Abstract
Many sensory systems involve multiple steps of signal amplification to produce a significant response. One such mechanism may be the clustering of transmembrane receptors. In bacterial chemotaxis, where a stoichiometric His-Asp phosphorelay from the kinase CheA to the response regulator CheY plays a central role, the chemoreceptors (methyl-accepting chemotaxis proteins) cluster together with CheA and the adaptor CheW, at a pole of a rod-shaped cell. This clustering led to a proposal that signal amplification occurs through an interaction between chemoreceptor homodimers. Here, by using in vivo disulfide crosslinking assays, we examined an interdimer interaction of the aspartate chemoreceptor (Tar). Two cysteine residues were introduced into Tar: one at the subunit interface and the other at the external surface of the dimer. Crosslinked dimers and higher oligomers (especially a deduced hexamer) were detected and their abundance depended on CheA and CheW. The ligand aspartate significantly reduced the amounts of higher oligomers but did not affect the polar localization of Tar-GFP. Thus, the binding of aspartate alters the rate of collisions between Tar dimers in assembled signaling complexes, most likely due to a change in the relative positions or trajectories of the dimers. These collisions could occur within a trimer-ofdimers predicted by crystallography, or between such trimers. These results are consistent with the proposal that the interaction of chemoreceptor dimers is involved in signal transduction.
Sensing and responding to extracellular signals are essential for any living cell. One of the most extensively studied sensing systems is the chemotaxis of Escherichia coli (1–4); i.e., migration toward or away from chemicals. The chemotactic signal transduction involves a His-Asp phosphate relay from the histidine kinase CheA to the response regulator CheY that is regulated by chemoreceptors or methyl-accepting chemotaxis proteins (MCPs). The aspartate chemoreceptor Tar has a very low threshold concentration (≈3 × 10-8 M) of l-aspartate for an attractant response (5–7). Moreover, E. coli responds to a very small change (<1%) in the receptor occupancy with aspartate (8). Therefore, an input signal has to be amplified to produce a significant response. The flagellar motor switching by phospho-CheY is a highly cooperative event that can account for at least some degree of signal amplification (9, 10). In addition, recent analyses by using fluorescence resonance energy transfer (11) suggested that much of the gain occurs at the receptor end of the signaling pathway. However, its mechanism remains to be elucidated: the gain could be achieved through the MCP–MCP interaction (12, 13) or the involvement of CheB (11, 14).
MCP forms a homodimer, regardless of its ligand occupancy state (15). Each subunit (≈60 kDa) consists of two transmembrane helices (TM1 and TM2), the ligand-binding domain in the periplasm, the signaling/adaptation domain in the cytoplasm, and the HAMP domain, which connects TM2 with the signaling/adaptation domain. The cytoplasmic domain forms a stable complex with the histidine kinase CheA and the adaptor protein CheW (16, 17) that forms a cluster at a cell pole (18–20), leading to a proposal that the lateral communication between MCP dimers in the cluster plays a critical role in signal amplification (12, 13). In fact, chemically synthesized multivalent ligands induce attractant responses of E. coli with lower thresholds than corresponding monovalent ligands presumably by promoting MCP clustering to increase sensitivity (21, 22).
A highly ordered structure of an MCP cluster has been proposed. The cytoplasmic fragment of the serine chemoreceptor Tsr crystallizes with a unit of a trimer of dimers (23). Genetic analyses support the putative contacts among three dimers (24). It has been proposed that trimer units of MCP dimers assemble into a lattice-like matrix, in which the excitation of an MCP dimer can be propagated to other dimers through their physical contacts (25, 26). Nevertheless, a direct physical interaction between MCP dimers has not been established, although Tar and Tsr can be crosslinked with a chemical crosslinker (24).
In this study, we performed in vivo disulfide crosslinking assays and showed that the interdimer crosslinking of Tar is sensitive to the attractant binding. We also showed that the attractant does not significantly alter the localization of Tar-GFP. Taken together, we suspect that the attractant may not induce a global redistribution of Tar in a cell and that the ligand may alter the arrangement of Tar dimers within a polar cluster.
Materials and Methods
Bacterial Strains and Plasmids. RP2859 (J. S. Parkinson, personal communication) lacks Tap, CheB, and CheR. HCB339 (27), HCB436 (28), and RP3098 (29) lack all four MCPs. In addition, HCB436 lacks CheB and CheR and RP3098 lacks all Che proteins.
The entire tar gene and its coding region were cloned into the low-copy-number vector pWSK29 (30) and the expression vector pBAD24 (31), yielding pOH251 and pHS5 (H. Sakamoto and I.K., unpublished work), respectively. The pBAD24 derivative with the P15A replicon (pBAD33) (31) was used as a vector compatible with the pBR322-based CheA/CheW-expressing plasmid pDV4 (32). Mutagenesis was performed essentially as described (33).
The tar-egfp fusion was constructed by replacing the tar stop codon by GCC (Ala; boxed) using the reverse primer: 5′-CGTTAGTAAATACTCGGGCAAATGTTTCC-3′ and igating the AvaI site (underlined) to that of the GFP-encoding vector pEGFP (Clontech). The tar-egfp coding region was placed downstream of the trc promoter of the vector pTrcHisC (Invitrogen).
Swarm Assay of Chemotaxis. Aliquots of fresh overnight cultures were spotted onto tryptone semisolid agar (1% tryptone, 0.5% NaCl, and 0.3% agar) supplemented with appropriate antibiotics, which was then incubated at 30°C.
In Vivo Disulfide Crosslinking Assay. Fresh overnight cultures were diluted with TG broth [1% tryptone, 0.5% NaCl, and 0.5% (wt/vol) glycerol] supplemented with appropriate antibiotics and/or inducers. After vigorous shaking at 30°C for 3.5 h, cells were harvested and suspended in SDS-loading buffer [35 mM Tris·HCl (pH 6.8)/6.7% glycerol/1% SDS/0.0007% bromophenol blue] supplemented with 2.5 mM N-ethylmaleimide (NEM) and 2.5 mM EDTA. Samples were boiled and subjected to nonreducing SDS/PAGE. Immunoblotting was carried out essentially as described (34), with the anti-Tar antibody raised against the C-terminal peptide [NH2-(C)PRLRIAEQDPNWETF-COOH] (Sawady Technology, Tokyo).
To examine the effect of an attractant, cells were washed twice with MLM [10 mM potassium phosphate buffer (pH 7.0)/0.1 mM EDTA/10 mM dl-lactate/0.1 mM methionine], resuspended in MLM, and divided into two aliquots. Aspartate or α-methylaspartate (1 mM) was added to one of the aliquots. The samples were incubated for 10 min at room temperature, and treated with 200 μM Cu(II)(o-phenanthroline)3 (Cu-phenanthroline) supplemented with or without 1 mM aspartate or methylaspartate for 10 min. The reactions were terminated with 2.5 mM NEM and 2.5 mM EDTA. The pH values of the samples were kept between 6.6 and 6.7 during preparation.
Fluorescence Microscopy. Microscopy was performed essentially as described (35). Cells were grown in TG broth containing appropriate antibiotics and inducers with vigorous shaking at 30°C, were harvested at late exponential phase, were washed twice, and were suspended in MLM supplemented with 10 mM aspartate, 10 mM serine, or none. A small aliquot of the cell suspension was spotted onto a slide glass coated with 0.5% agarose and was observed under an Olympus BX50 fluorescence microscope. The images were recorded and processed by using a C4742–95 digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) and IP LAB VER. 3.2 imaging software (Scanalytics, Billerica, MA).
Results
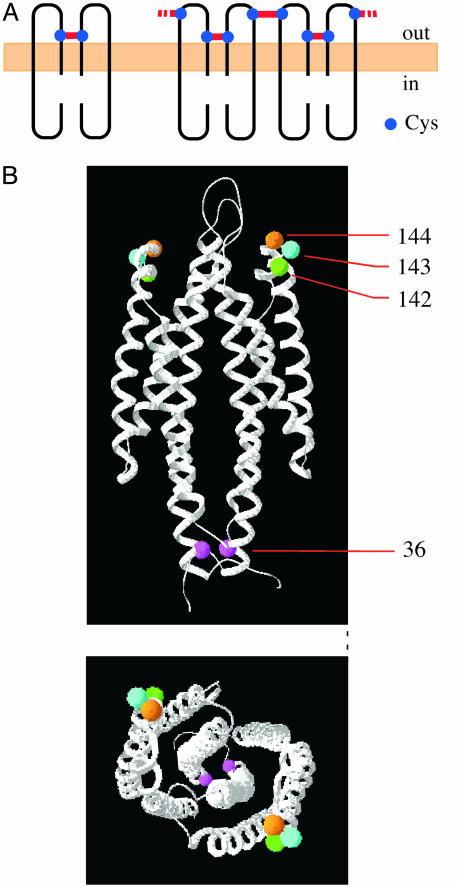
Construction and Expression of Cys-Replaced Tar Receptors. Hazelbauer and coworkers (36, 37) and Maruyama et al. (38) showed that introduction of Cys residues into appropriate positions of a chemoreceptor yields disulfides within or between its subunits in vivo. The rationale of detection of intra- and interdimer disulfides of the chemoreceptors was described by Bass and Falke (39). If a Cys residue is introduced at an appropriate position of the subunit interface within a Tar dimer, the two partner subunits will be crosslinked. For the S36C mutant, this result has been demonstrated (15, 40). If multiple Tar dimers are in close contact with each other, introduction of a Cys at the external surface of a Tar dimer would yield a disulfide between two subunits from neighboring dimers. Therefore, a mutant Tar protein with both inward and outward Cys residues would form a trimer, a tetramer, or higher oligomers by means of intersubunit disulfide bonds (Fig. 1A). Based on the crystal structure of the periplasmic fragment of Tar (41), we chose positions 142–144, which are far from position 36 (Fig. 1B), and do not directly involved in aspartate binding.
Fig. 1.
Introduction of Cys residues into Tar. (A) Strategy for detecting an interdimer interaction by using in vivo disulfide crosslinking. Wild-type Tar is devoid of Cys residue. Two subunits within a Tar homodimer can be crosslinked at position 36 (Left). If another Cys is introduced at an appropriate position on the external surface of the dimer, the double-mutant Tar protein (Right) would form a trimer, a tetramer, or higher oligomers by means of intersubunit disulfide bonds. (B) The three-dimensional structure of the periplasmic fragment of Tar of Salmonella typhimurium (41). (Upper) Side view. (Lower) Top view. Residues replaced by Cys are marked. Position 143 is Asn in S. typhimurium and Tyr in E. coli.
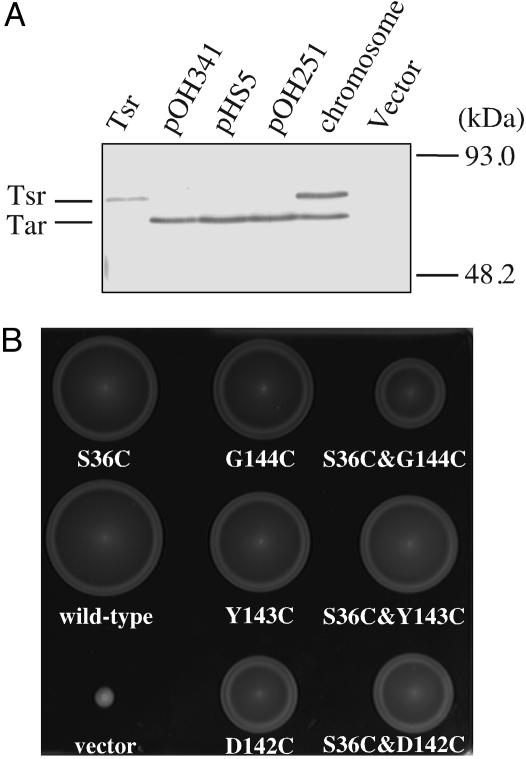
To prevent inappropriate crosslinking, overproduction of these mutant proteins should be carefully avoided. We constructed a low-copy-number plasmid carrying the tar gene with its native promoter (pOH251) and a plasmid carrying the tar-coding region downstream of the tightly regulatable araBAD promoter (pHS5), and introduced them into strain HCB436, which lacks any chemoreceptor, the methyltransferase CheR, and the methylesterase CheB (ΔMCP ΔCheRB). Immunoblotting with anti-Tar antibody revealed that the expression level of Tar from plasmid pOH251 was about twice as much as that of chromosome-encoded wild-type Tar and not more than the total amounts of chromosome-encoded Tar and Tsr (Fig. 2A). In the presence of 200 μM arabinose, the expression level of mutant Tar from plasmid pHS5 was similar to that of chromosome-encoded wild-type Tar (data not shown). The Cys mutation(s), S36C, D142C, Y143C, and/or G144C, were introduced into these plasmids. The resulting plasmids were introduced into strain HCB339 (ΔMCP). Transformant cells were tested for their swarming abilities in tryptone semisolid agar. HCB339 cells carrying any pOH251-derivative plasmid swarmed (Fig. 2B). In the presence of 200 μM arabinose, cells carrying any pHS5-derivative plasmid also swarmed (data not shown). These results indicate that the Cys substitutions per se did not affect receptor function. In the following experiments, we used the pOH251-derivative plasmids unless otherwise noted, but essentially similar results were obtained with the pHS5 derivatives.
Fig. 2.
Expression levels and swarming abilities of cells expressing the mutant Tar proteins. (A) Expression levels of the wild-type Tar protein from various plasmids. Whole-cell lysates were subjected to SDS/PAGE and immunoblotting with anti-Tar. HCB436 (ΔMCP ΔCheRB) was used as the plasmid host unless otherwise noted. Vector, pWSK29; Tsr, the Tsr-encoding plasmid pHS301; pOH341, the pBAD33-based plasmid (200 μM arabinose); pHS5, the pBAD24-based plasmid (200 μM arabinose); pOH251, the pWSK29-based plasmid; and chromosome, RP2859 (ΔTap, ΔCheRB) cells carrying pWSK29. (B) Swarming abilities of HCB339 cells (ΔMCP) expressing the mutant Tar proteins from the pWSK29-based plasmids. Overnight cultures were spotted onto tryptone semisolid agar supplemented with 50 μg/ml ampicillin, and the plate was then incubated at 30°C for 11 h. Mutated residues are indicated below the colonies. Vector, pWSK29.
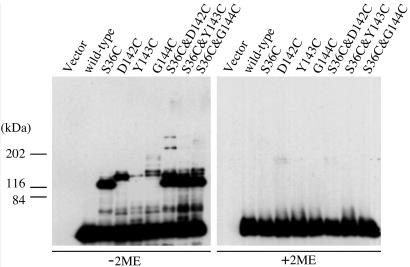
Detection of the Intra- and Interdimer Crosslinking Between Tar Monomers in Vivo. We examined the crosslinking of the Cys-substituted Tar proteins expressed in HCB436 cells by immunoblotting in the absence and presence of a reducing agent 2-mercaptoethanol (2ME) (Fig. 3). To avoid crosslinking during denaturation, the SDS-loading buffer was supplemented with 2.5 mM NEM and 2.5 mM EDTA. All of the mutants gave bands with apparent molecular masses of ≈60 kDa, which correspond to their monomers. In addition, Tar-S36C gave a band with an apparent molecular mass of ≈120 kDa, and the D142C, Y143C, and G144C proteins gave bands with apparent molecular masses of ≈150 kDa. Moreover, Tar-S36C&D142C gave two additional bands with higher apparent molecular masses: ≈240 and 360 kDa when roughly estimated by extrapolating the fitting curve of mobility of standard proteins. These additional bands may correspond to the intradimer crosslinked dimers, the interdimer crosslinked dimers, and the crosslinked oligomers, respectively, because they disappeared on treatment with 2ME. In the following experiments, we focused on the D142C mutation, which caused the most efficient interdimer crosslinking.
Fig. 3.
In vivo intra- and interdimer crosslinking of the Cys-replaced Tar proteins. Whole-cell lysates of HCB436 cells carrying pWSK29 (vector), pOH251 (wild-type), or its derivative (mutated residues are indicated) were subjected to nonreducing (-2ME) and reducing (+2ME) SDS/PAGE and immunoblotted with anti-Tar.
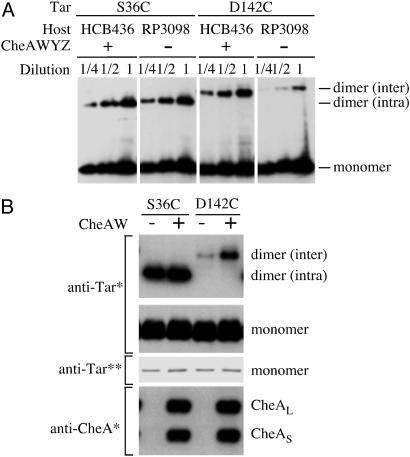
Effect of CheA and CheW on the Intra- and Interdimer Crosslinking. Because CheA and CheW are required for the optimal polar clustering of the MCPs (18, 19, 42), the lack of these proteins should affect the interdimer crosslinking. To avoid any complexity from the formation of oligomers, we used the single mutants and compared the intensities of the crosslinked dimers. Strain RP3098 was used as the host strain lacking CheA and CheW. Because no flagellar or chemotaxis promoter including the tar promoter is active in this strain, only the plasmids with the araBAD promoters can be used. Tar-D142C was less effectively crosslinked when expressed in RP3098 than in HCB436 (40–60% decrease), whereas the efficiency of crosslinking at position 36 was not significantly affected (Fig. 4A).
Fig. 4.
Effect of CheA and CheW on the intra- and interdimer crosslinking of the Cys-replaced Tar proteins. (A) Effect of cytoplasmic Che proteins on crosslinking. HCB436 (ΔMCP ΔCheRB) and RP3098 (ΔMCP ΔChe) cells expressing Tar-S36C or D142C were subjected to nonreducing SDS/PAGE, followed by immunoblotting with anti-Tar. (B) Effect of CheA and CheW on crosslinking. RP3098 cells expressing Tar-S36C or D142C were transformed with pDV4 (+)or pBR322 (-). Samples were subjected to reducing (anti-Tar**) or nonreducing (others) SDS/PAGE, followed by immunoblotting with anti-Tar (anti-Tar**) or anti-CheA antibodies as indicated. *, chemiluminescence; **, chemical staining.
We then examined whether the expression of CheA and CheW restores the interdimer crosslinking between Tar monomers in RP3098 (Fig. 4B). Strain RP3098 carrying each pBAD33-based plasmid (S36C or D142C) was transformed with pDV4 (CheA and CheW) or the vector pBR322. Crosslinking at position 142 was more effective when expressed in the presence of CheA and CheW than in their absence, whereas the efficiency of crosslinking at position 36 was not significantly affected. It should be noted that the total amount of either mutant Tar protein was not significantly varied. We therefore conclude that the interdimer crosslinking of Tar is not an artifact from sample preparation, but that it reflects a CheA/CheW-dependent interaction of Tar dimers in their native setting.
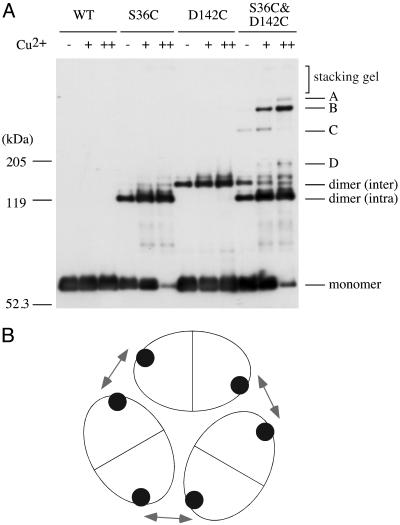
Detection of Higher Oligomers of Tar. To promote the formation of higher oligomers, we then used a catalyst for oxidation, Cu-phenanthroline (Fig. 5). HCB436 cells expressing each mutant Tar protein were treated with various concentrations of Cu-phenanthroline for 10 min, followed by the addition of the stop solution containing NEM and EDTA. Cu-phenanthroline enhanced crosslinking at position 36 and the efficiency reached almost 100% at 200 μM oxidant. Crosslinking at position 142 was also enhanced, but its efficiency was substantially less than that at position 36. For the S36C&D142C mutant, four major oligomers (named A, B, C, and D) were detected, with B and C corresponding to the 360- and 240-kDa bands described above. To estimate their sizes, the crosslinked dimer formed in the absence of NEM was excised from the gel, treated with Cu-phenanthroline, and subjected to SDS/PAGE again (data not shown). Two bands corresponding to oligomers B and C were detected, which led us to a tentative assignment of these oligomers as a hexamer and a tetramer, although other possibilities are not excluded. The deduced hexamer became predominant over other possible oligomers. This result is reminiscent of the crystal unit of a cytoplasmic fragment of Tsr that consists of a trimer of dimers (23). The deduced crosslinked hexamer might reflect the trimerization of MCP dimers in vivo. Moreover, Tar-S36C&D142C was less effectively crosslinked when expressed in strain RP3098 (ΔMCP ΔChe), although a small amount of the oligomer was detected in the presence of 50 μM or higher concentrations of Cu-phenanthroline (data not shown), suggesting that CheA and CheW enhance or stabilize, but are not prerequisite for, the receptor oligomerization.
Fig. 5.
Detection of higher oligomers. (A) Effect of an oxidant on crosslinking. Cells were treated with 0 (-), 20 (+), or 200 μM(++) Cu-phenanthroline for 10 min. The reaction was terminated by the addition of the stop solution supplemented with 2.5 mM NEM and 2.5 mM EDTA. Samples were subjected to nonreducing SDS/PAGE and immunoblotting with anti-Tar. Major oligomers are labeled A, B, C, and D. (B) Deduced orientation of the periplasmic domains of Tar in the putative hexamer unit. A view from outside the cell. Each Tar dimer (ellipse) is believed to be related by threefold symmetry. Filled circles, residue 142; arrows, interactions between dimers that are influenced by attractant binding.
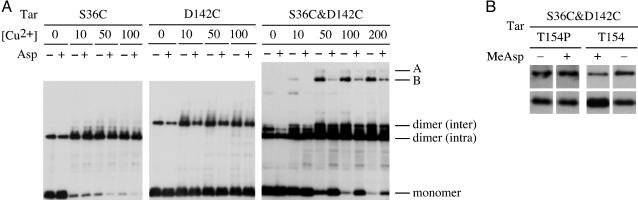
Effect of the Ligand Aspartate on the Interdimer Crosslinking and the Formation of the Deduced Hexamer. Is the interdimer interaction and/or the formation of the deduced hexamer of Tar involved in signaling? To address this question, we next examined whether a ligand has any effect on crosslinking. HCB436 cells expressing each mutant Tar protein were washed and divided into two aliquots (see Materials and Methods). The Tar-specific attractant aspartate was added to one of the aliquots. The samples were incubated for 10 min at 30°C, and for further treatment with or without Cu-phenanthroline. Tar-D142C was slightly less crosslinked when treated with aspartate in the absence and the presence of 10 μM Cu-phenanthroline, whereas the efficiency of crosslinking at position 36 was not significantly influenced by aspartate at any concentration of Cu-phenanthroline (Fig. 6A). Tar-S36C&D142C was clearly less crosslinked when treated with aspartate (Fig. 6A). Similar results were obtained with a nonmetabolizable analog, α-methyl-dl-aspartate (data not shown), but the effect of methylaspartate was lost when the T154P mutation, which abolishes aspartate binding (43), was introduced into Tar-S36C&D142C (Fig. 6B). These results indicate that the aspartate binding to Tar decreases the interdimer crosslinking and especially the formation of the higher crosslinked oligomers, and that it may decrease the interdimer interaction within and/or between trimer units.
Fig. 6.
Effects of the ligand aspartate on the intra- and interdimer crosslinking of the Cys-replaced Tar proteins. (A) Effect of aspartate on crosslinking. HCB436 cells expressing Tar-S36C, Tar-D142C, or Tar-S36C&D142C were washed with MLM, incubated in MLM supplemented with 1 mM aspartate (+) or none (-)at30°C for 10 min, and then treated with indicated concentrations (μM) of Cu-phenanthroline in the presence or absence of aspartate or serine at 30°C for 10 min. The reaction was terminated on ice by the addition of stop solution containing 2.5 mM NEM and 2.5 mM EDTA. Samples were subjected to nonreducing SDS/PAGE and immunoblotting with anti-Tar. (B) Effect of the T154P mutation, which abolishes aspartate-binding. HCB436 cells expressing Tar-S36C&D142C or Tar-S36C&D142C&T154P were incubated with or without 1 mM α-methyl-dl-aspartate (MeAsp) in the presence of 200 μM Cu-phenanthroline. (Upper) Oligomer B. (Lower) Monomer.
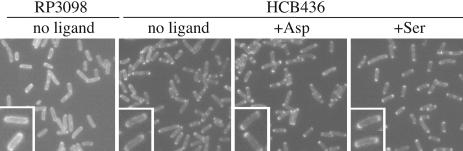
This effect of aspartate might result from its interference with polar localization of Tar. To address this issue, GFP was fused to the C terminus of Tar. Without induction, the fusion protein was expressed at a level similar to that of chromosome-encoded wild-type Tar and complemented the defect of a tar mutant (data not shown). Whereas the fluorescence signal was accumulated at poles of HCB436 cells (CheA+ CheW+), Tar-GFP was evenly distributed throughout the surface of RP3098 cells (CheA- CheW-; Fig. 7). This result is consistent with the report on the effects of CheA and CheW on the localization and clustering of MCPs (18, 19) and on the interdimer crosslinking of Tar (results described above), suggesting that the localization of Tar-GFP reflects that of wild-type Tar. By contrast, aspartate (up to 10 mM) did not significantly alter the localization of fluorescence signals at cell poles (Fig. 7). This finding suggests that the ligand-induced decreases in crosslinking of Tar does not result from global redistribution of the protein in a cell. The simplest explanation is that aspartate alters the relative positions or trajectories of Tar dimers in assembled signaling complexes, yielding changes in collision rates between dimers within or between trimers of dimers. The possibility that the aspartate effects arise from a slowing of subunit exchange is eliminated by our preliminary observation that aspartate increased crosslinking of some other Cys mutants (e.g., M110C; data not shown).
Fig. 7.
Subcellular localization of Tar-GFP in vivo. HCB436 (ΔMCP ΔCheRB) or RP3098 (ΔMCP ΔChe) cells carrying the plasmid encoding Tar-GFP were preincubated in MLM supplemented with 10 mM aspartate, 10 mM serine, or nothing. Cells were spotted onto a slide glass coated with 0.5% agarose and were observed under a fluorescent microscope.
Discussion
Disulfide crosslinking assays have been used to examine spatial proximity of residues in Tar (15, 40, 44), because the formation of a disulfide bond is highly sensitive to collisions between two thiol groups (during which the proximity should be 1.9 Å or less). This technique has also been used to probe the structures (and their changes on signaling) of the periplasmic domain (38, 45, 46), the transmembrane domain (36, 37, 47, 48), and of the cytoplasmic domain (39, 49–51) of MCPs. Some of these studies were carried out in vivo (36–38). Here, we extend this technique to probe the in vivo interaction between Tar homodimers, and the results are consistent with the model that interdimer interactions are involved in receptor signaling.
Considering that the structure of Tar should be fairly flexible (26), crosslinking at positions 142–144 may not necessarily imply that these positions constitute an interdimer contact site. In fact, Mehan et al. (52) recently showed that coupling of a bulky probe to Cys-139 that may be in direct contact with position 142 had no effect on kinase activation, providing strong evidence that this region does not constitute a contact surface between receptor dimers. Moreover, the effect of aspartate on the interdimer interaction in the periplasmic domain might not be direct, but rather, it might be exerted through the cytoplasmic contacts (23, 24, 26). A structural model for the intact MCP (26) proposes that the receptors in a trimer of dimers make contacts to other trimers in their periplasmic domains. Because this periplasmic contact is also trimeric, the deduced crosslinked hexamer could result from the interactions between, rather than within, trimers of dimers.
This study provides information about relative orientations or trajectories of the periplasmic domains of MCP dimers in their polar cluster that may be modulated by attractant binding. The effect of aspartate on disulfide bond formation between Cys-142 residues could result from (i) slowing of subunit exchange, (ii) slowing of collisions between dimers, (iii) altered trajectories of collisions between dimers, or (iv) altered relative positions of the dimers. Our preliminary observation that aspartate increased crosslinking of some other Cys mutants, including S36C&M110C (data not shown), argues against the first two possibilities. Therefore, we suspect that the ligand alters either the positions or trajectories of Tar dimers within a putative trimer of dimers, or between such trimers. The results seems to contradict the prediction of Kim et al. (26) that ligand binding simply enhances the dynamic motions of the dimers. However, if the trimer stability is reduced by the attractant, intratrimer crosslinking might be reduced. In this context, it is intriguing that effects of aspartate on the crosslinking were much more profound with the double Cys mutant protein than with the single Cys mutant protein (Fig. 6). The 36–36′ crosslink might reduce flexibility of the receptor dimer and might restrict the 142–142′ crosslink within the trimer unit. It should be noted that the double mutant produced the 36–36′ crosslinked dimers efficiently and this crosslinking was little affected by the addition of the attractant under the condition applied (Fig. 6).
Recent in vitro studies revealed rather low cooperativity for the kinase regulation by MCPs (53–55). Nevertheless, in vivo studies with fluorescence resonance energy transfer between CheY-YFP and CheZ-CFP revealed significant degrees of signal amplification especially in the presence of CheB (11). It should be possible to use our crosslinking assay to test whether receptor–receptor interactions are, indeed, important for signal amplification.
Acknowledgments
We thank Dr. John S. Parkinson (University of Utah, Salt Lake City) for providing strains and for critically reading the manuscript, Dr. Tohru Umemura for providing initial experimental setup assistance, and Dr. Toshiharu Yakushi for helpful discussions and encouragement. This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (to D.S. and I.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cu-phenanthroline, Cu(II)(o-phenanthroline)3; MCP, methyl-accepting chemotaxis protein; 2ME, 2-mercaptoethanol; NEM, N-ethylmaleimide.
References
- 1.Falke, J. J., Bass, R. B., Butler, S. L., Chervitz, S. A. & Danielson, M. A. (1997) Annu. Rev. Cell Dev. Biol. 13, 457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock, J. B. & Surette, M. G. (1996) in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingram, J. J., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), 2nd Ed., pp. 1103-1129.
- 3.Stock, J. & Levit, M. (2000) Curr. Biol. 10, R11-R14. [DOI] [PubMed] [Google Scholar]
- 4.Bourret, R. B. & Stock, A. M. (2002) J. Biol. Chem. 277, 9625-9628. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, S. & Koshland, D. E., Jr. (1979) J. Biol. Chem. 254, 9695-9702. [PubMed] [Google Scholar]
- 6.Hedblom, M. L. & Adler, J. (1983) J. Bacteriol. 155, 1463-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segall, J. E., Block, S. M. & Berg, H. C. (1986) Proc. Natl. Acad. Sci. USA 83, 8987-8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasuja, R., Lin, Y., Trentham, D. R. & Khan, S. (1999) Proc. Natl. Acad. Sci. USA 96, 11346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cluzel, P., Surette, M. & Leibler, S. (2000) Science 287, 1652-1655. [DOI] [PubMed] [Google Scholar]
- 10.Sourjik, V. & Berg, H. C. (2002) Proc. Natl. Acad. Sci. USA 99, 12669-12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sourjik, V. & Berg, H. C. (2002) Proc. Natl. Acad. Sci. USA 99, 123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray, D., Levin, M. D. & Morton-Firth, C. J. (1998) Nature 393, 85-88. [DOI] [PubMed] [Google Scholar]
- 13.Duke, T. A. J. & Bray. D. (1999) Proc. Natl. Acad. Sci. USA 96, 10104-10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkai, N., Alon, U. & Leibler, S. (2001) C. R. Acad. Sci. Ser. IV 2, 871-877. [Google Scholar]
- 15.Milligan, D. L. & Koshland, D. E., Jr. (1988) J. Biol. Chem. 263, 6268-6275. [PubMed] [Google Scholar]
- 16.Gegner, J. A., Graham, D. R., Roth, A. F. & Dahlquist, F. W. (1992) Cell 70, 975-982. [DOI] [PubMed] [Google Scholar]
- 17.Schuster, S. C., Swanson, R. V., Alex, L. A., Bourret, R. B. & Simon, M. I. (1993) Nature 365, 343-347. [DOI] [PubMed] [Google Scholar]
- 18.Maddock, J. R. & Shapiro, L. (1993) Science 259, 1717-1723. [DOI] [PubMed] [Google Scholar]
- 19.Skidmore, J. M., Ellefson, D. D., McNamara, B. P., Couto, M. M., Wolfe, A. J. & Maddock, J. R. (2000) J. Bacteriol. 182, 967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sourjik, V. & Berg, H. C. (2000) Mol. Microbiol. 37, 740-751. [DOI] [PubMed] [Google Scholar]
- 21.Gestwicki, J. E., Strong, L. E. & Kiessling, L. L. (2000) Chem. Biol. 7, 583-591. [DOI] [PubMed] [Google Scholar]
- 22.Gestwicki, J. E. & Kiessling, L. L. (2002) Nature 415, 81-84. [DOI] [PubMed] [Google Scholar]
- 23.Kim, K. K., Yokota, H. & Kim, S.-H. (1999) Nature 400, 787-792. [DOI] [PubMed] [Google Scholar]
- 24.Ames, P., Studdert, C. A., Reiser, R. H. & Parkinson, J. S. (2002) Proc. Natl. Acad. Sci. USA 99, 7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu, T. S., Le Novere, N., Levin, M. D., Beavil, A. J., Sutton, B. J. & Bray, D. (2000) Nat. Cell Biol. 2, 792-796. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S.-H., Wang, W. & Kim, K. K. (2002) Proc. Natl. Acad. Sci. USA 99, 11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfe, A. J., Conley, M. P., Kramer, T. J. & Berg, H. C. (1987) J. Bacteriol. 169, 1878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe, A. J. & Berg, H. C. (1989) Proc. Natl. Acad. Sci. USA 86, 6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slocum, M. K. & Parkinson, J. S. (1983) J. Bacteriol. 155, 565-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, R. F. & Kushner, S. R. (1991) Gene 100, 195-199. [PubMed] [Google Scholar]
- 31.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess, J. F., Oosawa, K., Matsumura, P. & Simon, M. I. (1987) Proc. Natl. Acad. Sci. USA 84, 7609-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umemura, T., Tatsuno, I., Shibasaki, M., Homma, M. & Kawagishi, I. (1998) J. Biol. Chem. 273, 30110-30115. [DOI] [PubMed] [Google Scholar]
- 34.Okumura, H., Nishiyama, S., Sasaki, A., Homma, M. & Kawagishi, I. (1998) J. Bacteriol. 180, 1862-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiomi, D., Zhulin, I. B., Homma, M. & Kawagishi, I. (2002) J. Biol. Chem. 277, 42325-42333. [DOI] [PubMed] [Google Scholar]
- 36.Lee, G. F., Lebert, R., Lilly, A. A. & Hazelbauer, G. L. (1995) Proc. Natl. Acad. Sci. USA 92, 3391-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughson, A. G. & Hazelbauer, G. L. (1996) Proc. Natl. Acad. Sci. USA 93, 11546-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maruyama, I. N., Mikawa, Y. G. & Maruyama, H. I. (1995) J. Mol. Biol. 253, 530-546. [DOI] [PubMed] [Google Scholar]
- 39.Bass, R. B. & Falke, J. J. (1999) Structure (London) 7, 829-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falke, J. J. & Koshland, D. E., Jr. (1987) Science 237, 1596-1600. [DOI] [PubMed] [Google Scholar]
- 41.Milburn, M. V., Prive, G. G., Milligan, D. L., Scott, W. G., Yeh, J., Jancarik, J., Koshland, D. E., Jr., & Kim, S. H. (1991) Science 254, 1342-1347. [DOI] [PubMed] [Google Scholar]
- 42.Lybarger, S. R. & Maddock, J. R. (1999) J. Bacteriol. 181, 5527-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, L. & Imae, Y. (1990) J. Bacteriol. 172, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falke, J. J., Dernburg, A. F., Sternberg, D. A., Zalkin, N., Milligan, D. L. & Koshland, D. E., Jr. (1988) J. Biol. Chem. 263, 14850-14858. [PubMed] [Google Scholar]
- 45.Chervitz, S. A. & Falke, J. J. (1995) J. Biol. Chem. 270, 24043-24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chervitz, S. A., Lin, C. M. & Falke, J. J. (1995) Biochemistry 34, 9722-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch, B. A. & Koshland, D. E., Jr. (1991) Proc. Natl. Acad. Sci. USA 88, 10402-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pakula, A. A. & Simon, M. I. (1992) Proc. Natl. Acad. Sci. USA 89, 4144-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danielson, M. A., Bass, R. B. & Falke, J. J. (1997) J. Biol. Chem. 272, 32878-32888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler, S. L. & Falke, J. J. (1998) Biochemistry 37, 10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bass, R. B. & Falke, J. J. (1998) J. Biol. Chem. 273, 25006-25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehan, R. S., White, N. C. & Falke, J. J. (2003) Biochemistry 42, 2952-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bornhorst, J. A. & Falke, J. J. (2000) Biochemistry 39, 9486-9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bornhorst, J. A. & Falke, J. J. (2001) J. Gen. Physiol. 118, 693-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levit, M. N. & Stock, J. B. (2002) J. Biol. Chem. 277, 36760-36765. [DOI] [PubMed] [Google Scholar]