Abstract
Metacaspases are evolutionarily distant homologs of caspases that are found outside the metazoan and are known to have key roles in programmed cell death (PCD). Two types of metacaspases (types I and II) have been defined in plants based on their domain structures; these have similarities to metazoan ‘initiator' and ‘executioner' caspases. However, we know little about metacaspases in unicellular organisms and even less about their roles in cell death. We identified a novel group of metacaspases in sequenced phytoplanktonic protists that show domain architectures distinct from either type I or II enzymes; we designate them as type III. Type III metacaspases exhibit a rearrangement of domain structures between N- and C-terminus. In addition, we found a group of metacaspase-like proteases in phytoplankton that show sequence homology with other metacaspases, but defy classification in conventional schemes. These metacaspase-like proteases exist in bacteria alongside a variant of type I metacaspases and we propose these bacterial metacaspases are the origins of eukaryotic metacaspases. Type II and III metacaspases were not detected in bacteria and they might be variants of bacterial type I metacaspases that evolved in plants and phytoplanktonic protists, respectively, during the establishment of plastids through the primary and secondary endosymbiotic events. A complete absence of metacaspases in protists that lost plastids, such as oömycetes and ciliates indicates the gene loss during the plastid-to-nucleus gene transfer. Taken together, our findings suggest endosymbiotic gene transfer (EGT) is a key mechanism resulting in the evolutionary diversity of cell death proteases.
Keywords: metacaspase, evolution, unicellular organism, phytoplankton, bacteria
Programmed cell death (PCD, or apoptosis) is widely identified as a characteristic of metazoans, but only recently has it been recognized that it occurs in virtually in all organisms, including plants, fungi, protists and bacteria.1, 2, 3, 4 Thus, PCD is a fundamental process in both eukaryotic and prokaryotic organisms and must have deep evolutionary roots.5, 6 A number of differences are clear in non-metazoan PCD, notably, the absence of the central cell-death proteases, the caspases. Instead distant homologs, metacaspases, are found.7, 8
Caspases are synthesized as inactive procaspases comprised of a prodomain, a large catalytic domain (p20) and a small domain (p10); they are grouped into ‘initiator' and ‘executioner' caspases based on their sequence similarities and the order of their activation during PCD.9 Initiator caspases have an N-terminal prodomain comprised of adapter domains such as a CARD (caspase activation and recruitment domain) or a DED (death effector domain); executioner caspases do not possess an extended prodomain.10 Initiator caspases are activated by proximity-induced dimerization via their N-terminal recruitment domain and consequently activate downstream executioner caspases by cleaving an interdomain linker region between the p20 and p10 domains, thus generating active caspase dimers.11 Active caspases recognize a tetrapeptide sequence within a broad spectrum of cellular targets and are responsible for the proteolytic cleavage, leading to cell death.
In plant lineages, the homologous metacaspases are classified into type I or type II on the basis of presence or absence of an N-terminal prodomain, similar to the classification of metazoan caspases into initiator and executioner caspases.7 Key features defining a prodomain in type I metacaspases are the presence of proline-rich repeats (PRR) and zinc-finger motifs (represented as a sequence of CXXC).12 Similar to the executioner caspases, type II metacaspases do not possess an extended prodomain, however unlike executioner caspases, they harbor a longer interdomain linker between the p20 and p10 domains (∼160 versus 30 aa).
In prokaryotes and most unicellular eukaryotes, the situation is less clear. Due to the lack of key domain structures in their sequence, no classification has been established and thus they are termed ‘metacaspases' or ‘metacaspase-like proteins' in most studies.1, 13, 14, 15, 16 These enzymes (subsequently referred to as metacaspases) bear the core peptide motifs of the caspase-hemoglobinase fold, validating their inclusion in the caspase family, however detailed differences have not been characterized and there have been few attempts at generalization.13
In contrast to traditional caspases, activation mechanisms of metacaspases remain elusive. Autocatalytic processing within an interdomain linker in types I and II recombinant metacaspases has been demonstrated but is not strictly required for their proteolytic activity.12, 17, 18 Recently, crystal structures of type I metacaspases were described in yeast and a parasitic protist revealing significant structural differences from other caspases, notably that they exist as monomers.19, 20 Considering that homodimerization is essential for caspase activation, the activation process of metacaspases might be different.11, 21 While many studies used caspase-specific fluorogenic substrates to define activity of metacaspases, metacaspases also have a different catalytic activity, cleaving preferentially after arginine or lysine instead of aspartate. This has led to the controversial suggestion that metacaspases are not responsible for caspase-like activities.12, 18
Evidence of roles for metacaspases that are not related to cell death is increasing as well. Yeast metacaspase, Yca1, is involved in the cell cycle regulation and protein quality control22, 23 and functions in cell cycle dynamics are reported for metacaspases from the parasitic protists, Trypanosoma brucei and Leishmania major.24, 25 Unlike the obvious functions of PCD in multicellular organisms, the selective advantages of cell death program in single cells are poorly established,1 thus evolutionary selection for metacapases may be based on functions unrelated to cell death.
Although metacaspases have been associated with cell death and other functions, detailed information about their diversity and mechanisms is lacking.22 While it is typically assumed that homologous proteases like caspases and metacaspases that share the same structural motifs also share the same functions, many putative proteases recently identified in sequenced genomes, have shown large variations in sequence and structures among homologous proteases and growing evidence indicates that structural variants have, in fact, different functions.26, 27
In this paper, we investigate metacaspases in unicellular organisms using sequence analyses to understand the diversity that is associated with structural features that may provide insights into origins and ultimately their diverse functions. We chose to concentrate on phytoplankton: unicellular photosynthetic algae and bacteria that occupy a similar niche across diverse marine and freshwater ecosystems.28
Results
Absence of a prodomain in type I metacaspases in phytoplankton
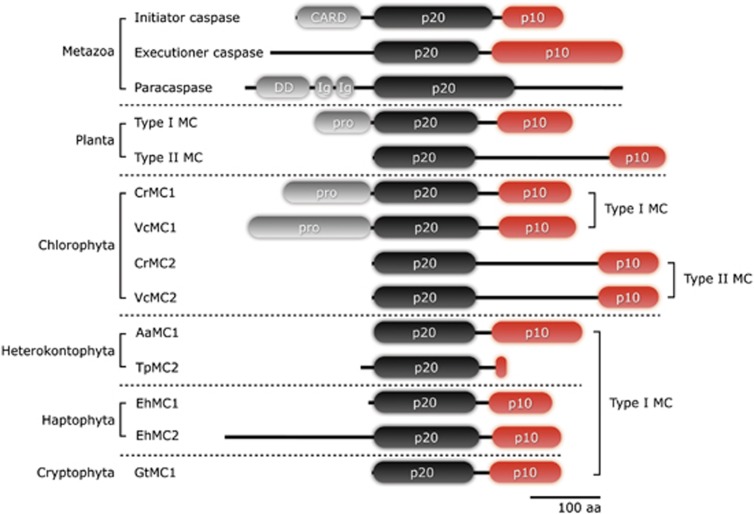
Multiple sequence alignments clearly indicated the presence of both type I and type II metacaspases in chlorophytes, Chlamydomonas reinhardtii (CrMC1 and CrMC2) and Volvox carteri (VcMC1 and VcMC2). The absence of a longer interdomain linker (161.3±32.9 aa in type II metacaspases versus 28.6±4.7 aa in type I metacaspases, Table 1) and presence of a prodomain indicated that CrMC1 and VcMC1 were type I metacaspases. Type II metacaspases were not found in green algal species other than C. reinhardtii and V. carteri (Figure 1). CvMC1 from another chlorophyte, Chlorella variabilis and CsMC1-3 from Coccomyxa subellipsoidea were all close to type I metacaspases.
Table 1. Average domain length for each type of metacaspases in phytoplankton (aa).
| Prodomain | p20 domain | Interdomain | p10 domain | |
|---|---|---|---|---|
| Type I | 95.2±83.3a | 147.4±2.3 | 28.6±4.7 | 97.6±30.0 |
| Type II | — | 144.5±0.5 | 161.3±32.9 | 81.6±2.9 |
| Type III | — | 147.0±1.2 | — | 95.5±4.0 |
| Metacaspase-like proteases | — | 152.0±20.5 | — | — |
High S.D. because of several type I metacaspases that do not have a clear prodomain region (AaMC1: 0 aa, GtMC1: 2 aa, EhMC1: 8 aa and TpMC2: 19 aa)
Figure 1.
Domain architecture of caspases in metazoans, a paracaspase in human, metacaspases in plants, and type I and type II metacaspases in phytoplankton. The catalytic domains are comprised of p20 and p10 domains and a prodomain, which possesses recruitment domains (for example, CARD or DED in initiator caspases, DD or Ig in a paracaspase and PRR or zinc-finger motifs in plant type I metacaspases). The prodomain is absent in several type I metacaspases in phytoplankton indicating the presence of a prodomain is not a definitive characteristics for type I metacaspases. Caspase-9 and -6 from human are shown as a representative initiator and executioner caspases and AtMC1 and AtMC4 from A. thaliana are presented as plant type I and type II metacaspases. The species abbreviations are: Aa, Aureococcus anophagefferens; Cr, Chlamydomonas. reinhardtii; Eh, Emiliania huxleyi; Gt, Guillardia theta; Tp, Thalassiosira pseudonana; Vc, Volvox carteri f. nagariensis
Sequence analyses indicated similar results for a number of metacaspases. Type II metacaspases were not identified in any of the heterokont, haptophyte or cryptophyte species examined, but metacaspases with close homology to Arabidopsis type I metacaspases were found in Aureococcus anophagefferens (AaMC1), Thalassiosira pseudonana (TpMC2), Emiliania huxleyi (EhMC1 and EhMC2) and Guillardia theta (GtMC1). The length of a prodomain was generally short (TpMC2: 19 aa and EhMC1: 8 aa compared with average of 95 aa) or absent (AaMC1 and GtMC1). The exception was one metacaspase from a haptophyte (EhMC2: 213 aa) that showed the extended N-terminus but two features defining a prodomain, presence of PRR and zinc-finger motifs, were not detected (Figure 1).
Discovery of type III metacaspases in phytoplankton
Unlike metacaspases in chlorophytes, most of the rest of the metacaspases in heterokonts (five out of six in T. pseudonana and all six metacaspases in Phaeodactylum tricornutum), a haptophyte (seven out of nine metacaspases in E. huxleyi) and a cryptophyte (nine out of eleven metacaspases in G. theta) resisted classification into type I or type II metacaspases because of the absence of homologous p10 domain at the C-terminus. However, several metacaspases displayed distinctly different sequences in their N-terminus, and re-examination by aligning the pruned N-terminal sequences revealed that these metacaspases had regions homologous to p10 domains (defined as a consensus sequence of SGCXDXQTSADV with 75% matching and other conserved short sequences, see Supplementary Figure 1) in their prodomain region. In other words, they showed evidence of distinct rearrangements of domain structures, in which the p10 domain was located in N-terminus instead of C-terminus (Figure 2).
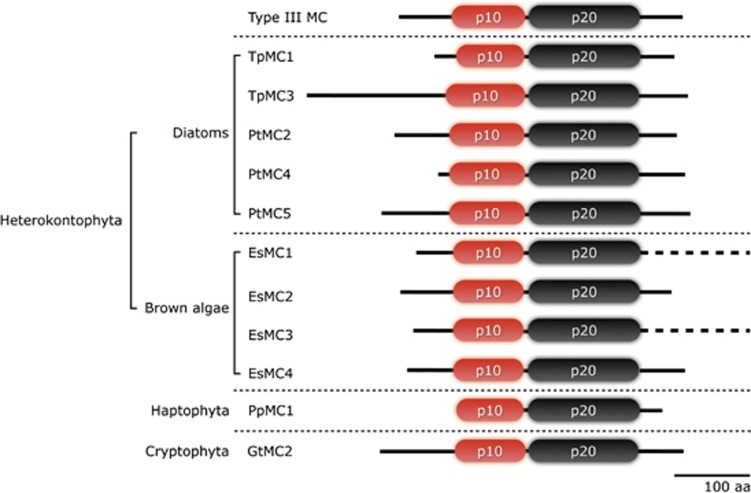
Figure 2.
Domain architecture of type III metacaspases identified in Heterokontophyta (Thalassiosira pseudonana, Phaeodactylum tricornutum and Ectocarpus siliculosus), Haptophyta, (Prymnesium parvum) and Cryptophyta, (Guillardia theta) exhibiting evidence of domain rearrangements. The p10 domain is found at the N-terminus instead of the C-terminus. EsMC1 and EsMC3 have extended C-termini (about 2500 aa) and the total length is not represented in this figure
This arrangement is prevalent in metacaspases from Heterokontophyta, (for example, TpMC1 and 3; PtMC2, 4 and 5), all metacaspases from the brown macroalga, Ectocarpus siliculosus (EsMC1-4) and metacaspases from Prymnesium parvum (PpMC1) and G. theta (GtMC2). Secondary structure predictions showed a common pattern of structural elements (Supplementary Figure 1) and overall sequence homology based on the pairwise % identity in conserved sequence elements (CSE), which additionally supported the evidence of domain rearrangements (Supplementary Table 2). Therefore, we designated this new group type III metacaspases.
Metacaspase-like proteases in phytoplankton and bacteria
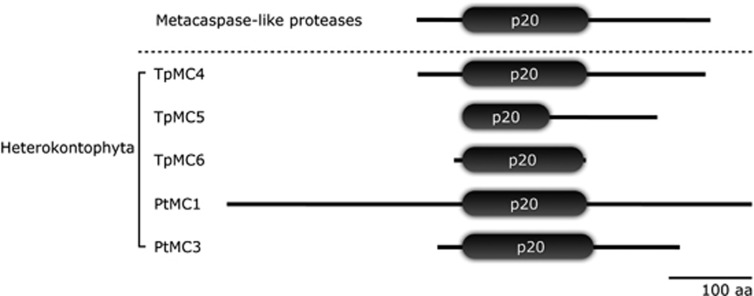
Many metacaspases in phytoplankton remained that defied classification as type I, II or III. In fact, we found that the unclassified metacaspases constituted a distinct clade in the phylogenetic tree. Metacaspases in this group did not show evidence of a p10 domain in either the N- and C-terminal region, nor a prodomain (Figure 3). No metacaspases from plants and green algal lineages showed this feature, but it was widespread in Heterokontophyta, Haptophyta and Cryptophyta (three in T. pseudonana, two in P. tricornutum, seven in E. huxleyi and nine in G. theta).
Figure 3.
Domain architecture of metacaspase-like proteases identified in two heterokonts (Thalassiosira pseudonana and Phaeodactylum tricornutum) that lacks a p10 domain at either N- or C-terminus. Metacaspase-like proteases are also found in a haptophyte (Emiliania Huxleyi) and a cryptophyte (Guillardia theta). (data not shown)
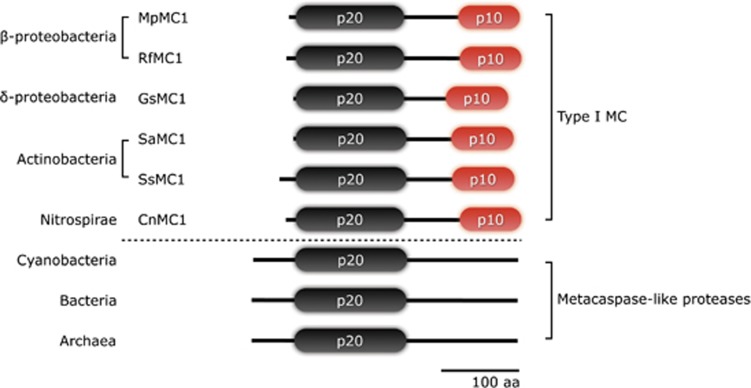
In order to understand the diversity of the metacaspases in phytoplankton, we expanded the pool of metacaspases to the putative ancestor of eukaryotic phytoplankton, the cyanobacteria. Type I, II and III metacaspases were not detected in any members of cyanobacteria, but we found that cyanobacterial metacaspases uniformly lacked the p10 domain. Expanding the analysis to the bacterial metacaspases, we found that most bacterial metacaspases did not have sequences homologous to p10 domains. The absence of conserved sequences representing a p10 domain was confirmed by the conserved domain database (CDD) search in the NCBI. Accepting the difficulty in creating a category based on absence of features, we have chosen to refer to this distinct group of enzymes, present in prokaryotes and eukaryotes, simply as metacaspase-like proteases. However, we also found a small number of metacaspases from Proteobacteria (β-Proteobacteria: DaMC1, Dechloromonas aromatica; LcMC1, Leptothrix cholodnii; MpMC1, Methylibium petroleiphilum; RfMC1, Rhodoferax ferrireducens and δ-Proteobacteria: GsMC1, Geobacter sulfurreducens PCA), Actinobacteria (SaMC1, Streptomyces avermitilis and SsMC1, Streptomyces scabiei) and Nitrospirae (CnMC1, Candidatus Nitrospira defluvii) that could be classified as type I metacaspases (having both p20 and p10 domains) (Figure 4 and Supplementary Figure 2), for example, a hypothetical protein GSU0716 from G. sulfurreducens PCA (GenBank accession number AAR34046). Type I metacaspases from bacteria did not have a prodomain as was the case for most phytoplankton but they did possess a slight longer interdomain linker (about 50 aa) compared with the type I metacaspases found in eukaryotes.
Figure 4.
Domain architecture of bacterial type I and metacaspase-like proteases identified in a wide range of bacterial groups. Metacaspase-like proteases are widespread in most bacterial groups, (including cyanobacteria and archaea) whereas type I metacaspases are found in fewer groups (including β-Proteobacteria, δ-Proteobacteria, Actinobacteria and Nitrospirae). Bacterial type I metacaspases exhibit a slightly longer interdomain linker than type I metacaspases in eukaryotes. Species abbreviations are: Cn, Candidatus Nitrospira defluvii; Gs, Geobacter sulfurreducens; Lc, Leptothrix cholodnii; Mp, Methylibium petroleiphilum; Rf, Rhodoferax ferrireducens; Sa, Streptomyces avermitilis; Ss, Streptomyces scabiei. The total lengths of metacaspase-like proteases are not represented because of high variance
Discussion
Ancestral forms of type I metacaspases do not possess a prodomain
Metacaspases clearly fit in the C14 family of caspases that belong to the CD clan of cysteine proteases, which also encompasses six other families: C11 clostripain, C13 legumain, C25 gingipain, C50 separase, C80 RTX self-cleaving toxin and C84 prtH peptidase.29 Two types of metacaspases (types I and II) are defined based on the presence of a prodomain analogous to the classification of caspases into initiator or executioner caspases.7 The molecular role of a prodomain in initiator caspases is the recruitment of caspases to multicomponent signaling complexes for caspase activation. Whether it also has this role in type I metacaspases is not clear, but the PRR and zinc-finger motifs found in type I metacaspase prodomains are known to be involved in protein-protein interactions, and zinc-finger domains in the metacaspase AtMC1 and LSD1 are essential for cell death function.30
However, phytoplankton metacaspases often lack prodomains (Figure 1), so we conclude that the prodomain is not a definitive feature. Further, because type II metacaspases are typically distinguished by the absence of a prodomain, this definition is not acceptable in a broader phylogenetic context. On the basis of our result, type II metacaspases should be defined by the presence of a longer interdomain linker and overall sequence homology.
The absence of a prodomain in most eukaryotic phytoplankton but its presence in plants and green algae suggests its emergence in these lineages. Our searches found an ancestral form of type I metacaspases (without prodomains) that have not been identified before in several groups of bacteria, including β-Proteobacteria, δ-Proteobacteria, Actinobacteria and Nitrospirae. In addition, a type I metacaspase with a prodomain was found in the predicted protein database in the Rhodophyta, C. tuberculosum (accession number IDg11585t1). Together with data from green lineages, our data indicate prodomains may have emerged after primary endosymbiosis. The absence of prodomains in Heterokontophyta, Haptophyta and Cryptophyta potentially indicate the loss of prodomains during secondary endosymbiotic events.
Two variants of type I metacaspases: Type II and III metacaspases in eukaryotes
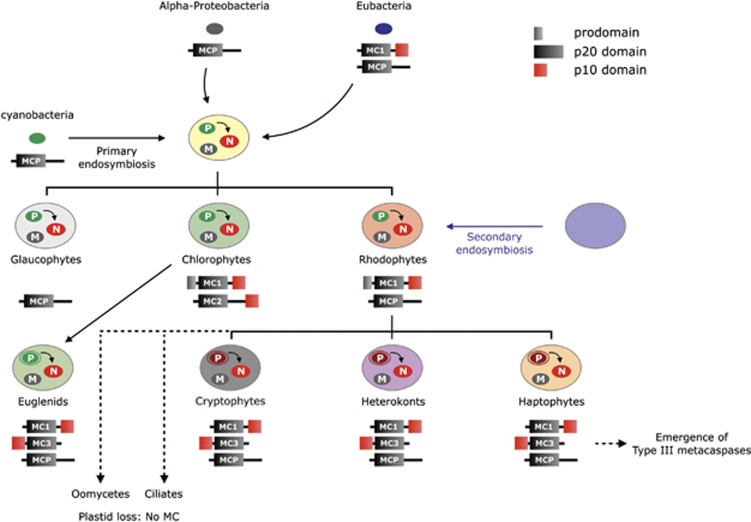
Type II metacaspases are exclusively found in plants and green algae. Previously, it had been suggested they were derived from type I metacaspases through horizontal gene transfer (HGT) during the establishment of plastid from endosymbiotic cyanobacteria.31 However, type II metacaspases are absent in most other phytoplankton groups that have plastids as a result of a primary endosymbiosis, while type III metacaspases (which might be derived from type I) are present in groups that arose from secondary endosymbiotic events. Another photosynthetic protist, Euglena gracilis that is known to have received its plastids from a green alga via secondary endosymbiosis has at least one type I (GenBank accession number EC674854) and one type III (GenBank accession number EC679812) metacaspase, further supporting the idea that type III metacaspases derived from type I, and while the secondary plastid-containing alga, Bigelowiella natans does not have type III metacaspases, neither does it have any type I metacaspases, perhaps due to systemic loss as part of genome reduction during endosymbiosis.32 Importantly, metacaspases are not present in oömycetes (for example, Phytophthora species), Heterokontophyta that secondarily lost their plastids33 nor are they found in ciliates, another group that lost plastids. Clearly, endosymbiotic gene transfer (EGT) might play key roles in metacaspase diversity.
The absence of type II metacaspases from some green algae, might be related to the complexity of the organisms' life history34 or elements like multicellularity and developmental programs. For example, multicellular plants have type II metacaspases, and as V. carteri exhibits a degree of multicellular organization, the presence of type II metacaspases is not unexpected.35 Type III metacaspases are absent in the heterokont, A. anophagefferens, which has only one type I metacaspase. A. anophagefferens is known to be especially tolerant of stressful conditions and may not need a diverse pool of metacaspases.36 All metacaspases in E. siliculosus are type III and this might be associated with the emergence of multicellularity in brown algae.37
Domain rearrangements found in type III metacaspases could affect protein function.38 We do not know whether type III metacaspases retain their activities, but at least some domain rearrangements in proteases have little or no effect on function.39 Indeed, recombinant caspase-3 and -6 that have a domain swapping like type III metacaspases are constitutively active and retain apoptotic functions.40 Circular permutation (CP) is a type of nonlinear domain rearrangement that may explain the mechanism for a new domain combination by which N- and C-terminal regions of a protein become exchanged.41 CP has been described for various types of proteins, such as DNA methyltransferases, ABC transporters, ribosomal proteins, histones and homeobox proteins, thus CP is widely spread in proteins.42, 43 Potential mechanisms of CP are unknown but domain rearrangements are a major source of evolutionary innovation and suggest active protein evolution in metacaspases.44
Metacaspase-like proteases in phytoplankton and their bacterial origin
Bacterial origins of PCD were suggested by Koonin and Aravind after finding components of cell death machinery in bacteria.8 Their widespread presence in non-metazoans and the subsequent finding of caspase-like proteases in a group of α-Proteobacteria (the Rhizobia) lead to the hypothesis of a mitochondrial endosymbiotic origin of eukaryotic metacaspases.5 Bacterial metacaspases, including caspase-like proteases in the Rhizobia do show a well-conserved p20 domain that contains catalytic active sites, but lack a p10 domain. We also found a group of proteases that lack a p10 domain in several groups of eukaryotic phytoplankton (Heterokontophyta, Haptophyta and Cryptophyta including their ancestor, Rhodophyta), and thus could represent a distinct feature compared with type I, II or III metacaspases.
For classical caspases, the p20 and p10 domains have a straightforward functional meaning: cleavage in the interdomain linker during the caspase activation results in products with masses of ∼20 kDa and ∼10 kDa, respectively.45, 46 The case for metacaspases is less clear; it has not been demonstrated that cleavage is needed for activation, as it is for caspases. We know that type I metacaspases in Yca1 and A. thaliana (AtMCP1b), undergo autoprocessing and yield at least a small (∼12 kDa) polypeptide.17, 18 One type II metacaspase in A. thaliana, Atmc9 also generates ∼22 and ∼15 kDa fragments after autoproteolytic processing, which correspond to p20 and p10 domains.12 No data are available yet, for newly identified type III metacaspases.
Metacaspase-like proteases, despite their apparent lack of a p10 domain, might have cleavages sites that could generate subunits, but our alignments and the secondary structure analyses have not identified an interdomain linker that contains a cleavage site. Alternatively, cleavage may not be needed; two recently-obtained crystal structures of yeast and trypanosome metacaspases indicate these proteases are monomers with different mechanisms to explain proteolytic activity.19, 20 Another class of caspase homologs, the paracaspases, are monomers and dimerization for proteolytic activity.47 Without better understanding, it seems wisest to maintain a more neutral category (metacaspase-like proteases) for this group (Figure 3).
Metacaspase-like proteases are widespread in α-Proteobacteria, which is consistent with the idea of a mitochondrial endosymbiotic origin.5 However, they are also found in most other bacterial groups, including all classes of Proteobacteria, Cyanobacteria and even Archaea, which might support acquisition through HGT. Metacaspase-like proteases are present in most eukaryotic phytoplankton, but not in plants and green algae, so it is more parsimonious to assume bacterial origins and a later gene loss in selected lineages. Why, then, would metacaspase-like proteases be lost?
The major characteristic of metacaspase-like proteases is the absence of a sequence homology in a p10 domain. Domains are structural units that frequently determine the function of proteins.48 In the case of caspases, one of the amino acids critical to determining substrate specificity is in the p10 domain.49 Absence of a p10 domain in metacaspase-like proteases therefore, could indicate non-function or quite different substrate specificity. In such cases, the protein might be redundant and loss could follow.
Our results provide evidence for origin of eukaryotic metacaspases from bacteria. The newly detected bacterial type I metacaspases (annotated as GSU0716 from G. sulfurreducens, in β-, δ-Proteobacteria, Actinobacteria and Nitrosporae but missing in α-Proteobacteria) have p20 and p10 domains but no prodomain; this is similar to type I metacaspases from phytoplankton. Therefore, we hypothesize that eukaryotic metacaspases originated from two bacterial metacaspases, type I metacaspases and metacaspase-like proteases (Figure 5).
Figure 5.
Simplified phylogenetic representation of hypothetical metacaspase evolution. Two proposed ancestral forms of metacaspases in bacteria are type I metacaspases (MC1) and a metacaspase-like protease lacking a p10 domain (MCP). Type I metacaspases are present in most eukaryotes. Type II metacaspases are found exclusively in higher plants and green algae, potentially derived from primary endosymbiosis (as they are absent in red algae and glaucophytes). Type III metacaspases are found in groups that underwent secondary endosymbiotic events (for example, heterokonts, haptophytes, cryptophytes and euglenids). Absent in oomycetes and ciliates (dashed line) is hypothesized to arise from a secondary loss, possibily associated with loss of plastids. The more poorly-defined metacaspase-like proteases are mostly present in bacterial groups and eukaryotes except green lineages, thus they have hypothetically been lost in green lineages
Materials and Methods
Database searches
Searches for metacaspase sequences in phytoplankton were carried out using the genome databases at DOE Joint Genome Institute (JGI: http://genome.jgi.doe.gov) using the current releases. Four species from Chlorophyta (Chlamydomonas reinhardtii v4.0, Chlorella variabilis NC64A, Coccomyxa subellipsoidea C-169 v2.0 and Volvox carteri f. nagariensis), three species from Heterokontophyta (Aureococcus anophagefferens, Phaeodactylum tricornutum v2.0 and Thalassiosira pseudonana), one species from Haptophyta (Emiliania huxleyi CCMP1516 v1.0) and one species from Cryptophyta (Guillardia theta CCMP2712 v1.0) were chosen and a total of 41 metacaspases were identified after extensive genome searches and used for sequence analyses. The recently-completed draft genome of Ectocarpus siliculosus was included to incorporate four metacaspases from brown algae (Bioinformatics and Systems Biology, http://bioinformatics.psb.ugent.be/webtools/bogas/overview/Ectsi). Detailed information is summarized in Supplementary Table 1. Additional metacaspases from the Rhodophyta, Calliarthron tuberculosum (http://dbdata.rutgers.edu/data/plantae/) and the Glaucophyta, Cyanophora paradoxa (http://cyanophora.rutgers.edu/cyanophora/home.php) were found from the predicted protein database. Nine metacaspases from Arabidopsis thaliana were obtained from The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org) and were used to construct a query protein set and further used as reference sequences for the following sequence analyses. The metacaspase sequences were also retrieved by TBLASTN, BLASTP and PSI-BLAST searches in GenBank to avoid missing any other metacaspase sequences.
Bacterial metacaspase sequences were obtained from the MEROPS database (http://merops.sanger.ac.uk) to incorporate metacaspases from genomes that had not been fully sequenced for extensive sequence comparisons. The MEROPS database is a collection of proteolytic enzymes grouped into homologous proteins based on the tertiary structure similarities thus provides solid metacaspase sequences.50 A total of 233 metacaspases from bacteria and eight from archaea were used. This includes all groups of Proteobacteria, Aquificae, Bacteroidetes, Chlamydiae, Chlorobi, Chloroflexi, Cyanobacteria, Deferribacteres, Fibrobacteres, Firmicutes, Nitrospirae, Planctomycetes, Spirochetes, Verrucomicrobia and Euryarchaeota.
Metacaspase sequence analyses
Metacaspase sequences obtained from JGI and MEROPS databases were examined by the inspection of caspase p20 domain (CASc superfamily, Caspase domain in the NCBI Conserved Domain Database) and used for the sequence alignments. All metacaspase sequences were aligned with well-defined type I (AtMC1-3) and type II (AtMC4-9) metacaspases from A. thaliana as reference sequences using the default parameters of the MUSCLE algorithm, implemented in Geneious, version 5.5.6 (Biomatters Ltd, Auckland, New Zealand). A consensus sequence for the p20 and p10 domains was established based on the results of alignment. Alignment results for the p10 domain were manually pruned using MacClade 4.08 based on the results of reference sequences and pruned sequences were then realigned to define different types of metacaspases.51 Secondary structure predictions and associated confidence values for metacaspases were made by using the PSIPRED protein structure prediction server (http://bioinf.cs.ucl.ac.uk/psipred).52
Glossary
- CARD
caspase activation and recruitment domain
- CDD
conserved domain database
- CP
circular permutation
- CSE
conserved sequence elements
- DED
death effector domain
- EGT
endosymbiotic gene transfer
- HGT
horizontal gene transfer
- PCD
programmed cell death
- PRR
proline-rich repeats
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by M Piacentini
Supplementary Material
References
- Franklin DJ, Brussaard CPD, Berges JA. What is the role and nature of programmed cell death in phytoplankton ecology. Eur J Phycol. 2006;41:1–14. [Google Scholar]
- Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- Madeo F, Herker E, Wissing S, Jungwirth H, Eisenberg T, Frohlich KU. Apoptosis in yeast. Curr Opin Microbiol. 2004;7:655–660. doi: 10.1016/j.mib.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM. Comparative genomics of phylogenetically diverse unicellular eukaryotes provide new insights into the genetic basis for the evolution of the programmed cell death machinery. J Mol Evol. 2009;68:256–268. doi: 10.1007/s00239-009-9201-1. [DOI] [PubMed] [Google Scholar]
- Uren AG, O'Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV, et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: Vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, et al. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem. 2004;279:45329–45336. doi: 10.1074/jbc.M406329200. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins. 2002;46:355–367. doi: 10.1002/prot.10060. [DOI] [PubMed] [Google Scholar]
- Bidle KD, Haramaty L, Ramos JBE, Falkowski P. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci USA. 2007;104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidle KD, Bender SJ. Iron starvation and culture age activate metacaspases and programmed cell death in the marine diatom Thalassiosira pseudonana. Eukaryot Cell. 2008;7:223–236. doi: 10.1128/EC.00296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman-Frank I, Bidle KD, Haramaty L, Falkowski PG. The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol Oceanogr. 2004;49:997–1005. [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J Biol Chem. 2005;280:14691–14699. doi: 10.1074/jbc.M413527200. [DOI] [PubMed] [Google Scholar]
- Wong AHH, Yan CY, Shi YG. Crystal structure of the yeast metacaspase Yca1. J Biol Chem. 2012;287:29251–29259. doi: 10.1074/jbc.M112.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLuskey K, Rudolf J, Proto WR, Isaacs NW, Coombs GH, Moss CX, et al. Crystal structure of a Trypanosoma brucei metacaspase. Proc Natl Acad Sci USA. 2012;109:7469–7474. doi: 10.1073/pnas.1200885109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- Lee REC, Puente LG, Kaern M, Megeney LA. A non-death role of the yeast metacaspase: Yca1p alters cell cycle dynamics. Plos One. 2008;3:e2956. doi: 10.1371/journal.pone.0002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee REC, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci USA. 2010;107:13348–13353. doi: 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms MJ, Ambit A, Appleton P, Tetley L, Coombs GH, Mottram JC. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J Cell Sci. 2006;119:1105–1117. doi: 10.1242/jcs.02809. [DOI] [PubMed] [Google Scholar]
- Ambit A, Fasel N, Coombs GH, Mottram JC. An essential role for the Leishmania major metacaspase in cell cycle progression. Cell Death Differ. 2008;15:113–122. doi: 10.1038/sj.cdd.4402232. [DOI] [PubMed] [Google Scholar]
- Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. Merops: the Peptidase Database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, et al. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases. J Cell Biol. 2007;179:375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Takeda H. Sugar Composition of the cell-wall and the taxonomy of Chlorella (Chlorophyceae) J Phycol. 1991;27:224–232. [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci USA. 2011;108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, Scornet D, Allen AE, Amoutzias G, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Bornberg-Bauer E, Beaussart F, Kummerfeld SK, Teichmann SA, Weiner J. The evolution of domain arrangements in proteins and interaction networks. Cell Mol Life Sci. 2005;62:435–445. doi: 10.1007/s00018-004-4416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr Opin Struct Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, MacFarlane M, Luo Z, Huang Z, Fernandes-Alnemri T, et al. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J Biol Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: Circularly permuted amino acid sequences. Proc Natl Acad Sci USA. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. Circular permutations in the molecular evolution of DNA methyltransferases. J Mol Evol. 1999;49:161–164. doi: 10.1007/pl00006529. [DOI] [PubMed] [Google Scholar]
- Uliel S, Fliess A, Unger R. Naturally occurring circular permutations in proteins. Protein Eng. 2001;14:533–542. doi: 10.1093/protein/14.8.533. [DOI] [PubMed] [Google Scholar]
- Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Yu JW, Jeffrey PD, Ha JY, Yang XL, Shi YG. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc Natl Acad Sci USA. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day WHE. Maddison WP, Maddison DR. MacClade, analysis of phylogeny and character evolution. Folia Primatol (Basel) 1989;54:190–202. doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.