanine aminotransferase (ALT) level of 65 IU/L, an aspartate aminotransferase (AST) level of 210 IU/L, an alkaline phosphatase level of 66 IU/L, and a direct bilirubin level of 1.7 mg/dL. He was admitted to the hospital for rehydration, and shortly after admission he was transferred to the intensive care unit and placed on broad-spectrum antibiotic therapy with levofloxacin and piperacillin-tazobactam. He also received intravenous fluids, vasopressors, and corticosteroids. The initial diagnosis was sepsis of unknown origin.
The patient’s renal function continued to deteriorate, his platelet count decreased further, and his liver enzyme levels increased (table 1). The patient required support in the intensive care unit for several days, and his condition remained stable. By October 31 (11 days after vaccination), the results of his liver function tests (AST, ALT, and direct bilirubin) peaked (at 318 IU/L, 82 IU/L, and 3.3 mg/dL, respectively), and the platelet count reached its lowest value, 30,000 cells/mm3 (table 1). The patient began to show improvement during the first days of November, and the laboratory findings continued to normalize. Cultures of blood, urine, and stool samples obtained prior to antibiotic administration yielded no bacterial growth, and tests for hepatitis A, B, and C were negative. After <3 weeks of hospitalization, the patient was discharged on November 15, 2004, in stable condition with residual weakness and an elevated creatinine level of 2.0 mg/dL, which eventually normalized.
On November 6, 2004 (17 days after vaccination), a blood sample was obtained, and the blood was refrigerated at the Centers for Disease Control and Prevention Division of Vector Borne Infectious Diseases (CDC) in Fort Collins, Colorado. On November 15 and 22, 2004 (27 and 34 days after vaccination, respectively), blood samples were obtained and sent to the Emory Vaccine Center in Atlanta, Georgia. PBMCs and serum were isolated for immunologic, virologic, and genetic analyses. The number of PBMCs was 107/mL 34 days after vaccination, which was 4-fold to 6-fold higher than the number observed in 10 unvaccinated control subjects who were matched for age and sex or in healthy vaccinees (data not shown).
Virologic and Immunologic Analyses
Virologic analyses
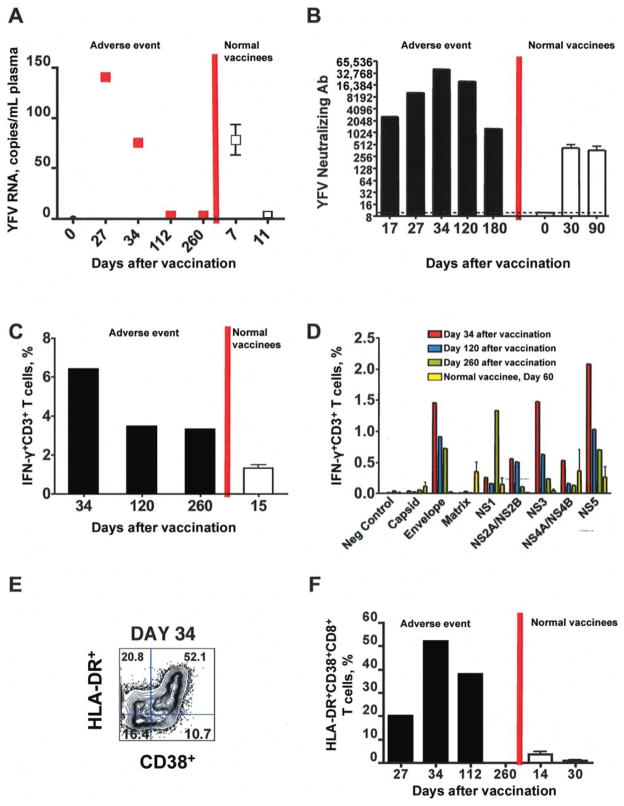
To measure the YFV load in the blood, plasma samples were tested for the presence of YFV RNA by real-time PCR [9]. YFV RNA was detected in the case patient’s plasma at 27 and 34 days after vaccination (figure 1A). This was remarkable, considering that in 5 healthy individuals vaccinated with YF-17D, viral RNA was detected at 7 days after vaccination and was cleared by 11 days after vaccination (figure 1A) [10]. These data suggest that prolonged viral persistence is highly unusual and possibly associated with the adverse events observed in this patient.
Figure 1.
Analysis of yellow fever virus (YFV) loads and adaptive immune responses in a patient with yellow fever vaccine–associated viscerotropic disease and healthy vaccinees. A, Viral loads in the case patient and healthy vaccinees. White square with error bars, mean and standard error for viral loads in 5 healthy vaccinees. B, Neutralizing antibody (Ab) titers in the case patient and healthy vaccinees. White bar, mean and standard error of titers from 3 normal vaccinees. C and D, Evaluation of T cell function. Peripheral blood mononuclear cells from the case patient or 2 healthy vaccinees were cultured with multiple vesicular stomatitis virus constructs expressing different YFV proteins, and the frequency of interferon (IFN)-γ+CD3+ T cells was evaluated. The magnitude (C) and breadth (D) of the response is shown. Mean and standard error for the percentage of IFN-γ+ T cells for 2 normal vaccinees is shown. E, Flow cytometric analysis of activated HLA-DR+CD38+CD8+ effector T cells at day 34 after vaccination. F, Kinetics of activated T cell expansion in the case patient and healthy vaccinees (average from 5 vaccinees). White bar, mean and standard error for the percentage of HLA-DR+CD38+ CD8+ T cells from 5 healthy vaccinees.
Immunologic analyses
The persistence of virus might have been caused by impaired immune responses to the virus. Thus, it was important to determine whether the vaccine induced effective adaptive immune responses. As indicated in figure 1B and table 1, the titers of neutralizing antibody against YFV at day 27 and 34 were 1/10,240 and 1/40,960, respectively, considerably higher than those observed in healthy vaccinees. The titer was also measured in the blood sample obtained 17 days after vaccination that had been refrigerated for several months at the CDC, and it was found to be >1/2,560. However, the accuracy of this value is questionable, given the fact that refrigeration of blood for several would have resulted in lysis of platelets, neutrophils, and other cells, as well as the concomitant release of proteases and enzymes that might have degraded proteins in the sample.
The functional capacity of YFV-specific T cells generated in response to vaccination was evaluated by determining the percentage of CD3+ T cells that also produced interferon (IFN)-γ after stimulation of PBMCs with various recombinant VSV-viral vectors expressing YFV proteins (figure 1C). Functional YFV-specific CD3+ T cells that produced IFN-γ in response to multiple YFV proteins were detected in patient samples through day 260 after vaccination (figure 1C). YFV-specific responses were also observed for CD4 T cells, although the CD8 T cell response was dominant (data not shown). In contrast, the magnitude of T cell responses in 2 healthy vaccinees at day 14 after vaccination, which is the peak of the normal response, was 5-fold to 10-fold lower than that observed in the case patient at day 34 after vaccination (figure 1C). However, the breadth of the T-cell response (which is a measure of the range of different T cell epitopes recognized by the T cells) in the case patient was similar to that in the healthy vaccinees (figure 1D).
We also assessed the activation status of CD8 T cells. A significantly large population of activated CD8 T cells, identified by their expression of CD3 and CD8 and by upregulation of CD38 and HLA-DR (figure 1E), were detected in the blood at 27 and 34 days after vaccination (figure 1F), indicating the presence of an ongoing CD8 T cell response. At day 34 after vaccination, >50% of CD8 T cells coexpressed HLA-DR and CD38 activation markers (figure 1F), as compared to 20% at day 27 after vaccination. In unvaccinated, healthy individuals, the frequency of this population is 1% (data not shown). This data implies that the population of activated CD8 T cells underwent profound expansion in the presence of persistent viremia due to YFV. This data is striking, given that the maximum expansion of the activated CD8 T cell population in 5 healthy vaccinees occurred at day 14 after vaccination and reached baseline levels by 30 days after vaccination (figure 1F). Furthermore, the activated CD8 T cell response to YFV in the healthy vaccinees was 5 to 10-fold lower than that observed in the case patient. The discrepancy between the frequency of HLA-DR+CD38+ T cells (50%; figure 1E) and the frequency of IFN-γ+CD3+ T cells (6%; figure 1C) suggests bystander activation. We have seen minimal evidence for bystander activation in healthy vaccinees who respond normally to YF-17D [17]. However, it is possible that bystander activation did occur in the individual who experienced this adverse event. Nevertheless, taken together, these data indicate that vaccination induced a robust magnitude of antigen-specific T and B cell responses, and thus persistent virus and viscerotropism appear not to have been caused by impairment in the magnitude of adaptive immunity.
This result raised the possibility that defects in other parts of the immune system, perhaps in the innate immune system, might have caused the disease. To address this question, we first performed a flow cytometric analysis of the cellular composition of PBMCs to determine whether there were changes in the numbers or activation status of various innate immune cells. There was a significant increase in the absolute numbers of plasmacytoid dendritic cells, which are involved in anti viral immunity, compared with the numbers observed in 4 unvaccinated control subjects and 4 healthy vaccinees (data not shown). However, there was no significant change in the frequency of myeloid dendritic cells (data not shown).
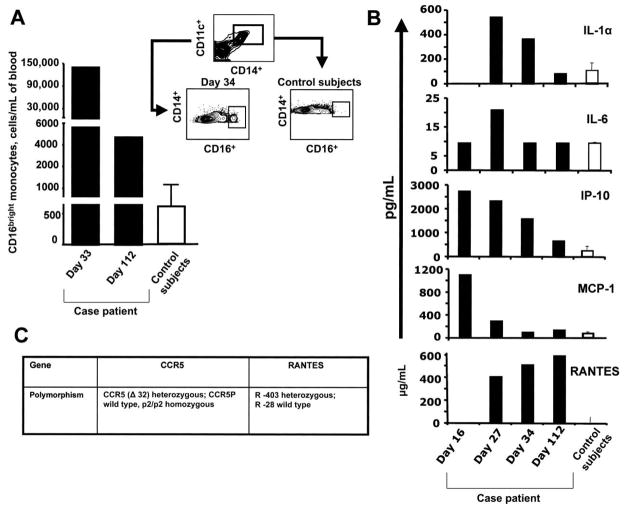
In addition, the subpopulation of CD14+CD16bright monocytes was increased about 200-fold 34 days after vaccination, compared with levels observed in 10 unvaccinated healthy control subjects matched for age and sex (figure 2A). There was no such increase in these cells in healthy vaccinees (data not shown). The CD14+CD16bright subset represent a very minor population of monocytes in normal peripheral blood, but it is expanded significantly in patients with HIV infection and rheumatoid arthritis [18–21]. These cells produce proinflammatory mediators, such as tumor necrosis factor (TNF)-α and RANTES and express the chemokine receptor CCR5, which is a receptor for RANTES and certain other chemokines [18–21, 22]. Analyses of cytokines and chemokines in the plasma revealed elevated levels of the proinflammatory mediators interleukin (IL)-1α, IL-6, IP-10, MCP-1, and RANTES, compared with the levels observed in unvaccinated control subjects matched for age and sex, at 27 days and, in some cases, at 34 days after vaccination (figure 2B and table 1). These increases were much higher than those observed in 5 healthy vaccinees (data not shown). The analysis of cytokines was also performed on the blood sample from day 17 after vaccination that had been refrigerated for several months. It showed elevated levels of IP-10 and MCP-1, but not IL-1α, IL-6, or RANTES. However, these data must be interpreted in light of the important caveat that refrigeration of whole blood for several months would have resulted in lysis of platelets, neutrophils, and other cells, as well as the concomitant release of proteases and enzymes that might have degraded proteins in the sample.
Figure 2.
Analysis of CD14+CD16bright monocytes, inflammatory mediators, and genetic polymorphisms in a patient with yellow fever vaccine–associated viscerotropic disease and unvaccinated control subjects. A, Flow cytometric analysis of CD14+CD16bright monocytes in case patient and in 6 unvaccinated control subjects matched for age and sex. B, Luminex analysis of proinflammatory cytokines and chemokines in case patient and 6 age and sex-matched, unvaccinated control subjects. C, Genetic polymorphisms in the case patient’s CCR5 and RANTES genes.
These flow cytometric and cytokine data suggested perturbations in CD14+ CD16bright innate immune cells as late as 34 days after vaccination. Because virus still persisted in the blood at this time, and because this virus might cause chronic and excessive stimulation of the immune system, it was unclear whether these changes were the cause, or the result, of impaired viral clearance. Several weeks after the patient’s recovery (113 days after vaccination), he returned to the clinic to provide blood samples for the evaluation of viral load and the status of the immune system. At this time point, virus was undetectable in the blood (figure 1A), and the number of PBMCs had returned to normal levels (approximately 106/mL). However, there was a persistent neutralizing antibody titer (1/20, 240; figure 1B) and robust reactivity of CD8+T cells to the following YFV-specific antigens: envelope, NS1, NS2A/2B, NS4A/4B, and NS5 (figure 1C–1E). In addition, the number of CD14+ CD16bright monocytes remained elevated, approximately 10-fold greater than that of unvaccinated control subjects matched for age and sex (figure 2A). Furthermore, analyses of plasma cytokines revealed a significantly higher level of RANTES, a chemokine that binds to and signals through the chemokine receptor CCR5, which is expressed on the CD14+ CD16bright monocytes and is thought to mediate the chemotaxis of such cells toward tissues [18–22] (figure 2B).
Genetic Analyses
Because this adverse event occurred within a week after vaccination—a time point at which adaptive responses cannot be detected—we examined the possibility of defects in early innate antiviral response genes. In mice, the alleles of the 2′5′ oligoadenylate synthetase 1b (Oas1b) gene determine virus yields and resistance or susceptibility to flavivirus-induced disease [13]. In humans, OAS1 and OAS2 gene products have been shown to function in antiviral pathways [14, 15]. In addition, Toll-like receptor 3 (TLR3)-deficient (Tlr3 −/−) mice were more resistant to lethal West Nile virus encephalitis, even though these animals showed a decreased cytokine response and increased virus levels in periphery. Virus entered the brain less efficiently in these animals [16]. Given that OAS1, OAS2, and TLR3 are linked to innate immunity and viral disease, the promoter regions—as well as all of the exons—for these genes were sequenced. No unique mutations were detected in the promoter regions or in any of the exons of these genes, and the patient was homozygous for the major allele of all known single nucleotide polymorphisms within these genes. Furthermore, the individual was wild type for DC-SIGN, which is known to recognize several viral pathogens [23].
The enhanced level of RANTES at day 113 after vaccination raised the question of whether there could be an alteration in the regulation of the expression of the gene encoding this chemokine. Importantly, previous studies suggest that CCR5, a receptor for RANTES is highly expressed on cells in the CD14+CD16bright monocyte subpopulation [18–22], which was greatly enhanced in this patient (figure 2A). This raised the issue of whether the observed anomalies might have been the result of genetic alterations in RANTES and CCR5. PCR was used to genotype the CCR5-Δ32 polymorphism [11]. Previous studies have shown that individuals who are homozygous for the CCR5 -Δ32 mutation are resistant to infection with HIV, and those who are heterozygous for this mutation exhibit delayed progression to AIDS [24, 25]. The patient was found to be heterozygous for this mutation (figure 2C).
With regards to RANTES, several single nucleotide polymorphisms in the RANTES gene have been reported to upregulate or downregulate RANTES gene activity. The most frequent of those polymorphic sites comprise RANTES-403 (G to A) and RANTES-28 (C to G) in the promoter region and RANTES-IN1.1 (T to C) in the first intron region [10, 26, 27]. Both promoter polymorphisms have been shown to increase RANTES transcription [26, 27]. Therefore, the regions of the RANTES promoter were sequenced, and this individual was found to be heterozygous for the RANTES-403 polymorphism and wild type for the RANTES-28 polymorphism (figure 2C).
DISCUSSION
The data presented for this case patient demonstrate the following: (1) persistent viremia that lasted at least through 34 days after vaccination, whereas in healthy vaccinees, viral loads in declined to baseline levels by day 11; (2) robust induction of T and B cell responses in the blood, by day 34 after vaccination; (3) a 200-fold increase in the numbers CD14+CD16bright monocytes in the blood, and a 20-fold increase that persisted at day 113, even when virus was no longer detectable in the blood; (4) heterozygosity for a genetic polymorphism in the CCR5-Δ32, which is known to reduce the level of CCR5 expression on cells [26, 27]; (5) heterozygosity for a genetic polymorphism in the RANTES-403G/A, which has been reported to be associated with enhanced expression of RANTES [12]; (6) constitutively elevated levels of plasma RANTES, which peaked at day 113 after vaccination.
The induction of robust adaptive immune responses suggests that the persistent viremia was unlikely to be due to suboptimal adaptive immunity. On the contrary, the robust magnitude of the adaptive immune response may well have been a result of the persistent viremia. Importantly, even in the presence of high titers of neutralizing antibodies, viral load levels remained very high, suggesting that antibodies were not sufficient to control the virus. This is of interest, because it has long been suggested that, for YF-17D vaccination, antibodies are the correlate of protection [3–5]. Because the innate immune system plays a critical role in the induction of adaptive immunity, it is likely that innate immune events such as the “sensing” of the YFV through Toll-like receptors [28], capture of viral antigens by dendritic cells, and the initial stimulation of antigen-specific T and B cells occurred normally in this case patient. However, it is conceivable that other aspects of innate immune function involved in directly controlling viral replication or mediating viral clearance might have been compromised. However, such putative defects did not lie with the genes encoding OAS1, OAS2, TLR3, or DC-SIGN, because this individual was wild type for these genes.
The significance of the polymorphisms in CCR5 and RANTES is at present unclear. The increased numbers of CCR5-expressing cells in the blood suggests a defect in their trafficking into virally infected tissues, perhaps resulting in impaired immunity. Consistent with this, migration of activated CD8+ T cells from the lymph nodes to tissues, and to the liver in particular, is thought to be mediated by CCR5 [28, 29]. Therefore, it is possible that the reduced expression of CCR5, due to polymorphism in this gene, may have impaired the responsiveness of cells to ligands such as RANTES and consequently affected their migration into tissues.
In addition, the constitutive expression of RANTES could have also contributed to excessive CCR5 activation and desensitization, and the systemic expression of RANTES could also have disrupted the chemokine gradient, which might normally have guided the CCR5-mediated migration of activated T cells and CD14+CD16bright monocytes to tissues. Consequently a “treadmill” situation might have resulted, in which despite a robust immune response in the blood, impaired migration of effector cells resulted in persistent viral replication in specific tissues, such as the liver. These results are consistent with a recent report that demonstrates that CCR5 deficiency enhances risk of symptomatic infection with West Nile virus, another flavivirus [30]. Clearly, further animal model studies are required to substantiate this hypothesis.
It is important to stress however, that while these polymorphisms may have contributed to the development of adverse events in this patient, other host defects could be responsible for the development of adverse events in other patients after YF-17D vaccination. Furthermore, it is entirely possible that other, as yet undetermined, lesions that affect the innate immune system, might have contributed to viscerotropic disease in this individual.
Acknowledgments
Financial support: National Institutes of Health (grants U19 AI05726601 to R.A. and B.P., R01 AI048638 to B.P., R01 DK057665 to B.P., U54 AI057157 to B.P., and U01 AI-50019 to B.P.).
Footnotes
Presented in part: 55th Annual Scientific Meeting of the American Society of Tropical Medicine and Hygiene, Atlanta, Georgia, 14 November 2006 (Symposium 57 on yellow fever vaccine).
Potential conflicts of interest: D.T. is employed by Sanofi Pasteur. B.P., R.A., C.d.R, and R.A. received funding from a research grant from Sanofi Pasteur. P.B. serves on the speakers’ bureau for Gilead, Glaxo SmithKline, Bristol Myers Squibb, Boehringer Ingelheim, and Roche.
References
- 1.Monath TP. Milestones in the conquest of yellow fever. In: Koprowski H, Oldstone MBA, editors. Microbe hunters: then and now. Lansing, MI: Medi-Ed Press; 1996. pp. 95–112. [Google Scholar]
- 2.Theiler M, Smith HH. The use of yellow fever virus modified by in vitro cultivation for human immunization. J Exp Med. 1937;65:787–800. doi: 10.1084/jem.65.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pugachev KV, Guirakhoo F, Monath TP. New developments in flavivirus vaccines with special attention to yellow fever. Curr Opin Infect Dis. 2005;18:387–94. doi: 10.1097/01.qco.0000178823.28585.ad. [DOI] [PubMed] [Google Scholar]
- 4.Monath TP. Yellow fever vaccine. Expert Rev Vaccines. 2005;4:553–74. doi: 10.1586/14760584.4.4.553. [DOI] [PubMed] [Google Scholar]
- 5.Khromava AY, Eidex RB, Weld LH, et al. Yellow fever vaccine: an updated assessment of advanced age as a risk factor for serious adverse events. Vaccine. 2005;23:3256–63. doi: 10.1016/j.vaccine.2005.01.089. [DOI] [PubMed] [Google Scholar]
- 6.Doblas A, Domingo C, Bae HG, et al. Yellow fever vaccine-associated viscerotropic disease and death in Spain. J Clin Virol. 2006;36:156–8. doi: 10.1016/j.jcv.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Kitchener S. Viscerotropic and neurotropic disease following vaccination with the 17D yellow fever vaccine, ARILVAX. Vaccine. 2004;22:2103–5. doi: 10.1016/j.vaccine.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 8.Galler R, Pugachev V, Santos LS, Ocran SW, Monath TP. Phenotypic and molecular analyses of yellow fever 17DD vaccine viruses associated with serious adverse events in Brazil. Virology. 2001;290:309–319. doi: 10.1006/viro.2001.1168. [DOI] [PubMed] [Google Scholar]
- 9.Bae HG, Nitsche A, Teichmann A, Biel SS, Niedrig M. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J Virol Methods. 2003;110:185–91. doi: 10.1016/s0166-0934(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 10.Reinhardt B, Jaspert R, Niedrig M, Kostner C, L’Age-Stehr J. Development of viremia and humoral and cellular parameters of immune activation after vaccination with yellow fever virus strain 17D: a model of human flavivirus infection. J Med Virol. 1998;56:159–67. doi: 10.1002/(sici)1096-9071(199810)56:2<159::aid-jmv10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 11.Kristiansen TB, Knudsen TB, Ohlendorff S, Eugen-Olsen J. A new multiplex PCR strategy for the simultaneous determination of four genetic polymorphisms affecting HIV-1 disease progression. J Immunol Methods. 2001;252:147–51. doi: 10.1016/s0022-1759(01)00349-0. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Chao D, Nakayama EE, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci USA. 1999;96:4581–5. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perelygin AA, Scherbik SS, Zhulin IB, Stockman BM, Li Y, Brinton MA. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci U S A. 2002;99:9322–7. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh A, Desai SY, Sarkar SN, et al. Effects of mutating specific residues present near the amino terminus of 2′–5′-oligoadenylate synthetase. J Biol Chem. 1997;272:15452–8. doi: 10.1074/jbc.272.24.15452. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh A, Sarkar SN, Guo W, Bandyopadhyay S, Sen GC. Enzymatic activity of 2′–5′-oligoadenylate synthetase is impaired by specific mutations that affect oligomerization of the protein. J Biol Chem. 1997;272:33220–6. doi: 10.1074/jbc.272.52.33220. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Town T, Alexopoulou L, Andersen JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1294–5. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 17.Miller JD, et al. Using the smallpox and yellow fever vaccines to study human effector and memory CD8 T cell responses to acute viral infection. Immunity. 2008;28:710–22. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Tacke F, Randolph GJ. Migratory fate and differentiation of blood monocyte subsets. Immunobiology. 2006;211:609–18. doi: 10.1016/j.imbio.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Grage-Griebenow E, Flad HD, Ernst M. Heterogeneity of human peripheral blood monocyte subsets. J Leukoc Biol. 2001;69:11–20. [PubMed] [Google Scholar]
- 20.Weber C, Belge KU, von Hundelshausen P, et al. Differential chemokine receptor expression and function in human monocyte subpopulations. J Leukoc Biol. 2000;67:699–704. doi: 10.1002/jlb.67.5.699. [DOI] [PubMed] [Google Scholar]
- 21.Frankenberger M, Sternsdorf T, Pechumer H, Pforte A, Ziegler-Heitbrock HW. Differential cytokine expression in human blood monocyte subpopulations: a polymerase chain reaction analysis. Blood. 1996;87:373–7. [PubMed] [Google Scholar]
- 22.Braciak TA, Bacon K, Xing Z, et al. Overexpression of RANTES using a recombinant adenovirus vector induces the tissue-directed recruitment of monocytes to the lung. J Immunol. 1996;157:5076–84. [PubMed] [Google Scholar]
- 23.van Kooyk Y, Geijtenbeek TB. DC-SIGN: escape mechanism for pathogens. Nat Rev Immunol. 2003;3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Hwangbo Y, Holte S, et al. Analysis of genetic polymorphisms in CCR5, CCR2, stromal cell-derived factor-1, RANTES, and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin in seronegative individuals repeatedly exposed to HIV-1. J Infect Dis. 2004;190:1055–8. doi: 10.1086/423209. [DOI] [PubMed] [Google Scholar]
- 25.Hladik F, Liu H, Speelmon E, et al. Combined effect of CCR5-Δ32 heterozygosity and the CCR5 promoter polymorphism -2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79:11677–84. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An P, Nelson GW, Wang L, et al. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci U S A. 2002;99:10002–7. doi: 10.1073/pnas.142313799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickel RG, Casolaro V, Wahn U, et al. Atopic dermatitis is associated with a functional mutation in the promoter of the C-C chemokine RANTES. J Immunol. 2000;164:1612–6. doi: 10.4049/jimmunol.164.3.1612. [DOI] [PubMed] [Google Scholar]
- 28.Querec T, Bennouna S, Alkan S, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203:413–24. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai M, Yoneyama H, Harada A, et al. Active participation of CCR5(+)CD8(+) T lymphocytes in the pathogenesis of liver injury in graft-versus-host disease. J Clin Invest. 1999;104:49–57. doi: 10.1172/JCI6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass WG, McDermott DH, Lim JK, et al. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med. 2006;203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]